Elevated Serum Levels of miRNA-155 in Children with Atopic Dermatitis: A Potential Biomarker of Disease
Abstract
1. Introduction
2. Results
2.1. Group Characteristics
2.2. Evaluation of miRNA in the Pathogenesis of AD
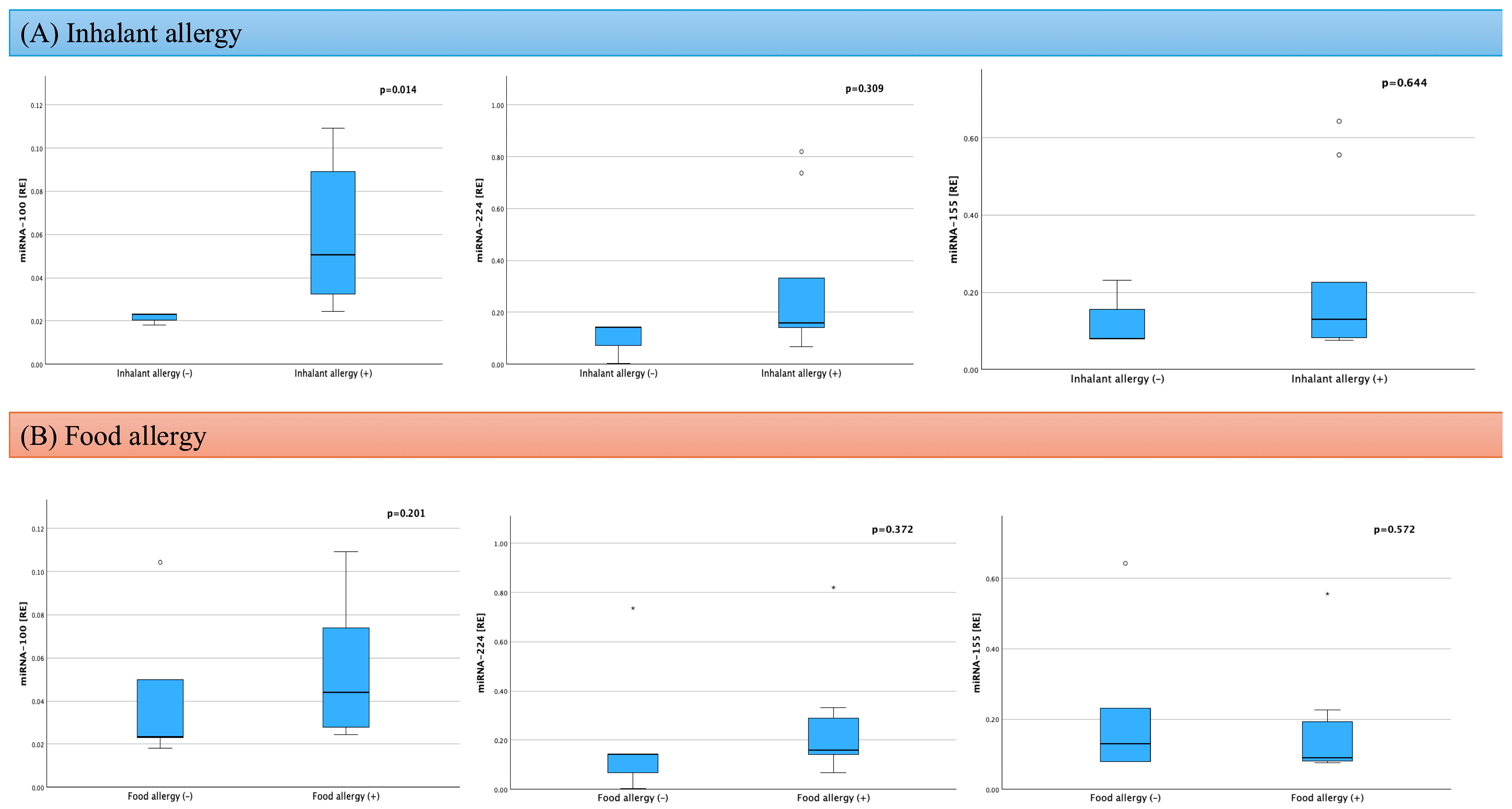
2.3. Assessment of Factors Influencing miRNA in AD
3. Discussion
4. Material and Methods
4.1. Study Population
4.2. Blood Serum Collection
4.3. miRNA Detection in the Serum
4.4. Cytokines Detection in the Serum
4.5. Statistical Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Atopic Dermatitis |
| BOD | Burden of disease |
| CDK9 | Cyclin-dependent kinase 9 |
| CLDN5 | Keratinocytes lead to a deficiency of claudin-5 |
| CTLA-4 | Cytotoxic T lymphocyte-associated protein 4 |
| DALY | Disability-adjusted life years |
| IgE | Immunoglobulin E |
| IL-4 | Interleukin 4 |
| IL-5 | Interleukin 5 |
| IL-6 | Interleukin 6 |
| IL-10 | Interleukin 10 |
| IL-13 | Interleukin 13 |
| miRNAs | MicroRNAs |
| PBMCs | Blood mononuclear cells |
| RE | Relative expression |
| RT | Room temperature |
| SCORAD | Severity of skin lesions |
| tCI | Topical calcineurin inhibitors |
| tCS | Topical corticosteroids |
| TLR2 | Targeting toll-like receptor 2 |
| WBC | Leukocyte count |
References
