Brassinosteroid Synthesis and Perception Differently Regulate Phytohormone Networks in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
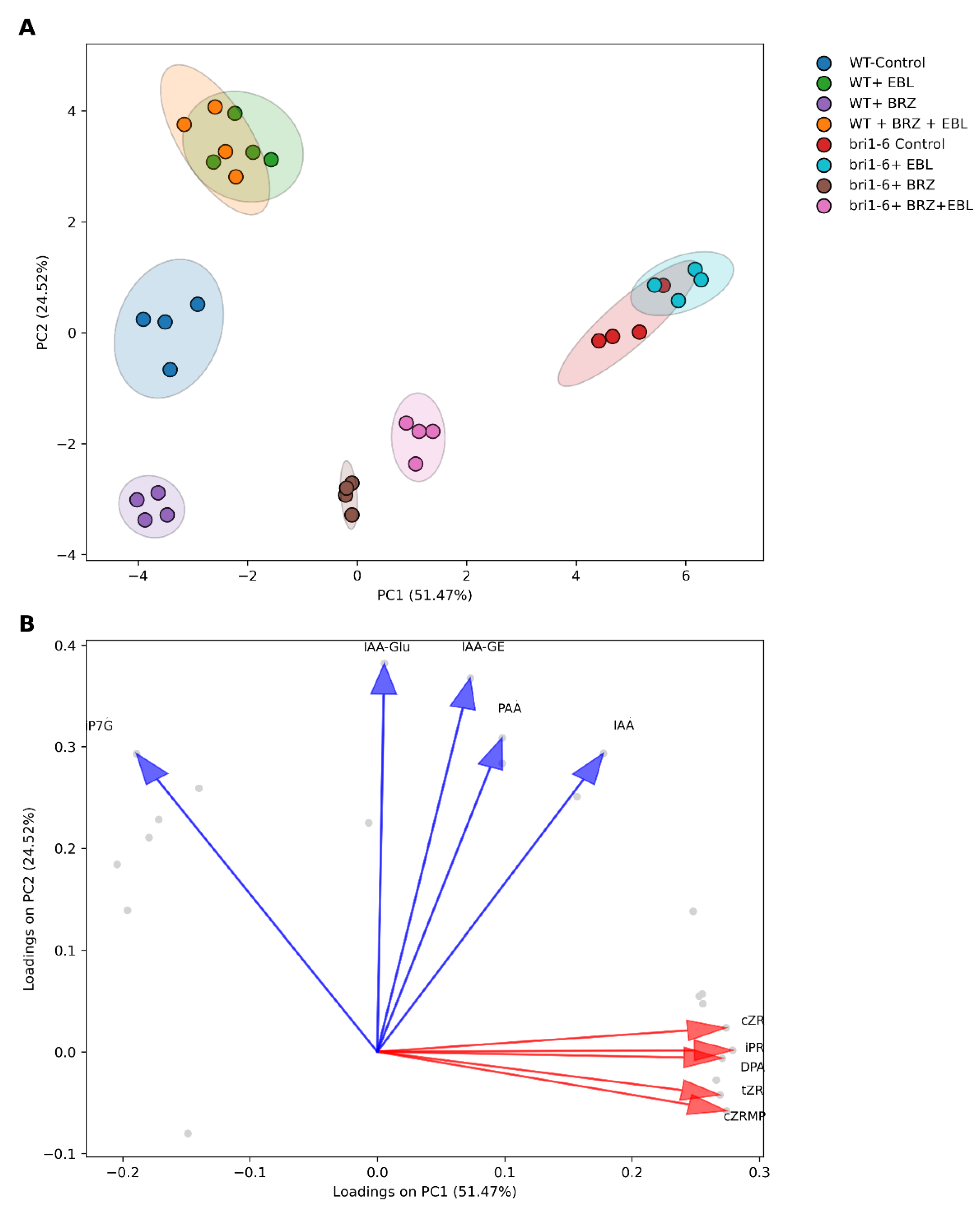
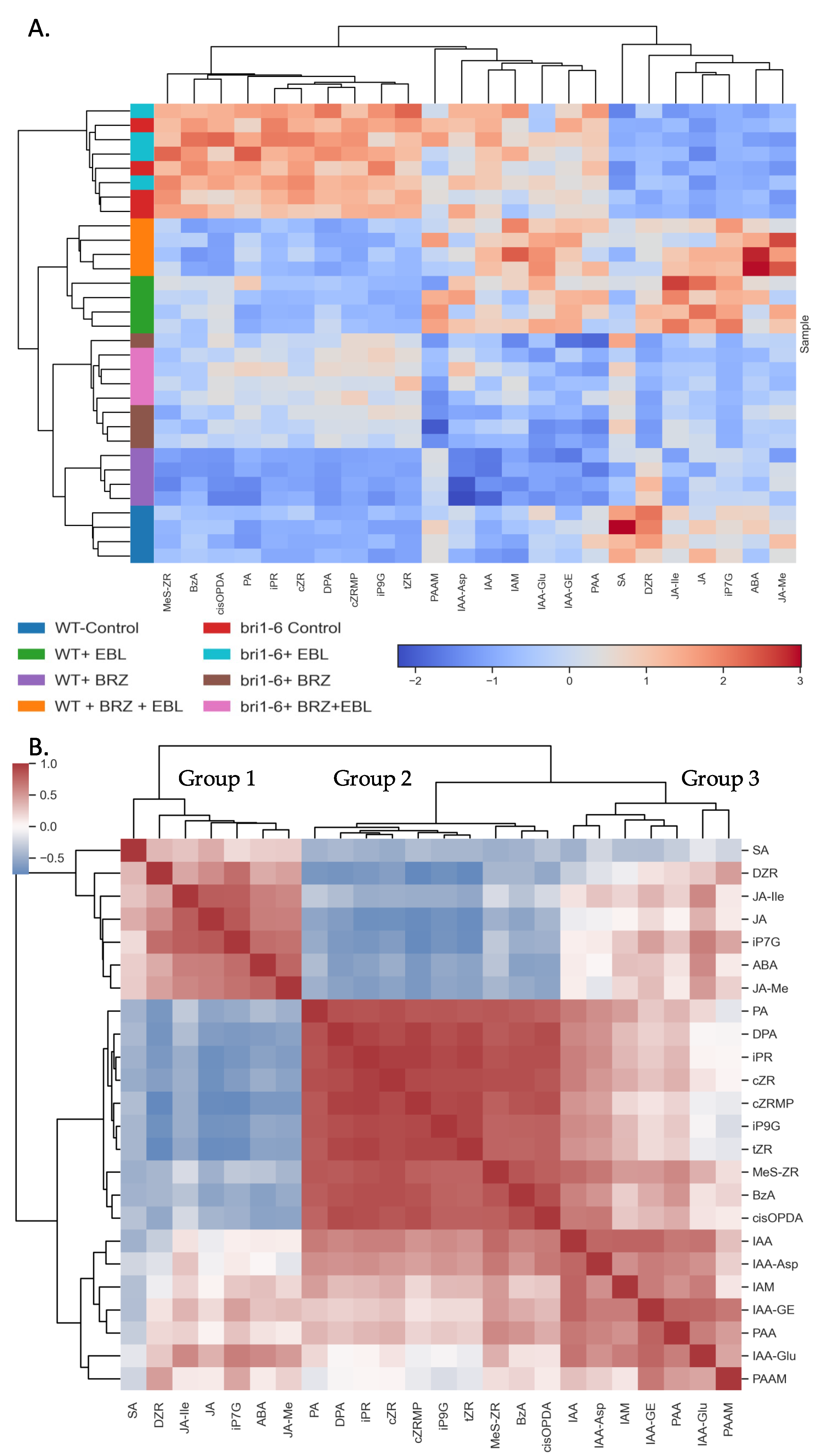
2.1. Overall Survey of the Effect of EBL and BRZ on the Two Genotypes
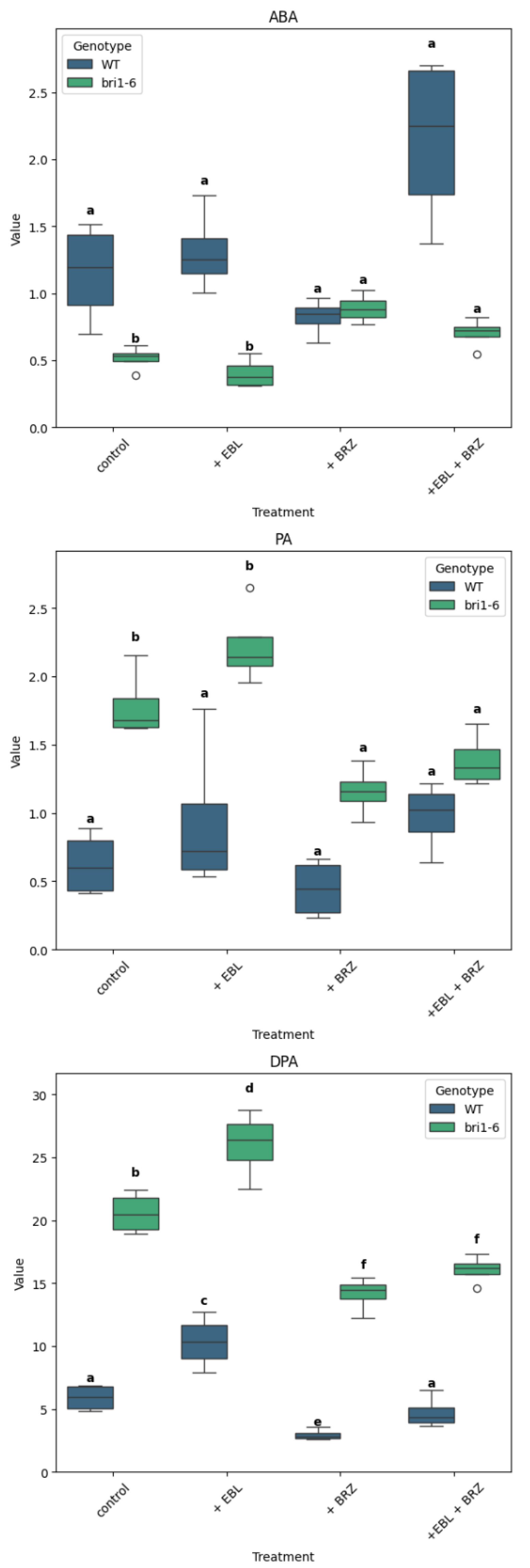
2.2. The Impact of EBL and Brassinazole (BRZ) on ABA and Its Metabolites
2.3. The Impact of EBL and Brassinazole (BRZ) on SA and Its Metabolites
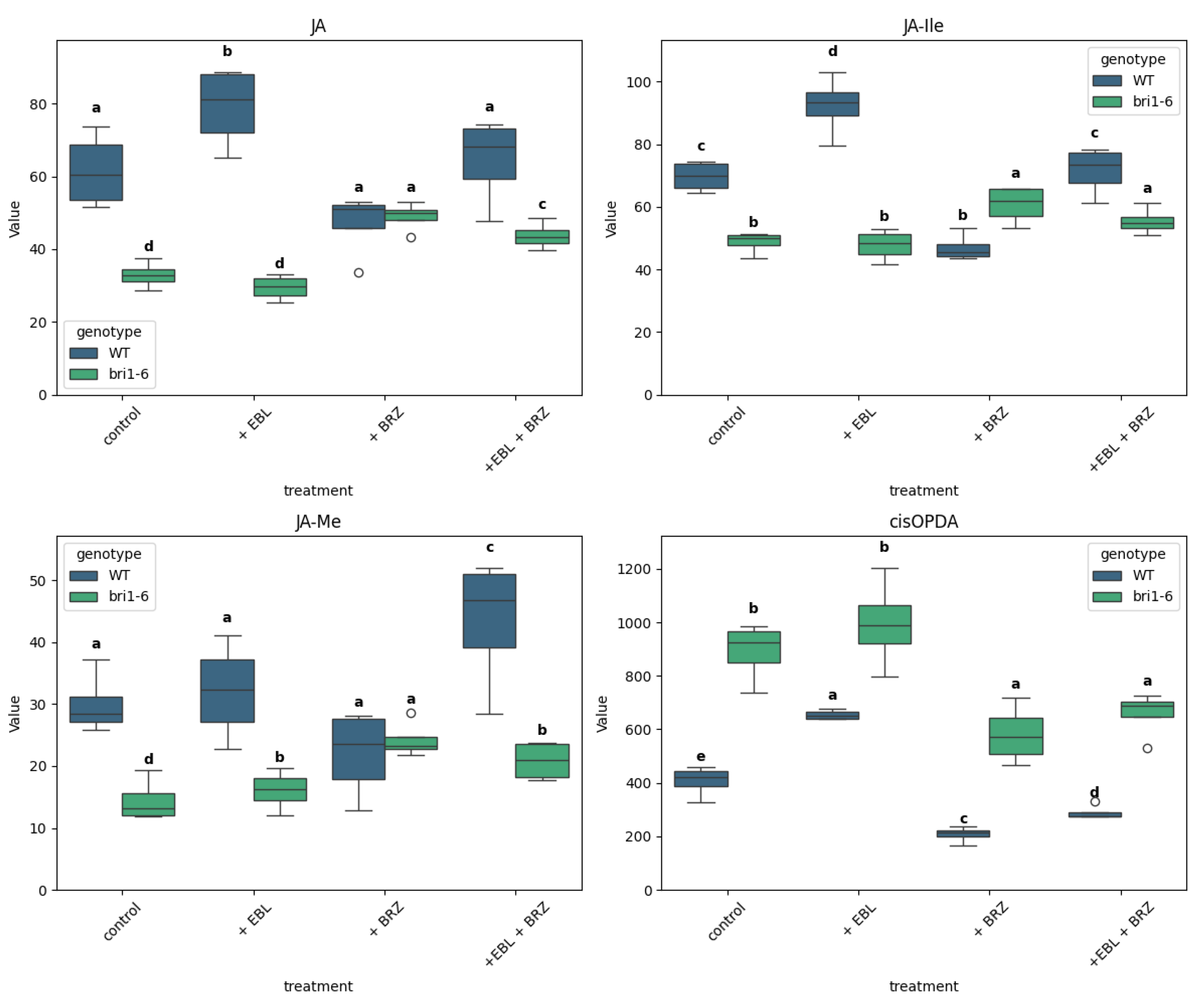
2.4. The Impact of EBL and Brassinazole (BRZ) on JA and Related Metabolites
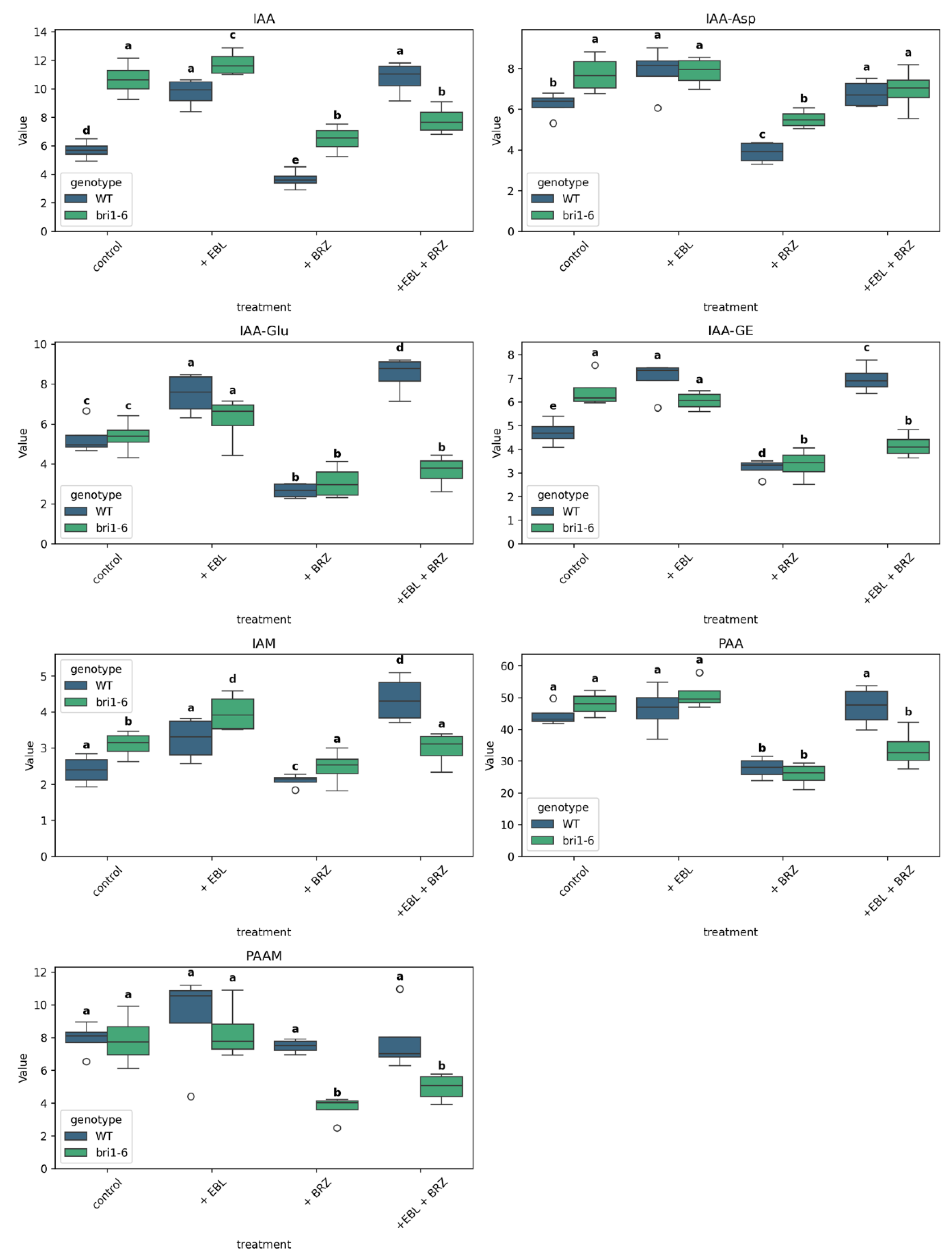
2.5. The Impact of EBL and Brassinazole (BRZ) on Auxins
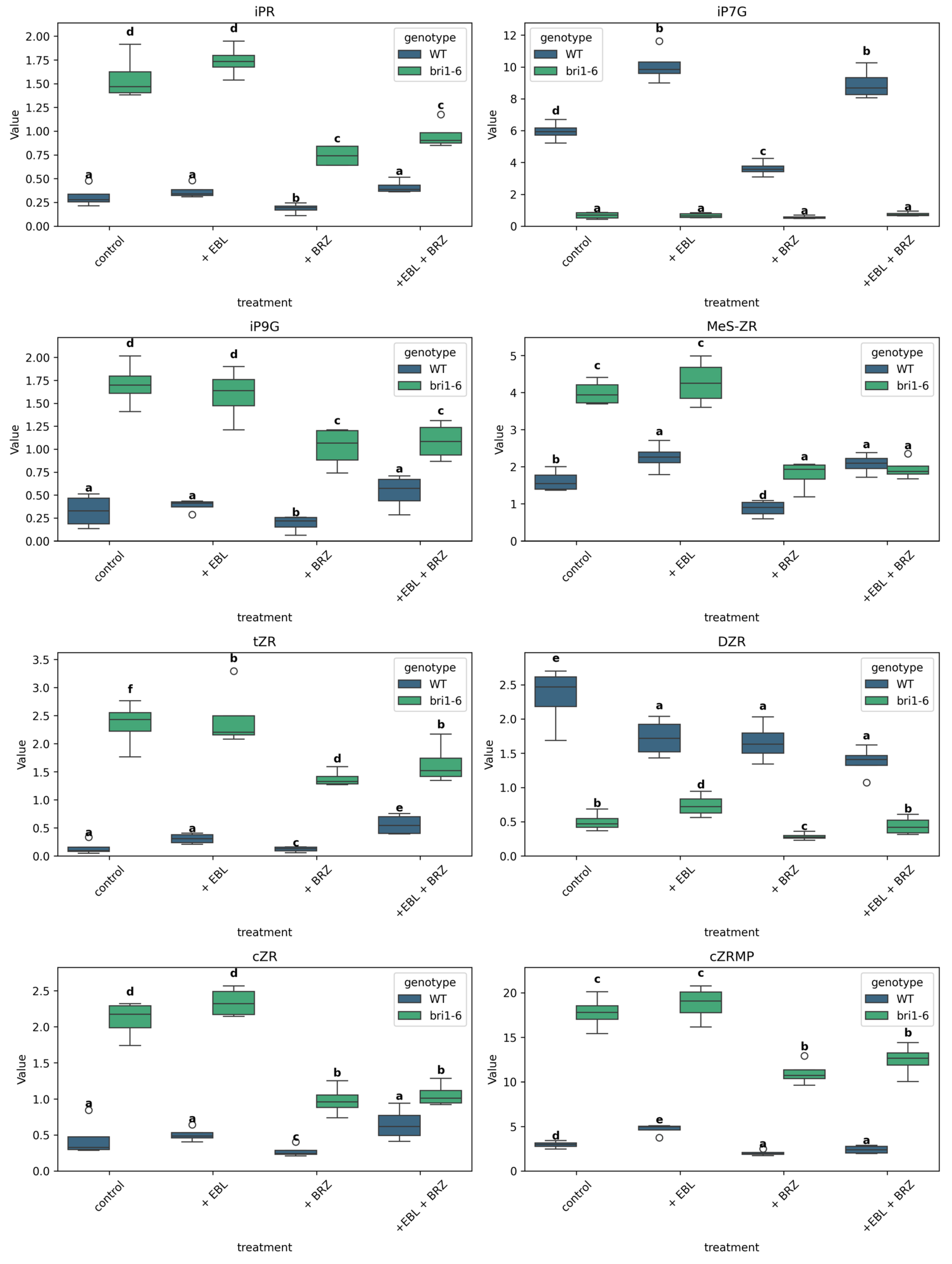
2.6. The Impact of EBL and Brassinazole (BRZ) on Cytokinins
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Preparation of Chemical Stocks
4.3. Hormone Extraction and Quantification
4.4. Statistical Analysis
4.4.1. Analysis of Variance (ANOVA) and Post Hoc Tests
4.4.2. Boxplots
4.4.3. Principal Component Analysis (PCA)
4.4.4. Heatmap
4.4.5. Software and Code Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| ABA-GE | Abscisic Acid Glucose Ester |
| BAK1 | Brassinosteroid-Insensitive 1-Associated Receptor Kinase 1 |
| BES1 | Brassinosteroid-Insensitive 1-EMS-Suppressor 1 |
| BzA | Benzoic Acid |
