Downregulation of Mitophagy, Complex I Biogenesis, and Signaling by ROBO Receptors—Implications for Psoriasis Pathogenesis
Abstract
1. Introduction
2. Results
2.1. Differentially Expressed Genes
2.2. GSEA Results
2.3. Upregulated Pathways
2.3.1. Lesional Skin Compared to Control Skin
2.3.2. Non-Lesional Skin Compared to Control Skin
2.3.3. Lesional Skin Compared to Non-Lesional Skin
2.4. Downregulated Pathways
2.4.1. Lesional Skin Compared to Control Skin
2.4.2. Non-Lesional Skin Compared to Control Skin
2.4.3. Lesional Skin Compared to Non-Lesional Skin
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Sample Collection and Skin Biopsy Procedure
4.3. RNA Extraction and Sequencing
4.4. Bioinformatic Processing and Statistical Analysis
4.5. Gene Set Analysis, Network Analysis, and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nestle, F.O.; Kaplan, D.H.; Barker, J. Psoriasis. N. Engl. J. Med. 2009, 361, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Patrizi, A.; Raone, B.; Ravaioli, G.M. Management of atopic dermatitis: Safety and efficacy of phototherapy. Clin. Cosmet. Investig. Dermatol. 2015, 8, 511–520. [Google Scholar] [CrossRef]
- Gladman, D.D.; Antoni, C.; Mease, P.; Clegg, D.O.; Nash, P. Psoriatic arthritis: Epidemiology, clinical features, course, and outcome. Ann. Rheum. Dis. 2005, 64 (Suppl. 2), ii14–ii17. [Google Scholar] [CrossRef]
- Gottlieb, A.B.; Dann, F. Comorbidities in patients with psoriasis. Am. J. Med. 2009, 122, 1150.e1–1150.e9. [Google Scholar] [CrossRef]
- Grozdev, I.; Korman, N.; Tsankov, N. Psoriasis as a systemic disease. Clin. Dermatol. 2014, 32, 343–350. [Google Scholar] [CrossRef]
- Najarian, D.J.; Gottlieb, A.B. Connections between psoriasis and Crohn’s disease. J. Am. Acad. Dermatol. 2003, 48, 805–821; quiz 822–824. [Google Scholar] [CrossRef]
- Prodanovich, S.; Kirsner, R.S.; Kravetz, J.D.; Ma, F.; Martinez, L.; Federman, D.G. Association of psoriasis with coronary artery, cerebrovascular, and peripheral vascular diseases and mortality. Arch. Dermatol. 2009, 145, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Schon, M.P.; Boehncke, W.H. Psoriasis. N. Engl. J. Med. 2005, 352, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chang, C.; Lu, Q. The Inflammatory Response in Psoriasis: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 50, 377–389. [Google Scholar] [CrossRef]
