Integrated Meta-Analysis of Scalp Transcriptomics and Serum Proteomics Defines Alopecia Areata Subtypes and Core Disease Pathways
Abstract
1. Introduction
2. Results
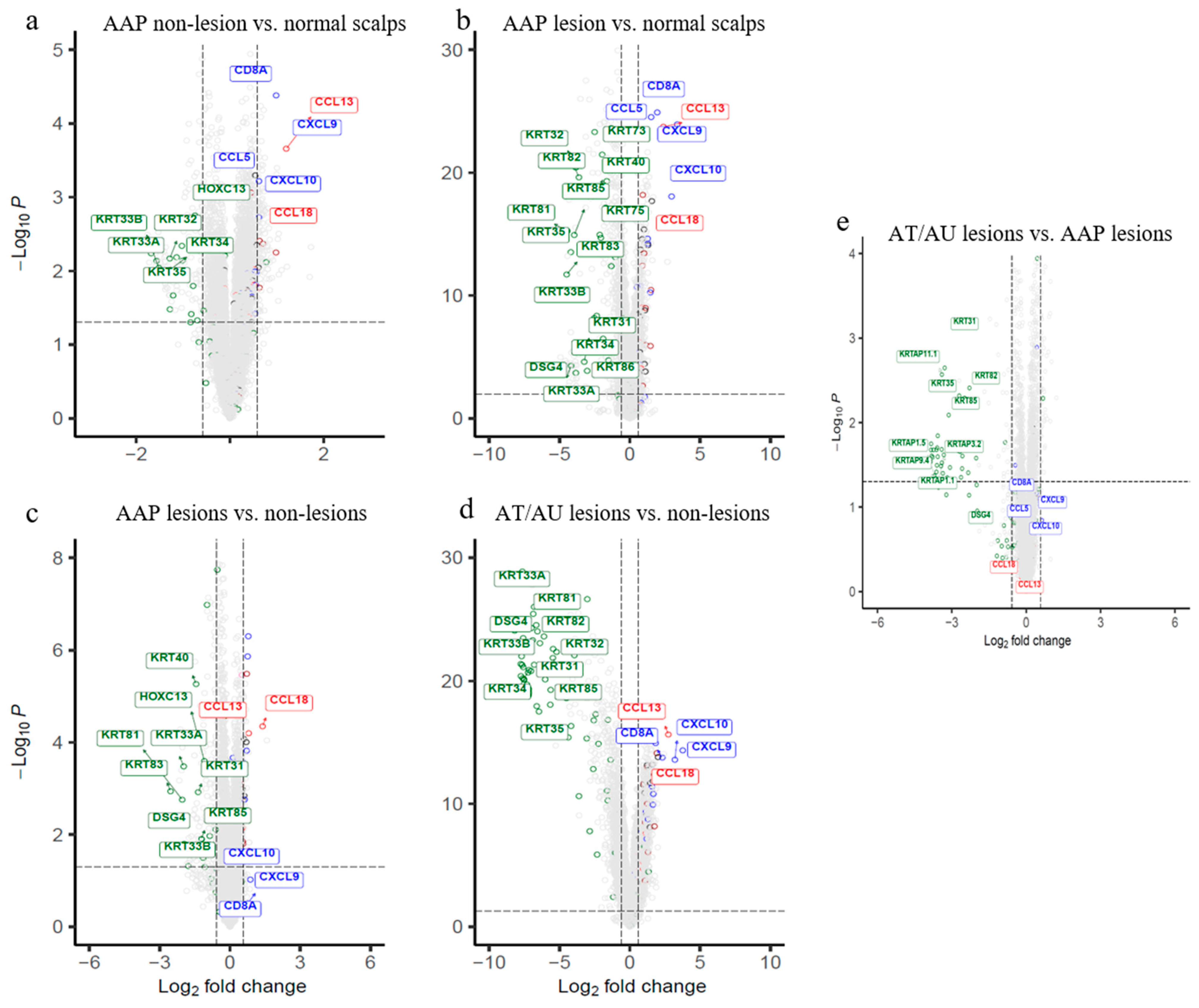
2.1. Subclinical Immune Activation and Early Keratin Changes in Non-Lesional AAP Scalp
2.2. Lesional AAP Scalp Shows Enhanced Immune Activation and Downregulation of Follicular Structural Genes
2.3. AT/AU Lesions Show Amplified Inflammation and Deeper Keratin Loss Compared to AAP Lesions
2.4. Evaluation of Ferroptosis-Associated Genes and IRS (Inner Root Sheath) Keratin Genes
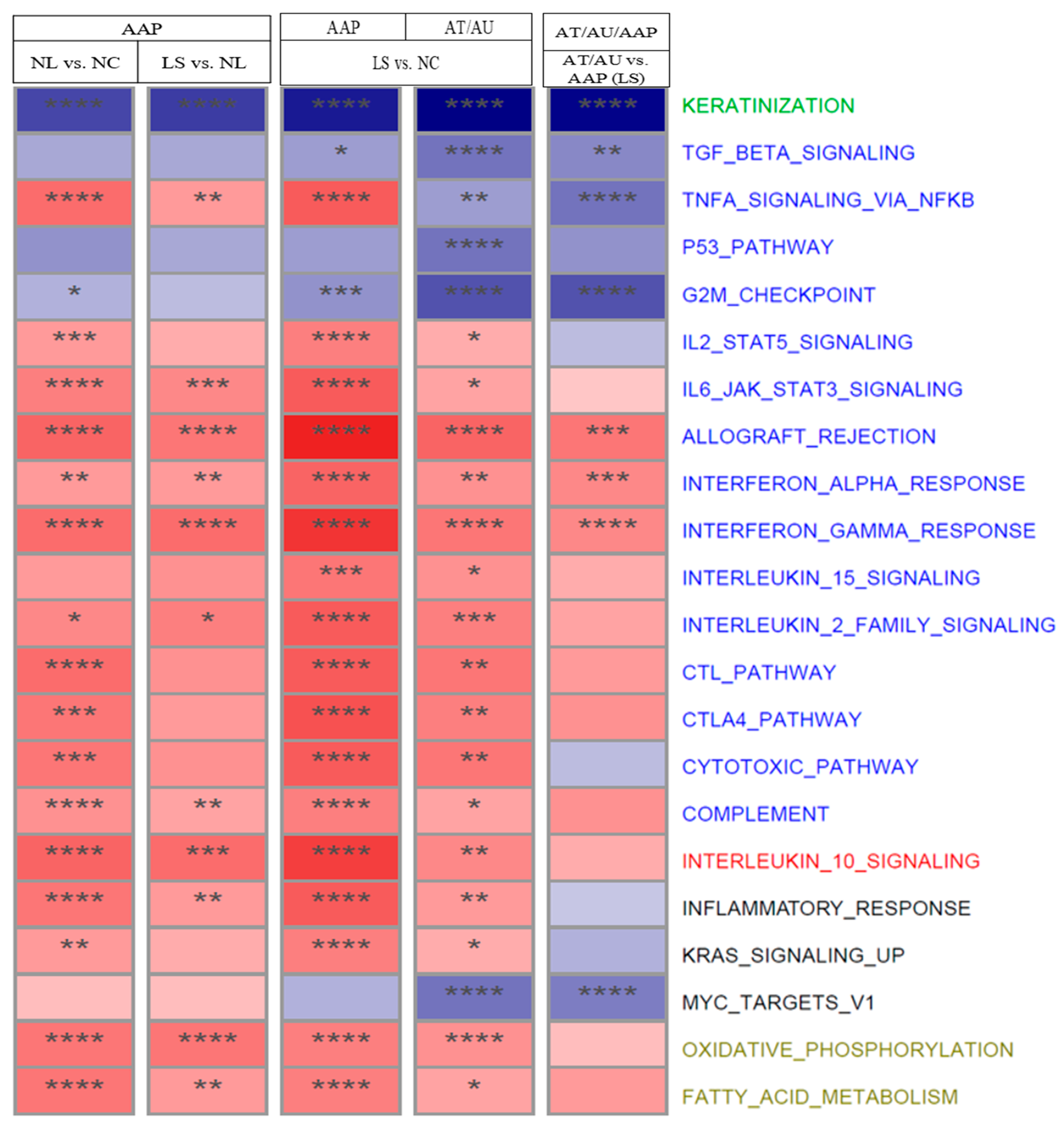
2.5. Pathway and Systemic Correlates Across Disease Stages
2.6. Concordant Serum Protein Changes
3. Discussion
3.1. Non-Lesional AAP Scalp Vs. Normal Scalps
3.2. Lesional AAP Vs. Normal Controls/Non-Lesional AAP
3.3. Lesional AT/AU Vs. AAP
3.4. Ferroptosis-Associated Genes and IRS (Inner Root Sheath) Keratin Genes
3.5. Pathway Enrichment Analysis Across AA Subtypes
3.6. Proteomic Profiles Across AA Subtypes
4. Materials and Methods
4.1. Transcriptomic Datasets
4.2. Serum Proteomics
4.3. Data Processing and Differential Expression Analysis
4.4. Meta-Analysis
4.5. Pathway Enrichment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Strazzulla, L.C.; Wang, E.H.C.; Avila, L.; Sicco, K.L.; Brinster, N.; Christiano, A.M.; Shapiro, J. Alopecia areata: Disease characteristics, clinical evaluation, and new perspectives on pathogenesis. J. Am. Acad. Dermatol. 2018, 78, 1–12. [Google Scholar] [CrossRef]
- Yamaguchi, H.L.; Yamaguchi, Y.; Peeva, E. Pathogenesis of alopecia areata and vitiligo: Commonalities and differences. Int. J. Mol. Sci. 2024, 25, 4409. [Google Scholar] [CrossRef] [PubMed]
- Guttman-Yassky, E.; Pavel, A.B.; Diaz, A.; Zhang, N.; Del Duca, E.; Estrada, Y.; King, B.; Banerjee, A.; Banfield, C.; Cox, L.A.; et al. Ritlecitinib and brepocitinib demonstrate significant improvement in scalp alopecia areata biomarkers. J. Allergy Clin. Immunol. 2022, 149, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Glickman, J.W.; Dubin, C.; Dahabreh, D.; Han, J.; Del Duca, E.; Estrada, Y.D.; Zhang, N.; Kimmel, G.W.; Singer, G.; Krueger, J.G.; et al. An integrated scalp and blood biomarker approach suggests the systemic nature of alopecia areata. Allergy 2021, 76, 3053–3065. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, A.; Cerise, J.E.; Chen, J.C.; Mackay-Wiggan, J.; Duvic, M.; Price, V.; Hordinsky, M.; Norris, D.; Clynes, R.; Christiano, A.M. Molecular signatures define alopecia areata subtypes and transcriptional biomarkers. EBioMedicine 2016, 7, 240–247. [Google Scholar] [CrossRef]
- Xing, L.; Dai, Z.; Jabbari, A.; Cerise, J.E.; Higgins, C.A.; Gong, W.; De Jong, A.; Harel, S.; DeStefano, G.M.; Rothman, L.; et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 2014, 20, 1043–1049. [Google Scholar] [CrossRef]
- Xu, W.; Wei, D.; Song, X. Identification of SLC40A1, LCN2, CREB5, and SLC7A11 as ferroptosis-related biomarkers in alopecia areata through machine learning. Sci. Rep. 2024, 14, 3800. [Google Scholar] [CrossRef]
- Wang, Z.; Wong, P.; Langbein, L.; Schweizer, J.; Coulombe, P.A. Type II Epithelial Keratin 6hf (K6hf) Is Expressed in the Companion Layer, Matrix, and Medulla in Anagen-Stage Hair Follicles. J. Investig. Dermatol. 2003, 121, 1276–1282. [Google Scholar] [CrossRef]
- Langbein, L.; Rogers, M.A.; Praetzel-Wunder, S.; Böckler, D.; Schirmacher, P.; Schweizer, J. Novel Type I Hair Keratins K39 and K40 Are the Last to Be Expressed in Differentiation of the Hair: Completion of the Human Hair Keratin Catalog. J. Investig. Dermatol. 2007, 127, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, A.; Sansaricq, F.; Cerise, J.; Chen, J.C.; Bitterman, A.; Ulerio, G.; Borbon, J.; Clynes, R.; Christiano, A.M.; Mackay-Wiggan, J. An open-label pilot study to evaluate the efficacy of tofacitinib in moderate to severe patch-type alopecia areata, totalis, and universalis. J. Investig. Dermatol. 2018, 138, 1539–1545. [Google Scholar] [CrossRef]
- Petukhova, L.; Duvic, M.; Hordinsky, M.; Norris, D.; Price, V.; Shimomura, Y.; Kim, H.; Singh, P.; Lee, A.; Chen, W.V.; et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 2010, 466, 113–117. [Google Scholar] [CrossRef]
- Zainodini, N.; Hassanshahi, G.; Arababadi, M.K.; Khorramdelazad, H.; Mirzaei, A. Differential expression of CXCL1, CXCL9, CXCL10 and CXCL12 chemokines in alopecia areata. Iran. J. Immunol. 2013, 10, 40–46. [Google Scholar]