- Zhou, N.Y.; Nili, A.; Blackwell, C.K.; Ogbuefi, N.; Cummings, P.; Lai, J.; Griffith, J.W.; Paller, A.S.; Wakschlag, L.S.; Fishbein, A.B. Parent report of sleep health and attention regulation in a cross-sectional study of infants and preschool-aged children with atopic dermatitis. Pediatr. Dermatol. 2022, 39, 61–68. [Google Scholar] [CrossRef]
- Nakahara, T.; Kido-Nakahara, M.; Tsuji, G.; Furue, M. Basics and recent advances in the pathophysiology of atopic dermatitis. J. Dermatol. 2021, 48, 130–139. [Google Scholar] [CrossRef]
- Kowalska-Oledzka, E.; Czarnecka, M.; Baran, A. Epidemiology of atopic dermatitis in Europe. J. Drug Assess. 2019, 8, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Hanifin, J.M.; Rajka, G. Diagnostic features of atopic dermatitis. Acta Dermatovener 1980, 92, 44–47. [Google Scholar] [CrossRef]
- Laughter, M.R.; Maymone, M.B.C.; Mashayekhi, S.; Arents, B.W.M.; Karimkhani, C.; Langan, S.M.; Dellavalle, R.P.; Flohr, C. The global burden of atopic dermatitis: Lessons from the Global Burden of Disease Study 1990–2017. Br. J. Dermatol. 2021, 184, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Metrics, G.H. Atopic Dermatitis—Level 4 Cause: Institute for Health Metrics and Evaluation. Available online: https://www.healthdata.org/research-analysis/diseases-injuries-risks/factsheets/2021-atopic-dermatitis-level-4-disease (accessed on 18 March 2025).