- Luo, Y.; Luo, Y.; Chang, J.; Xiao, Z.; Zhou, B. Identification of candidate biomarkers and pathways associated with psoriasis using bioinformatics analysis. Hereditas 2020, 157, 30. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhao, L.; Ding, H.; Song, J.; Zhang, Q.; Yu, S.; Wang, Y.; Wang, H. Interleukins and Psoriasis. J. Cutan. Med. Surg. 2024, 28, NP19–NP35. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Duffin, K.C.; Helms, C.; Ding, J.; Stuart, P.E.; Goldgar, D.; Gudjonsson, J.E.; Li, Y.; Tejasvi, T.; Feng, B.J.; et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009, 41, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Krueger, J.G.; Fretzin, S.; Suárez-Fariñas, M.; Haslett, P.A.; Phipps, K.M.; Cameron, G.S.; McColm, J.; Katcherian, A.; Cueto, I.; White, T.; et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. J. Allergy Clin. Immunol. 2012, 130, 145–154.e9. [Google Scholar] [CrossRef]
- Chen, W.; Xie, K.; Liu, X.; Chen, H. Identification of key pathways and genes in psoriasis via gene microarray analysis. Mol. Med. Rep. 2016, 13, 2327–2337. [Google Scholar] [CrossRef] [PubMed]
- Sevimoglu, T.; Arga, K.Y. Computational Systems Biology of Psoriasis: Are We Ready for the Age of Omics and Systems Biomarkers? Omics A J. Integr. Biol. 2015, 19, 669–687. [Google Scholar] [CrossRef]
- Fan, F.; Huang, Z.; Chen, Y. Integrated analysis of immune-related long noncoding RNAs as diagnostic biomarkers in psoriasis. PeerJ 2021, 9, e11018. [Google Scholar] [CrossRef]
- Zou, A.; Kong, Q.; Sang, H. Identification of key apoptosis-related genes and immune infiltration in the pathogenesis of psoriasis. Hereditas 2022, 159, 26. [Google Scholar] [CrossRef]
- Liu, S.; Zhuang, Z.; Liu, F.; Yuan, X.; Zhang, Z.; Liang, X.; Li, X.; Chen, Y. Identification of potential biomarkers and infiltrating immune cells from scalp psoriasis. Gene 2024, 893, 147918. [Google Scholar] [CrossRef]