| BR | Brassinosteroids |
| BRZ | Brassinazole (Brassinosteroid Biosynthesis Inhibitor) |
| BZR1 | Brassinosteroid-Resistant 1 |
| BRI1 | Brassinosteroid Insensitive 1 Receptor Kinase |
| BR6ox | Brassinosteroid-6-Oxidase |
| DPA | Dihydrophaseic Acid |
| DZR | Dihydrozeatin Riboside |
| EBL | 24-Epibrassinolide |
| JA | Jasmonic Acid |
| JA-Ile | Jasmonic Acid-Isoleucine |
| JA-Me | Jasmonic Acid Methyl Ester |
| IAA | Indole-3-Acetic Acid |
| IAA-Asp | Indole-3-Acetic Acid Aspartate |
| IAA-GE | Indole-3-Acetic Acid Glucose Ester |
| IAA-Glu | Indole-3-Acetic Acid Glutamine |
| IAM | Indole-3-Acetamide |
| iP | Isopentenyl Adenine |
| iP7G – | Isopentenyl Adenine-7-Glucoside |
| iP9G | Isopentenyl Adenine-9-Glucoside |
| iPR | Isopentenyl Adenine Riboside |
| MeS-ZR | 2-Methylthio Zeatin Ribosides of Trans-Zeatin |
| PCA | Principal Component Analysis |
| PA | Phaseic Acid |
| PAA | Phenylacetic Acid |
| PAA-M | Phenylacetamide |
| SA | Salicylic Acid |
| tZR | Trans-Zeatin Riboside |
| tZ | Trans-Zeatin |
| tZRMP | Trans-Zeatin Riboside Monophosphate |
| ZR | Zeatin Riboside |
References
- Sun, S.; Zhao, X.; Shi, Z.; He, F.; Qi, G.; Li, X.; Niu, Y.; Zhou, W. Exogenous 24-Epibrassinolide Improves Low-Temperature Tolerance of Maize Seedlings by Influencing Sugar Signaling and Metabolism. Int. J. Mol. Sci. 2025, 26, 585. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Vukašinović, N.; Wang, Y.; Vanhoutte, I.; Fendrych, M.; Guo, B.; Kvasnica, M.; Jiroutová, P.; Oklestkova, J.; Strnad, M.; Russinova, E. Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 2021, 7, 619–632, Erratum in Nat. Plants 2024, 10, 1052. [Google Scholar] [CrossRef]
- Naveen, N.; Kumari, N.; Avtar, R.; Jattan, M.; Ahlawat, S.; Rani, B.; Malik, K.; Sharma, A.; Singh, M. Evaluation of Effect of Brassinolide in Brassica juncea Leaves under Drought Stress in Field Conditions. Horticulturae 2021, 7, 514. [Google Scholar] [CrossRef]
- Sun, S.; Yao, X.; Liu, X.; Qiao, Z.; Liu, Y.; Li, X.; Jiang, X. Brassinolide can improve drought tolerance of maize seedlings under drought stress: By inducing the photosynthetic performance, antioxidant capacity and ZmMYB gene expression of maize seedlings. J. Soil Sci. Plant Nutr. 2022, 22, 2092–2104. [Google Scholar] [CrossRef]
- Lu, K.; Gu, Y.; Du, Y.; Yao, Y.; Tan, X.; Wu, L.; Zhou, J.; Yuan, J. NAC transcription factors are key regulators of Brassinolide-Enhanced drought tolerance in Camellia oil tree. BMC Plant Biol. 2025, 25, 625. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, D.; Zhang, Y.; Du, Y.; Wang, M.; Liu, J.; Chu, J.; Yao, X. Brassinolide alleviated drought stress faced by bulbil formation of Pinellia ternata by reducing ROS metabolism and enhancing AsA-GSH cycle. Sci. Hortic. 2024, 323, 112525. [Google Scholar] [CrossRef]