- Czarnowicki, T.; He, H.; Krueger, J.G.; Guttman-Yassky, E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol. 2019, 143, 1–11. [Google Scholar] [CrossRef]
- Thyssen, J.P.; Vestergaard, C.; Barbarot, S.; Deleuran, M.; Bruin-Weller, M.S.; Bieber, T.; Taïeb, A.; Seneschal, J.; Cork, M.J.; Paul, C.; et al. European Task Force on Atopic Dermatitis: Position on atopic dermatitis pathophysiology and biomarker use for targeted treatment in adults. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1937–1947. [Google Scholar] [CrossRef]
- Choy, E.H.; De Benedetti, F.; Takeuchi, T.; Hashizume, M.; John, M.R.; Kishimoto, T. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020, 16, 335–345. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Neurath, M.F.; Finotto, S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011, 22, 83–89. [Google Scholar] [CrossRef]
- Gubernatorova, E.O.; Gorshkova, E.A.; Polinova, A.I.; Drutskaya, M.S. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020, 53, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Huang, J.; Zhong, H.; Shen, N.; Faggioni, R.; Fung, M.; Yao, Y. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol. Ther. 2014, 141, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Asadullah, K.; Sterry, W.; Volk, H.D. Interleukin-10 therapy—Review of a new approach. Pharmacol. Rev. 2003, 55, 241–269. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, N.; Smith, S.D. Improving Psychological Health Outcomes in Children with Atopic Dermatitis. Clin. Cosmet. Investig. Dermatol. 2023, 16, 2821–2827. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, C.; Zhuo, F.; Guo, Y.; Wang, S.; Zhang, K.; Li, X.; Dai, W.; Dou, X.; Yu, B. Skin microbiota: Pathogenic roles and implications in atopic dermatitis. Front. Cell. Infect. Microbiol. 2025, 14, 1518811. [Google Scholar] [CrossRef]
- Flowers, L.; Grice, E.A. The skin microbiota: Balancing risk and reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- Gołuchowska, N.; Ząber, A.; Będzichowska, A.; Tomaszewska, A.; Rustecka, A.; Kalicki, B. The Role of MicroRNA in the Pathogenesis of Atopic Dermatitis. Int. J. Mol. Sci. 2025, 26, 5846. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Specjalski, K.; Jassem, E. MicroRNAs: Potential Biomarkers and Targets of Therapy in Allergic Diseases? Arch. Immunol. Ther. Exp. 2019, 67, 213–223. [Google Scholar] [CrossRef]
- Lynam-Lennon, N.; Maher, S.G.; Reynolds, J.V. The roles of microRNA in cancer and apoptosis. Biol. Rev. 2009, 84, 55–71. [Google Scholar] [CrossRef]
- Lind, E.F.; Ohashi, P.S. Mir-155, a central modulator of T-cell responses. Eur. J. Immunol. 2014, 44, 11–15. [Google Scholar] [CrossRef]