- Gao, Z.; Jin, Y.Q.; Wu, W. SOCS3 treatment prevents the development of alopecia areata by inhibiting CD8⁺ T cell-mediated autoimmune destruction. Oncotarget 2017, 8, 33432–33443. [Google Scholar] [CrossRef]
- Dai, Z.; Xing, L.; Cerise, J.; Wang, E.H.; Jabbari, A.; de Jong, A.; Petukhova, L.; Christiano, A.M.; Clynes, R. CXCR3 blockade inhibits T cell migration into the skin and prevents development of alopecia areata. J. Immunol. 2016, 197, 1089–1099. [Google Scholar] [CrossRef]
- Lin, X.; Zhu, L.; He, J. Morphogenesis, growth cycle and molecular regulation of hair follicles. Front. Cell Dev. Biol. 2022, 10, 899095. [Google Scholar] [CrossRef] [PubMed]
- Trüeb, R.M.; Dias, M.F.R.G. Alopecia areata: A comprehensive review of pathogenesis and management. Clin. Rev. Allergy Immunol. 2018, 54, 68–87. [Google Scholar] [CrossRef] [PubMed]
- Barbulescu, C.C.; Goldstein, N.B.; Roop, D.R.; Norris, D.A.; Birlea, S.A. Harnessing the power of regenerative therapy for vitiligo and alopecia areata. J. Investig. Dermatol. 2020, 140, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.L.; Yamaguchi, Y.; Peeva, E. Hair regrowth in alopecia areata and re-pigmentation in vitiligo in response to treatment: Commonalities and differences. J. Eur. Acad. Dermatol. Venereol. 2024, 39, 498–511. [Google Scholar] [CrossRef]
- Peterle, L.; Sanfilippo, S.; Borgia, F.; Cicero, N.; Gangemi, S. Alopecia Areata: Oxidative Stress, Biomarkers, and Novel Therapeutic Approaches. Antioxidants 2023, 12, 135. [Google Scholar] [CrossRef]
- Reznik, A.S.; Vats, K.; Singh, K.; Mizes, A.; Jaklitsch, E.; Bayır, H.; Kagan, V.E.; Bunimovich, Y.L. Emerging Roles of Ferroptosis in Skin Pathophysiology. J. Investig. Dermatol. 2025, in press. [CrossRef]
- Langbein, L.; Rogers, M.A.; Winter, H.; Praetzel, S.; Beckhaus, U.; Rackwitz, H.R.; Schweizer, J. The Catalog of Human Hair Keratins: I. Expression of the Nine Type I Members in the Hair Follicle. J. Biol. Chem. 1999, 274, 19874–19884. [Google Scholar] [CrossRef] [PubMed]