- El-Mashad, A.A.A.; Mohamed, H.I. Brassinolide alleviates salt stress and increases antioxidant activity of cowpea plants (Vigna sinensis). Protoplasma 2012, 249, 625–635. [Google Scholar] [CrossRef]
- Mu, D.; Feng, N.; Zheng, D.; Zhou, H.; Liu, L.; Chen, G.; Mu, B. Physiological mechanism of exogenous brassinolide alleviating salt stress injury in rice seedlings. Sci. Rep. 2022, 12, 20439. [Google Scholar] [CrossRef]
- Sanchez, F.W.; Crane, J.H.; Bayabil, H.K.; Sarkhosh, A.; Shahid, M.A.; Schaffer, B. Brassinosteroid priming mitigates negative physiological responses of Garcina humilis to flooding and salinity. Plant Stress 2025, 16, 100892. [Google Scholar] [CrossRef]
- Tao, Y.; Yu, Q.-X.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Yu, J.-Q.; Xia, X.-J. Application of 24-epibrassinolide decreases the susceptibility to cucumber mosaic virus in zucchini (Cucurbita pepo L). Sci. Hortic. 2015, 195, 116–123. [Google Scholar] [CrossRef]
- Liu, W.; Guo, X.; Chen, Q.; Fu, L.; Wei, R.; Ali, M.M.; Shah, M.S. Brassinolide-induced resistance enhances antioxidant defense and metabolic pathways against anthracnose in Camellia sinensis ‘Fuding Dabaicha’. Sci. Hortic. 2025, 349, 114258. [Google Scholar] [CrossRef]