- Okoye, I.S.; Czieso, S.; Ktistaki, E.; Roderick, K.; Coomes, S.M.; Pelly, V.S.; Kannan, Y.; Perez-Lloret, J.; Zhao, J.L.; Baltimore, D.; et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. USA 2014, 111, E3081–E3090. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.D.; Feng, S.Y.; Huang, A.F. Role of miR-155 in inflammatory autoimmune diseases: A comprehensive review. Inflamm. Res. 2022, 71, 1501–1517. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Janson, P.; Majuri, M.L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589.e20. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, G.; Du, H.; Shi, Y. Role and potential mechanisms of miR-100 in different diseases (Review). Oncol. Rep. 2025, 54, 91. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Z.; He, L.; Yan, L.; Tan, B.; Ma, L.; He, G.; Dai, Z.; Sun, R.; Li, C. Hydrogels treat atopic dermatitis by transporting marine-derived miR-100-5p-abundant extracellular vesicles. ACS Biomater. Sci. Eng. 2024, 10, 7667–7682. [Google Scholar] [CrossRef]
- Shaoqing, Y.; Ruxin, Z.; Guojun, L.; Zhiqiang, Y.; Hua, H.; Shudong, Y.; Jie, Z. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am. J. Rhinol. Allergy 2011, 25, e242–e246. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, L.; Hu, H.; Dong, P. MiR-224 ameliorates inflammation and symptoms in mouse model of allergic rhinitis by targeting CDK9. Allergol. Immunopathol. 2021, 49, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, J.; Guo, F.; Zheng, B.; Zhang, X. A novel inhibitory role of microRNA-224 in particulate matter 2.5-induced asthmatic mice by inhibiting TLR2. J. Cell. Mol. Med. 2020, 24, 3040–3052. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zitzer, N.C.; Snyder, K.; Meng, X.; Taylor, P.A.; Efebera, Y.A.; Devine, S.M.; Blazar, B.R.; Garzon, R.; Ranganathan, P. MicroRNA-155 Modulates Acute Graft-versus-Host Disease by Impacting T Cell Expansion, Migration, and Effector Function. J. Immunol. 2018, 200, 4170–4179. [Google Scholar] [CrossRef]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of tabela bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Daniel, E.; Roff, A.; Hsu, M.H.; Panganiban, R.; Lambert, K.; Ishmael, F. Effects of allergic stimulation and glucocorticoids on miR-155 in CD4+T-cells. Am. J. Clin. Exp. Immunol. 2018, 7, 57–66. [Google Scholar] [PubMed]
- Zhang, Y.; Sun, E.; Li, X.; Zhang, M.; Tang, Z.; He, L.; Lv, K. MiR-155 contributes to Df1-induced asthma by increasing the prolifera- tive response of Th cells via CTLA-4 downregulation. Cell. Immunol. 2017, 314, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Malmhäll, C.; Alawieh, S.; Lu, Y.; Sjöstrand, M.; Bossios, A.; Eldh, M.; Rådinger, M. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 2014, 133, 1429–1438. [Google Scholar] [CrossRef]