- Liu, S.; Yuan, X.; Su, H.; Liu, F.; Zhuang, Z.; Chen, Y. ZNF384: A Potential Therapeutic Target for Psoriasis and Alzheimer’s Disease Through Inflammation and Metabolism. Front. Immunol. 2022, 13, 892368. [Google Scholar] [CrossRef]
- Xing, L.; Wu, T.; Yu, L.; Zhou, N.; Zhang, Z.; Pu, Y.; Wu, J.; Shu, H. Exploration of Biomarkers of Psoriasis through Combined Multiomics Analysis. Mediat. Inflamm. 2022, 2022, 7731082. [Google Scholar] [CrossRef]
- Tokuyama, M.; Mabuchi, T. New Treatment Addressing the Pathogenesis of Psoriasis. Int. J. Mol. Sci. 2020, 21, 7488. [Google Scholar] [CrossRef] [PubMed]
- Albanesi, C.; Madonna, S.; Gisondi, P.; Girolomoni, G. The Interplay Between Keratinocytes and Immune Cells in the Pathogenesis of Psoriasis. Front. Immunol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Levine, B. Autophagy balances inflammation in innate immunity. Autophagy 2018, 14, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Rodriguez-Enriquez, S.; Lemasters, J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007, 462, 245–253. [Google Scholar] [CrossRef]
- Bonam, S.R.; Mastrippolito, D.; Georgel, P.; Muller, S. Pharmacological targets at the lysosomal autophagy-NLRP3 inflammasome crossroads. Trends Pharmacol. Sci. 2024, 45, 81–101. [Google Scholar] [CrossRef]
- Li, L.; Lu, H.; Zhang, Y.; Li, Q.; Shi, S.; Liu, Y. Effect of Azelaic Acid on Psoriasis Progression Investigated Based on Phosphatidylinositol 3-Kinase (PI3K)/Protein Kinase B (AKT) Signaling Pathway. Clin. Cosmet. Investig. Dermatol. 2022, 15, 2523–2534. [Google Scholar] [CrossRef]
- Bai, S.; Cheng, H.; Li, H.; Bo, P. Integrated bioinformatics analysis identifies autophagy-associated genes as candidate biomarkers and reveals the immune infiltration landscape in psoriasis. Autoimmunity 2024, 57, 2259137. [Google Scholar] [CrossRef]
- Douroudis, K.; Kingo, K.; Traks, T.; Reimann, E.; Raud, K.; Rätsep, R.; Mössner, R.; Silm, H.; Vasar, E.; Kõks, S. Polymorphisms in the ATG16L1 gene are associated with psoriasis vulgaris. Acta Derm. Venereol. 2012, 92, 85–87. [Google Scholar] [CrossRef]