- Silva, A.P.S.D.; Alencar, A.A.D.S.; Sudré, C.P.; Araújo, M.D.S.B.D.; Lobato, A.K.D.S. Brassinosteroids: Relevant Evidence Related to Mitigation of Abiotic and Biotic Stresses in Plants. Agronomy 2024, 14, 840. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, F.; Xie, Q. Balancing growth and adaptation to stress: Crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 2020, 43, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Park, C.-H.; Son, S.-H.; Youn, J.-H.; Kim, S.-K. Endogenous level of abscisic acid down-regulated by brassinosteroids signaling via BZR1 to control the growth of Arabidopsis thaliana. Plant Signal. Behav. 2021, 16, 1926130. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Choe, S. Brassinosteroid biosynthesis anddwarf mutants. J. Plant Biol. 2005, 48, 1–15. [Google Scholar] [CrossRef]
- Zebosi, B.; Vollbrecht, E.; Best, N.B. Brassinosteroid biosynthesis and signaling: Conserved and diversified functions of core genes across multiple plant species. Plant Commun. 2024, 5, 100982. [Google Scholar] [CrossRef]
- Vukašinović, N.; Russinova, E. BRexit: Possible Brassinosteroid Export and Transport Routes. Trends Plant Sci. 2018, 23, 285–292. [Google Scholar] [CrossRef]
- Starodubtseva, A.; Kalachova, T.; Iakovenko, O.; Stoudková, V.; Zhabinskii, V.; Khripach, V.; Ruelland, E.; Martinec, J.; Burketová, L.; Kravets, V. BODIPY Conjugate of Epibrassinolide as a Novel Biologically Active Probe for In Vivo Imaging. Int. J. Mol. Sci. 2021, 22, 3599. [Google Scholar] [CrossRef]