- Qiu, L.; Zhang, Y.; Do, D.C.; Ke, X.; Zhang, S.; Lambert, K.; Kumar, S.; Hu, C.; Zhou, Y.; Ishmael, F.T.; et al. miR-155 Modulates Cockroach Allergen– and Oxidative Stress–Induced Cyclooxygenase-2 in Asthma. J. Immunol. 2018, 201, 916–929. [Google Scholar] [CrossRef]
- Hammad, N.M.; Nabil, F.; Elbehedy, E.M.; Sedeek, R.; Gouda, M.I.; Arafa, M.A. Role of microRNA-155 as a potential biomarker for allergic rhinitis in children. Can. Respir. J. 2021, 2021, 5554461. [Google Scholar] [CrossRef]
- Comer, B.S.; Camoretti-Mercado, B.; Kogut, P.C.; Halayko, A.J.; Solway, J.; Gerthoffer, W.T. Cyclooxygenase-2 and microRNA-155 expression are elevated in asthmatic airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 2015, 52, 438–447. [Google Scholar] [CrossRef]
- Ma, L.; Xue, H.; Wang, F.; Shu, C.-M.; Zhang, J.-H. MicroRNA-155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin. Exp. Immunol. 2015, 181, 142–149. [Google Scholar] [CrossRef]
- El-Korashi, L.A.; Nafea, O.E.; Nafea, A.E.; Elkholy, B.M.; Elhawy, L.L.; Abdelhadi, A.A. MicroRNA-155 is a potential predictive tool for atopic dermatitis severity in children: A preliminary study. Egypt. J. Immunol. 2024, 31, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Zhou, B.; Wei, Z.; Luo, Y. IL-32 promotes the occurrence of atopic dermatitis by activating the JAK1/microRNA-155 axis. J. Transl. Med. 2022, 20, 207. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rosenberg, H.; Dyer, K.; Foster, P. Eosinophils: Changing perspectives in health and disease. Nat. Rev. Immunol. 2013, 13, 9–22. [Google Scholar] [CrossRef]
- Simon, D.; Braathen, L.R.; Simon, H.U. Eosinophils and atopic dermatitis. Allergy 2004, 59, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.A.; Socorro, A.I.; Galvan, F.H.; Cazorla, R.S.; Lemes, C.A.; Cabrera, L.C. Eosinophils: Old cells, new directions. Front. Med. 2025, 11, 1470381. [Google Scholar] [CrossRef]
- Hagino, T.; Hamada, R.; Yoshida, M.; Fujimoto, E.; Saeki, H.; Kanda, N. Total eosinophil count as a biomarker for therapeutic effects of upadacitinib in atopic dermatitis over 48 weeks. Front. Immunol. 2024, 15, 1365544. [Google Scholar] [CrossRef] [PubMed]