- Yoshihara, N.; Ueno, T.; Takagi, A.; Oliva Trejo, J.A.; Haruna, K.; Suga, Y.; Komatsu, M.; Tanaka, K.; Ikeda, S. The significant role of autophagy in the granular layer in normal skin differentiation and hair growth. Arch. Dermatol. Res. 2015, 307, 159–169. [Google Scholar] [CrossRef]
- Peral de Castro, C.; Jones, S.A.; Cheallaigh, C.N.; Hearnden, C.A.; Williams, L.; Winter, J.; Lavelle, E.C.; Mills, K.H.; Harris, J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J. Immunol. 2012, 189, 4144–4153. [Google Scholar] [CrossRef]
- Varshney, P.; Saini, N. PI3K/AKT/mTOR activation and autophagy inhibition plays a key role in increased cholesterol during IL-17A mediated inflammatory response in psoriasis. Biochim. Et Biophys. Acta Mol. Basis Dis. 2018, 1864, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Rajawat, Y.; Hilioti, Z.; Bossis, I. Autophagy: A target for retinoic acids. Autophagy 2010, 6, 1224–1226. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Wang, S.; Xu, M.; Liu, M.; Liao, M.; Frank, J.A.; Adhikari, S.; Bower, K.A.; Shi, X.; et al. GSK3β signaling is involved in ultraviolet B-induced activation of autophagy in epidermal cells. Int. J. Oncol. 2012, 41, 1782–1788. [Google Scholar] [CrossRef]
- Silvagni, E.; Missiroli, S.; Patergnani, S.; Boncompagni, C.; D’Ugo, C.; Garaffoni, C.; Ciliento, M.S.; Lanza, G.; Bonora, M.; Gafà, R.; et al. Tofacitinib restores psoriatic arthritis fibroblast-like synoviocytes function via autophagy and mitochondrial quality control modulation. J. Autoimmun. 2024, 143, 103159. [Google Scholar] [CrossRef]
- Mimaki, M.; Wang, X.; McKenzie, M.; Thorburn, D.R.; Ryan, M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Et Biophys. Acta 2012, 1817, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Scacco, S.; Petruzzella, V.; Budde, S.; Vergari, R.; Tamborra, R.; Panelli, D.; van den Heuvel, L.P.; Smeitink, J.A.; Papa, S. Pathological mutations of the human NDUFS4 gene of the 18-kDa (AQDQ) subunit of complex I affect the expression of the protein and the assembly and function of the complex. J. Biol. Chem. 2003, 278, 44161–44167. [Google Scholar] [CrossRef]