- Nagata, N.; Asami, T.; Yoshida, S. Brassinazole, an Inhibitor of Brassinosteroid Biosynthesis, Inhibits Development of Secondary Xylem in Cress Plants (Lepidium sativum). Plant Cell Physiol. 2001, 42, 1006–1011. [Google Scholar] [CrossRef]
- Rozhon, W.; Akter, S.; Fernandez, A.; Poppenberger, B. Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction. Molecules 2019, 24, 4372. [Google Scholar] [CrossRef]
- Bajguz, A.; Chmur, M.; Gruszka, D. Comprehensive Overview of the Brassinosteroid Biosynthesis Pathways: Substrates, Products, Inhibitors, and Connections. Front. Plant Sci. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Chmur, M.; Bajguz, A. Brassinolide Enhances the Level of Brassinosteroids, Protein, Pigments, and Monosaccharides in Wolffia arrhiza Treated with Brassinazole. Plants 2021, 10, 1311. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A Putative Leucine-Rich Repeat Receptor Kinase Involved in Brassinosteroid Signal Transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Xu, S.; Li, J. BAK 1 Directly Regulates Brassinosteroid Perception and BRI 1 Activation. J. Integr. Plant Biol. 2013, 55, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wei, Y.; Shen, B.; Liu, L.; Mao, J. Interaction of the Transcription Factors BES1/BZR1 in Plant Growth and Stress Response. Int. J. Mol. Sci. 2024, 25, 6836. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383, Erratum in Nature 2001, 411, 219. [Google Scholar] [CrossRef]
- Wang, W.; Bai, M.-Y.; Wang, Z.-Y. The brassinosteroid signaling network—A paradigm of signal integration. Curr. Opin. Plant Biol. 2014, 21, 147–153. [Google Scholar] [CrossRef]
- Noguchi, T.; Fujioka, S.; Choe, S.; Takatsuto, S.; Yoshida, S.; Yuan, H.; Feldmann, K.A.; Tax, F.E. Brassinosteroid-Insensitive Dwarf Mutants of Arabidopsis Accumulate Brassinosteroids. Plant Physiol. 1999, 121, 743–752. [Google Scholar] [CrossRef]
- Zada, A.; Lv, M.; Li, J. Molecular Lesions in BRI1 and Its Orthologs in the Plant Kingdom. Int. J. Mol. Sci. 2024, 25, 8111. [Google Scholar] [CrossRef]
- Yang, X.; Bai, Y.; Shang, J.; Xin, R.; Tang, W. The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 2016, 39, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D. Brassinosteroid/Abscisic Acid Antagonism in Balancing Growth and Stress. Dev. Cell 2016, 38, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, Z.; Xie, S.; Li, Z.; Zhou, Y.; Duan, L. Jasmonate mimic modulates cell elongation by regulating antagonistic bHLH transcription factors via brassinosteroid signaling. Plant Physiol. 2024, 195, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Peng, Y.