- Torres, T.; Mendes-Bastos, P.; Cruz, M.J.; Duarte, B.; Filipe, P.; Lopes, M.J.P.; Gonçalo, M. Interleukin-4 and Atopic Dermatitis: Why Does it Matter? A Narrative Review. Dermatol. Ther. 2025, 15, 579–597. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Altara, R.; Manca, M.; Hermans, K.C.; Daskalopoulos, E.P.; Brunner-La Rocca, H.P.; Hermans, R.J.; Struijker-Boudier, H.A.; Blankesteijn, M.W. Diurnal rhythms of serum and plasma cytokine profiles in healthy elderly individuals assessed using membrane based multiplexed immunoassay. J. Transl. Med. 2015, 13, 129. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nilsonne, G.; Lekander, M.; Åkerstedt, T.; Axelsson, J.; Ingre, M. Diurnal Variation of Circulating Interleukin-6 in Humans: A Meta-Analysis. PLoS ONE 2016, 11, e0165799. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kiecolt-Glaser, J.K. Stress, food, and inflammation: Psychoneuroimmunology and nutrition at the cutting edge. Psychosom. Med. 2010, 72, 365–369. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, L.; Wu, W.J.; Lyu, L.C.; Tu, Y.; Gu, H.; Chen, X.F.; Chai, Y.J.; Man, M.Q.; He, L. MiRNA-224-5p regulates the defective permeability barrier in sensitive skin by targeting claudin-5. Skin Res. Technol. 2024, 30, e13720. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Variable | Study Group (n = 12) | Control Group (n = 9) | p-Value |
|---|---|---|---|
| Age [years]; mean (±SD) | 5.83 (±3.51) | 8.56 (±4.27) | pSt = 0.125 |
| Sex; n (%) | |||
| Females | 8 (66.7) | 8 (66.7) | pchi2 = 1.00 |
| Males | 4 (33.3) | 4 (33.3) | |
| Height [cm]; mean (±SD) | 115.79 (25.02) | 131.89 (19.45) | pSt = 0.126 |
| Height [pc]; mean (±SD) | 50.00 (29.88) | 55.22 (25.02) | pSt = 0.668 |
| Weight [kg]; median (IQR) | 20.03 (13.49–27.62) | 31.30 (17.40–45.30) | pUMW = 0.111 |
| Weight [pc]; mean (±SD) | 41.83 (28.02) | 62.22 (23.30) | pSt = 0.085 |
| WBC [×109/L]; median (IQR) | 8.71 (7.72–13.44) | 6.42 (5.27–7.55) | pUMW = 0.009 |
| Eosinophils [%]; median (IQR) | 6.40 (3.40–2.70) | 4.10 (3.25–8.40) | pUMW = 0.345 |
| Eosinophils [×103/μL]; median (IQR) | 0.66 (0.26–1.50) | 0.27 (0.16–0.69) | pUMW = 0.148 |
| IgE [IU/mL]; median (IQR) | 487.50 (123.75–1151.00) | 45.00 (34.50–66.00) | pUMW = 0.012 |
| Vitamin D [ng/mL]; mean (±SD) | 32.96 (10.15) | 26.22 (9.05) | pSt = 0.154 |
| SCCORAD [points]; median (IQR) | 60.00 (56.00–72.00) | - | - |
| Comorbidities [n], % | - | - | |
| Respiratory allergies | 9 (75) | - | - |
| Food allergies | 7 (58) | - | - |
| Asthma | 3 (25) | - | - |
| Allergic Rhinitis | 8 (67) | - | - |
| Variable | Study Group (n = 12) | Control Group (n = 9) | p-Value |
|---|---|---|---|
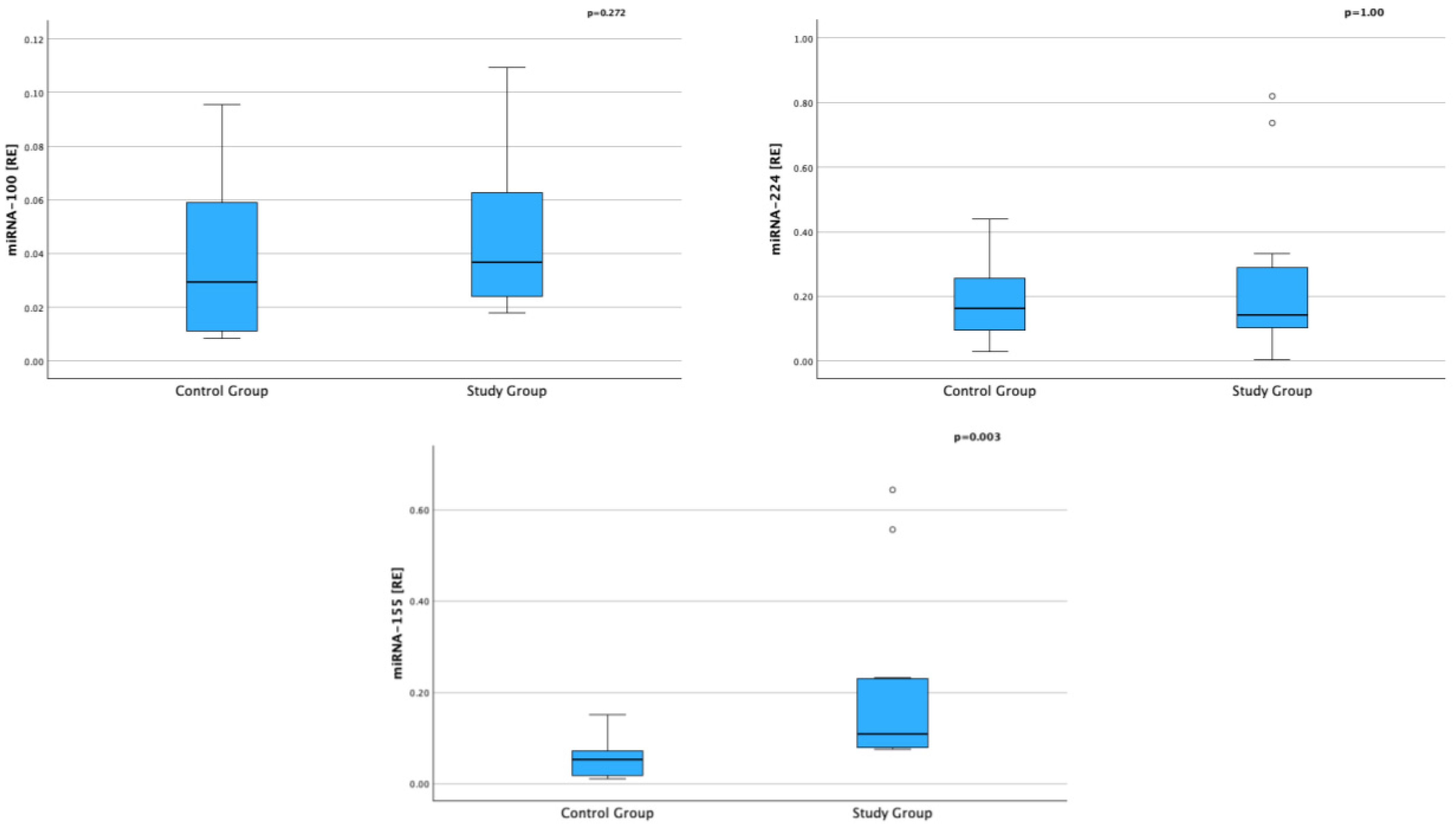
| miRNA-100 [RE]; median (IQR) | 0.039 (0.023–0.073) | 0.029 (0.010–0.067) | pUMW = 0.272 |
| miRNA-224 [RE]; median (IQR) | 0.143 (0.086–0.311) | 0.162 (0.075–0.0297) | pUMW = 1.000 |
| miRNA-155 [RE]; median (IQR) | 0.109 (0.079–0.230) | 0.052 (0.015–0.109) | pUMW = 0.003 |
| IL-4 [ng/mL]; median (IQR) | 0.00 (0.00–26.05) | 13.60 (0.00–38.50) | pUMW = 0.473 |