- Poursadegh Zonouzi, A.; Ghorbian, S.; Abkar, M.; Poursadegh Zonouzi, A.A.; Azadi, A. Mitochondrial complex I gene variations; as a potential genetic risk factor in pathogenesis of multiple sclerosis. J. Neurol. Sci. 2014, 345, 220–223. [Google Scholar] [CrossRef]
- Xiang, H.; Tang, H.; He, Q.; Sun, J.; Yang, Y.; Kong, L.; Wang, Y. NDUFA8 is transcriptionally regulated by EP300/H3K27ac and promotes mitochondrial respiration to support proliferation and inhibit apoptosis in cervical cancer. Biochem. Biophys. Res. Commun. 2024, 693, 149374. [Google Scholar] [CrossRef]
- Zhou, Y.; Zou, J.; Xu, J.; Zhou, Y.; Cen, X.; Zhao, Y. Recent advances of mitochondrial complex I inhibitors for cancer therapy: Current status and future perspectives. Eur. J. Med. Chem. 2023, 251, 115219. [Google Scholar] [CrossRef]
- Söderman, J.; Almer, S. Discerning endoscopic severity of inflammatory bowel disease by scoping the peripheral blood transcriptome. Gastro Hep Adv. 2024, 3, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Söderman, J.; Berglind, L.; Almer, S. Inverse and Concordant Mucosal Pathway Gene Expressions in Inflamed and Non-Inflamed Ulcerative Colitis Patients: Potential Relevance to Aetiology and Pathogenesis. Int. J. Mol. Sci. 2022, 23, 6944. [Google Scholar] [CrossRef]
- Zhu, J.; Han, L.; Liu, R.; Zhang, Z.; Huang, Q.; Fang, X.; Yang, K.; Huang, G.; Zheng, Z.; Yawalkar, N.; et al. Identification of proteins associated with development of psoriatic arthritis in peripheral blood mononuclear cells: A quantitative iTRAQ-based proteomics study. J. Transl. Med. 2021, 19, 331. [Google Scholar] [CrossRef]
- Calabrese, C.; Nolte, H.; Pitman, M.R.; Ganesan, R.; Lampe, P.; Laboy, R.; Ripa, R.; Fischer, J.; Polara, R.; Panda, S.K.; et al. Mitochondrial translocation of TFEB regulates complex I and inflammation. EMBO Rep. 2024, 25, 704–724. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, J.; Kim, S.K.; Park, H.; Kim, J.E.; Lee, S. Autophagy: Guardian of Skin Barrier. Biomedicines 2022, 10, 1817. [Google Scholar] [CrossRef] [PubMed]