- Glickman, J.W.; Dubin, C.; Renert-Yuval, Y.; Dahabreh, D.; Kimmel, G.W.; Auyeung, K.; Estrada, Y.D.; Singer, G.; Krueger, J.G.; Pavel, A.B.; et al. Cross-sectional study of blood biomarkers of patients with moderate to severe alopecia areata reveals systemic immune and cardiovascular biomarker dysregulation. J. Am. Acad. Dermatol. 2021, 84, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Lind, L.; Ärnlöv, J.; Lindahl, B.; Siegbahn, A.; Sundström, J.; Ingelsson, E. Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis 2015, 242, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Irizarry, R.A.; Gentleman, R.; Martinez-Murillo, F.; Spencer, F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004, 99, 909–917. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hartung, J.; Knapp, G. Statistical Meta-Analysis with Applications; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2008. [Google Scholar]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2019. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, L.; Peeva, E.; Yamaguchi, Y.; Ye, Z.; Hyde, C.L.; Guttman-Yassky, E. Integrated Meta-Analysis of Scalp Transcriptomics and Serum Proteomics Defines Alopecia Areata Subtypes and Core Disease Pathways. Int. J. Mol. Sci. 2025, 26, 9662. https://doi.org/10.3390/ijms26199662
Xi L, Peeva E, Yamaguchi Y, Ye Z, Hyde CL, Guttman-Yassky E. Integrated Meta-Analysis of Scalp Transcriptomics and Serum Proteomics Defines Alopecia Areata Subtypes and Core Disease Pathways. International Journal of Molecular Sciences. 2025; 26(19):9662. https://doi.org/10.3390/ijms26199662
Chicago/Turabian StyleXi, Li, Elena Peeva, Yuji Yamaguchi, Zhan Ye, Craig L. Hyde, and Emma Guttman-Yassky. 2025. "Integrated Meta-Analysis of Scalp Transcriptomics and Serum Proteomics Defines Alopecia Areata Subtypes and Core Disease Pathways" International Journal of Molecular Sciences 26, no. 19: 9662. https://doi.org/10.3390/ijms26199662
APA StyleXi, L., Peeva, E., Yamaguchi, Y., Ye, Z., Hyde, C. L., & Guttman-Yassky, E. (2025). Integrated Meta-Analysis of Scalp Transcriptomics and Serum Proteomics Defines Alopecia Areata Subtypes and Core Disease Pathways. International Journal of Molecular Sciences, 26(19), 9662. https://doi.org/10.3390/ijms26199662