| IL-13 [ng/mL]; median (IQR) | 118.50 (23.56–825.11) | 475.55 (65.75–618.55) | pUMW = 0.536 |
| IL-5 [ng/mL]; median (IQR) | 0.00 (0.00–00.62) | 0.00 (0.00–0.00) | pUMW = 0.536 |
| IL-6 [ng/mL]; median (IQR) | 0.00 (0.00–28.30) | 12.44 (0.00–310.1555) | pUMW = 0.295 |
| IL-10 [ng/mL]; median (IQR) | 0.00 (0.00–58.74) | 0.85 (0.00–25.80) | pUMW = 0.657 |
| Variable | tCS/tCI (+) (n = 7) | tCS/tCI (−) (n = 5) | p-Value |
|---|---|---|---|
| miRNA-100 [RE]; median (IQR) | 0.026 (0.022–0.056) | 0.051 (0.030–0.106) | pUMW = 0.201 |
| miRNA-224 [RE]; median (IQR) | 0.143 (0.141–0.737) | 0.067 (0.035–0.246) | pUMW = 0.167 |
| miRNA-155 [RE]; median (IQR) | 0.089 (0.078–0.178) | 0.159 (0.079–0.555) | pUMW = 0.465 |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Diagnosis of atopic dermatitis based on Hanifin and Rajka criteria | Other skin diseases |
| Moderate to severe form of AD (SCORAD ≥ 25 points) | Active infectious disease |
| Consent to participate in the study | Autoimmune diseases |
| Congenital or acquired immunodeficiency | |
| Lack of consent to participate in the study |
| Step | Temperature | Time | Cycle |
|---|---|---|---|
| Enzyme activation | 95 °C | 20 s | 1 |
| Denature | 95 °C | 1 s | 40 |
| Anneal/Extend | 60 °C | 20 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gołuchowska, N.; Ząber, A.; Walczewska, S.; Będzichowska, A.; Brodaczewska, K.; Majewska, A.; Kalicki, B.; Tomaszewska, A. Elevated Serum Levels of miRNA-155 in Children with Atopic Dermatitis: A Potential Biomarker of Disease. Int. J. Mol. Sci. 2025, 26, 9689. https://doi.org/10.3390/ijms26199689
Gołuchowska N, Ząber A, Walczewska S, Będzichowska A, Brodaczewska K, Majewska A, Kalicki B, Tomaszewska A. Elevated Serum Levels of miRNA-155 in Children with Atopic Dermatitis: A Potential Biomarker of Disease. International Journal of Molecular Sciences. 2025; 26(19):9689. https://doi.org/10.3390/ijms26199689
Chicago/Turabian StyleGołuchowska, Natalia, Aldona Ząber, Sylwia Walczewska, Agata Będzichowska, Klaudia Brodaczewska, Aleksandra Majewska, Bolesław Kalicki, and Agata Tomaszewska. 2025. "Elevated Serum Levels of miRNA-155 in Children with Atopic Dermatitis: A Potential Biomarker of Disease" International Journal of Molecular Sciences 26, no. 19: 9689. https://doi.org/10.3390/ijms26199689
APA StyleGołuchowska, N., Ząber, A., Walczewska, S., Będzichowska, A., Brodaczewska, K., Majewska, A., Kalicki, B., & Tomaszewska, A. (2025). Elevated Serum Levels of miRNA-155 in Children with Atopic Dermatitis: A Potential Biomarker of Disease. International Journal of Molecular Sciences, 26(19), 9689. https://doi.org/10.3390/ijms26199689