- Bisiak, F.; McCarthy, A.A. Structure and Function of Roundabout Receptors. Sub-Cell. Biochem. 2019, 93, 291–319. [Google Scholar] [CrossRef]
- Wang, B.; Xiao, Y.; Ding, B.B.; Zhang, N.; Yuan, X.; Gui, L.; Qian, K.X.; Duan, S.; Chen, Z.; Rao, Y.; et al. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell 2003, 4, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, T.; Wani, W.Y.; Kamal, M.A.; Kaur, R. Role of Angiogenic Growth Factors in Psoriasis: A Review. Curr. Drug Metab. 2018, 19, 910–916. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Assarsson, M.; Söderman, J.; Duvetorp, A.; Mrowietz, U.; Skarstedt, M.; Seifert, O. Narrowband UVB treatment induces expression of WNT7B, WNT10B and TCF7L2 in psoriasis skin. Arch. Dermatol. Res. 2019, 311, 535–544. [Google Scholar] [CrossRef]
- Teh, M.T.; Blaydon, D.; Ghali, L.R.; Briggs, V.; Edmunds, S.; Pantazi, E.; Barnes, M.R.; Leigh, I.M.; Kelsell, D.P.; Philpott, M.P. Role for WNT16B in human epidermal keratinocyte proliferation and differentiation. J. Cell Sci. 2007, 120, 330–339. [Google Scholar] [CrossRef]
- Wang, W.; Yu, X.; Wu, C.; Jin, H. IL-36γ inhibits differentiation and induces inflammation of keratinocyte via Wnt signaling pathway in psoriasis. Int. J. Med. Sci. 2017, 14, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Romhányi, D.; Szabó, K.; Kemény, L.; Sebestyén, E.; Groma, G. Transcriptional Analysis-Based Alterations Affecting Neuritogenesis of the Peripheral Nervous System in Psoriasis. Life 2022, 12, 111. [Google Scholar] [CrossRef]
- Cai, M.; Zheng, Q.; Chen, Y.; Liu, S.; Zhu, H.; Bai, B. Insights from the neural guidance factor Netrin-1 into neurodegeneration and other diseases. Front. Mol. Neurosci. 2024, 17, 1379726. [Google Scholar] [CrossRef]
- Fu, J.; Guo, O.; Zhen, Z.; Zhen, J. Essential Functions of the Transcription Factor Npas4 in Neural Circuit Development, Plasticity, and Diseases. Front. Neurosci. 2020, 14, 603373. [Google Scholar] [CrossRef]
- Metcalf, D.; Isaacs, A.M. The role of ESCRT proteins in fusion events involving lysosomes, endosomes and autophagosomes. Biochem. Soc. Trans. 2010, 38, 1469–1473. [Google Scholar] [CrossRef] [PubMed]
- Veleri, S.; Punnakkal, P.; Dunbar, G.L.; Maiti, P. Molecular Insights into the Roles of Rab Proteins in Intracellular Dynamics and Neurodegenerative Diseases. Neuromolecular Med. 2018, 20, 18–36. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, J.; Strunk, B.S.; Weisman, L.S. PI5P and PI(3,5)P(2): Minor, but Essential Phosphoinositides. Cell Struct. Funct. 2017, 42, 49–60. [Google Scholar] [CrossRef]
- Fredriksson, T.; Pettersson, U. Severe psoriasis–oral therapy with a new retinoid. Dermatologica 1978, 157, 238–244. [Google Scholar] [CrossRef]

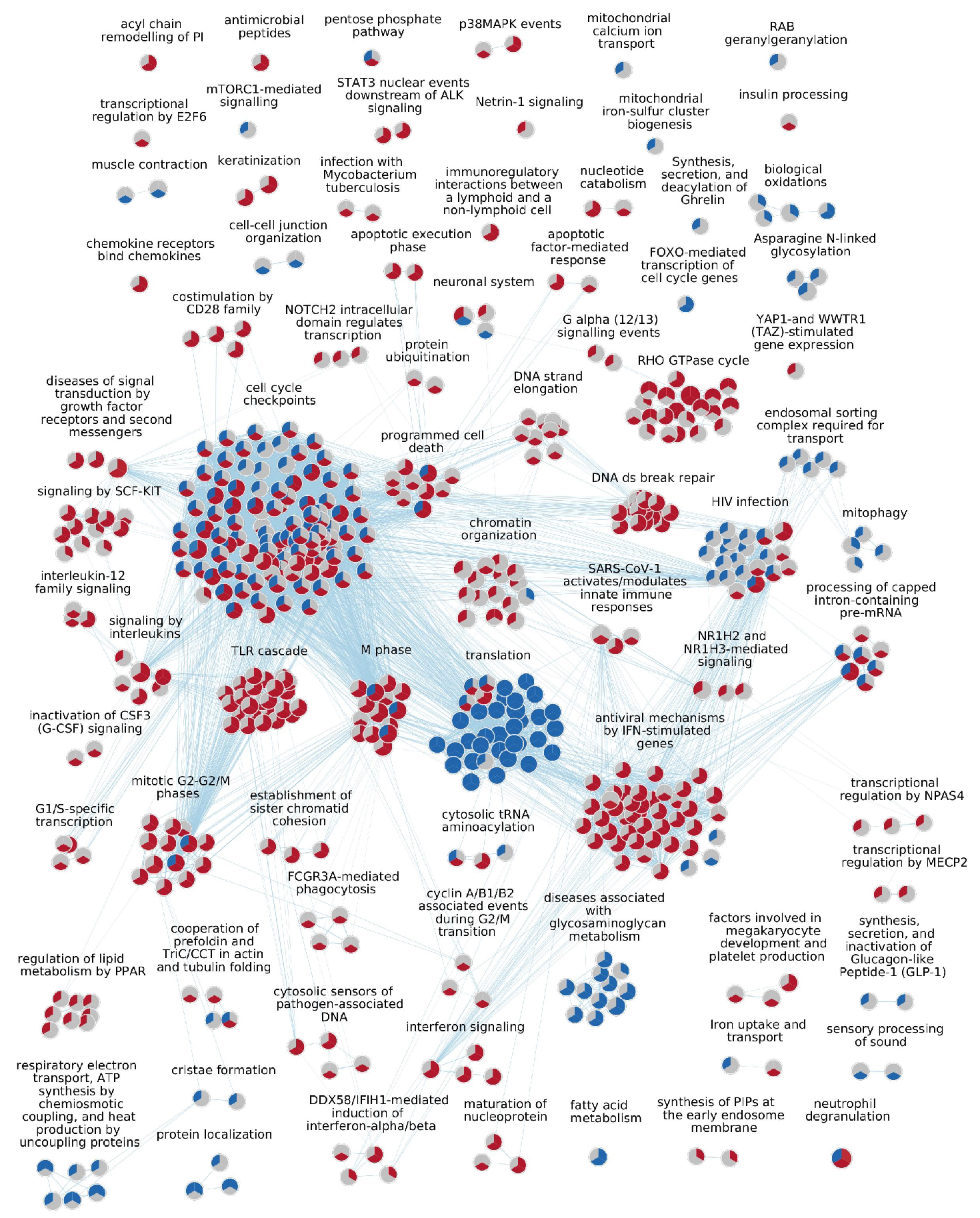
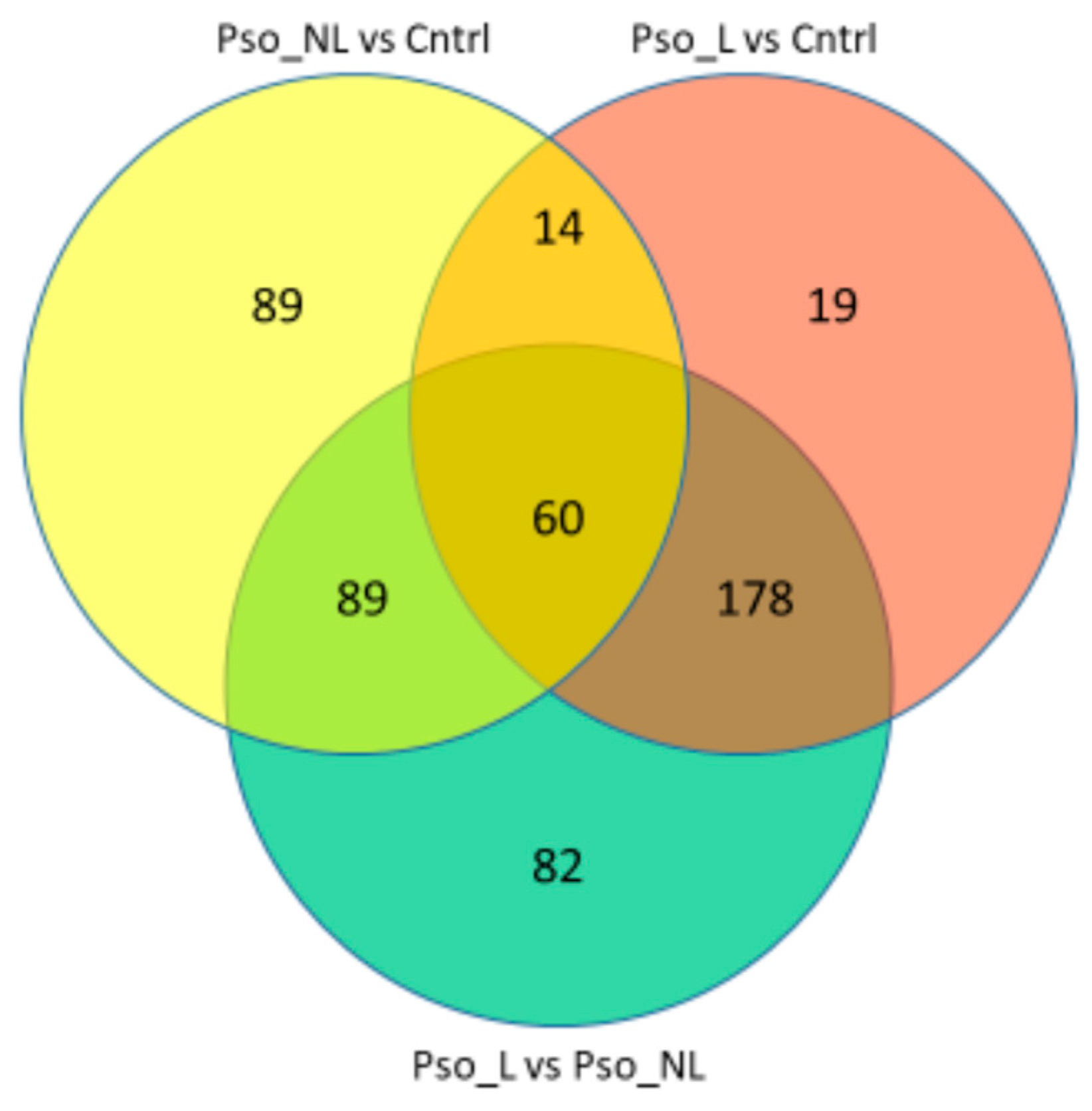
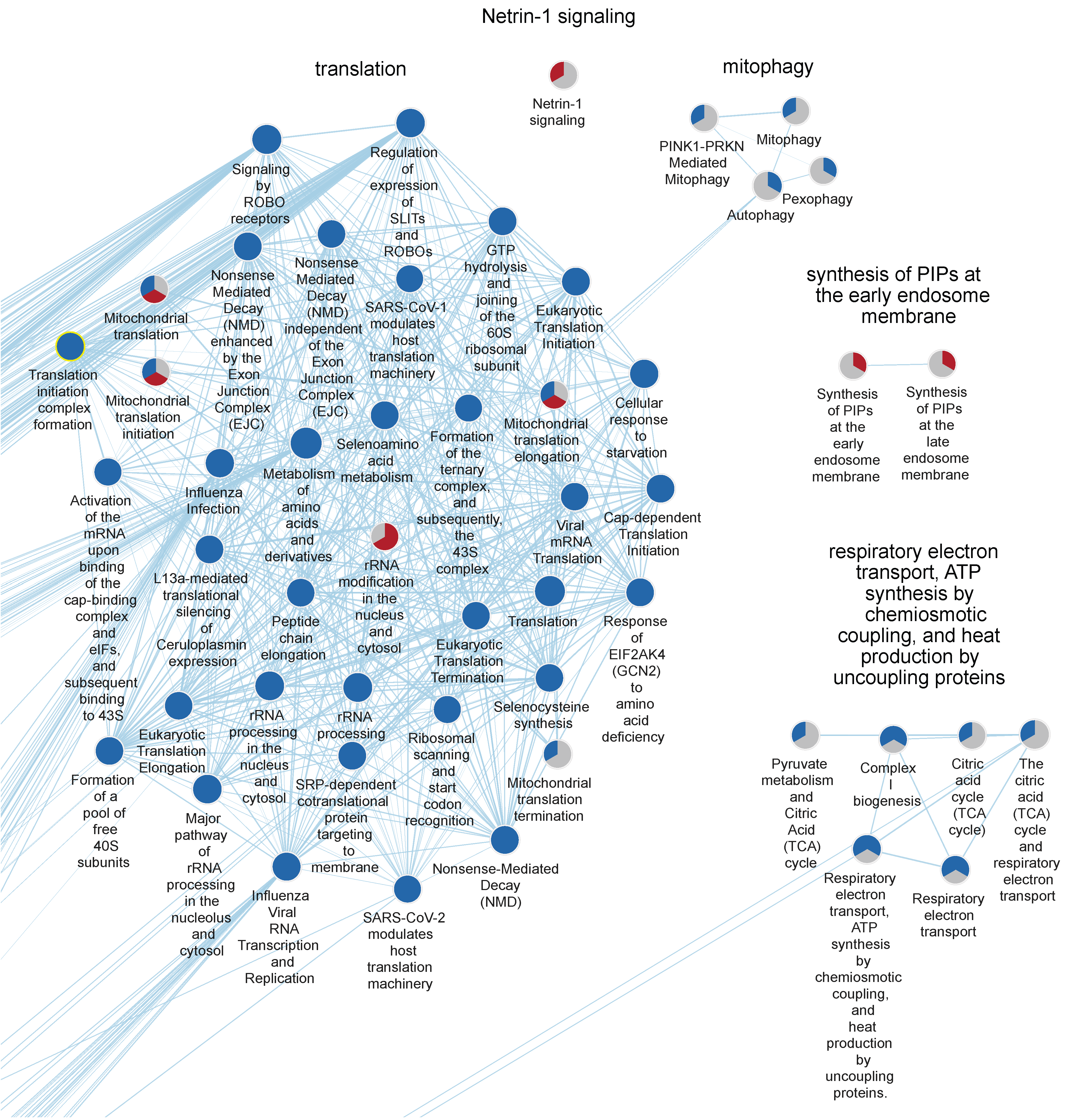
| Cluster Name | Median NES | Median % CEGs | Comb. FDR Bonferroni |
|---|---|---|---|
| UPREGULATED | |||
| lesional vs. control | |||
| M phase | 2.07 | 42 | 6.6 × 10−13 |
| Interferon signaling | 2.02 | 42.5 | 5.3 × 10−11 |
| Cell cycle checkpoints | 1.65 | 33 | 1.7 × 10−9 |
| Signaling by interleukins | 1.73 | 55 | 9.9 × 10−7 |
| Antiviral mechanism by IFN-stimulated genes | 1.96 | 50 | 1.8 × 10−6 |
| non-lesional vs. control | |||
| Chromatin organization | 1.69 | 21 | 4.4 × 10−5 |
| RHO GTPase cycle | 1.63 | 32 | 2.3 × 10−4 |
| G alpha (12/13) signaling events | 1.76 | 38 | 1.0 × 10−3 |
| Transcriptional regulation by NPAS4 | 1.81 | 37 | 2.3 × 10−3 |
| Netrin-1 signaling | 1.72 | 20 | 8.1 × 10−3 |
| lesional vs. non-lesional | |||
| Antiviral mechanism by IFN-stimulated genes | 1.98 | 56 | 7.4 × 10−7 |
| Keratinization | 1.92 | 16 | 3.7 × 10−6 |
| HIV infection | 1.70 | 38 | 6.1 × 10−6 |
| Antimicrobial peptides | 2.20 | 39 | 1.7 × 10−5 |
| DNA double-strand break repair | 2.04 | 41 | 3.1 × 10−5 |
| DOWNREGULATED | |||
| lesional vs. control | |||
| Translation | −2.30 | 64 | 3.8 × 10−21 |
| Protein localization | −1.78 | 49.5 | 2.5 × 10−3 |
| Fatty acid metabolism | −1.53 | 33 | 8.0 × 10−3 |
| Biological oxidations | −1.63 | 43 | 8.6 × 10−3 |
| Diseases associated with glycosaminoglycan metabolism | −1.72 | 62 | 1.2 × 10−2 |
| non-lesional vs. control | |||
| Translation | −2.30 | 48 | 9.4 × 10−27 |
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins | −2.52 | 55 | 6.1 × 10−19 |
| Protein localization | −2.22 | 59 | 2.9 × 10−9 |
| Cell cycle checkpoints | −2.19 | 58 | 3.9 × 10−7 |
| Processing of capped intron-containing pre-mRNA | −1.79 | 39 | 1.8 × 10−6 |
| lesional vs. non-lesional | |||
| Translation | −2.18 | 55 | 3.7 × 10−15 |
| Diseases associated with glycosaminoglycan metabolism | −1.81 | 61 | 8.9 × 10−3 |
| Sensory processing of sound | −1.69 | 35 | 9.1 × 10−3 |
| F cell cycle genes | −1.79 | 50 | 1.0 × 10−2 |
| Muscle contraction | −1.59 | 46 | 1.2 × 10−2 |
| Psoriasis | Control | |
|---|---|---|
| Number of subjects | 23 | 25 |
| Age, years, mean ± SD a (range) | 53.7 ± 14.3 (22–71) | 47.0 ± 18.1 (19–86) |
| Male/female ratio, n | 12:11 | 11:14 |
| PASI, mean ± SD (range) | 5.3 ± 3.3 (0.5–11.3) | - |
| Family history of psoriasis, % | 69.6 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Assarsson, M.; Söderman, J.; Dienus, O.; Seifert, O. Downregulation of Mitophagy, Complex I Biogenesis, and Signaling by ROBO Receptors—Implications for Psoriasis Pathogenesis. Int. J. Mol. Sci. 2025, 26, 5546. https://doi.org/10.3390/ijms26125546
Assarsson M, Söderman J, Dienus O, Seifert O. Downregulation of Mitophagy, Complex I Biogenesis, and Signaling by ROBO Receptors—Implications for Psoriasis Pathogenesis. International Journal of Molecular Sciences. 2025; 26(12):5546. https://doi.org/10.3390/ijms26125546
Chicago/Turabian StyleAssarsson, Malin, Jan Söderman, Olaf Dienus, and Oliver Seifert. 2025. "Downregulation of Mitophagy, Complex I Biogenesis, and Signaling by ROBO Receptors—Implications for Psoriasis Pathogenesis" International Journal of Molecular Sciences 26, no. 12: 5546. https://doi.org/10.3390/ijms26125546
APA StyleAssarsson, M., Söderman, J., Dienus, O., & Seifert, O. (2025). Downregulation of Mitophagy, Complex I Biogenesis, and Signaling by ROBO Receptors—Implications for Psoriasis Pathogenesis. International Journal of Molecular Sciences, 26(12), 5546. https://doi.org/10.3390/ijms26125546