-J.; Yuan, L.-B.; Dai, Y.-S.; Chen, Q.-F.; Yu, L.-J.; Bai, M.-Y.; Zhang, W.-Q.; Xie, L.-J.; Xiao, S. Brassinosteroids Antagonize Jasmonate-Activated Plant Defense Responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 2020, 182, 1066–1082. [Google Scholar] [CrossRef]
- Sánchez-Parra, B.; Frerigmann, H.; Alonso, M.-M.; Loba, V.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants 2014, 3, 324–347. [Google Scholar] [CrossRef]
- Perez, V.C.; Zhao, H.; Lin, M.; Kim, J. Occurrence, Function, and Biosynthesis of the Natural Auxin Phenylacetic Acid (PAA) in Plants. Plants 2023, 12, 266. [Google Scholar] [CrossRef]
- Huang, J.; Van Der Hoorn, R.A.L. The ancestral salicylic acid biosynthesis pathway in plants. Trends Plant Sci. 2025, S1360138525002304. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.S.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.-H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; Liu, J.; Zhang, P.; Kudoyarova, G.; Liu, C.-J.; Zhang, K. Spatially distributed cytokinins: Metabolism, signaling, and transport. Plant Commun. 2024, 5, 100936. [Google Scholar] [CrossRef]
- Asami, T.; Mizutani, M.; Fujioka, S.; Goda, H.; Min, Y.K.; Shimada, Y.; Nakano, T.; Takatsuto, S.; Matsuyama, T.; Nagata, N.; et al. Selective Interaction of Triazole Derivatives with DWF4, a Cytochrome P450 Monooxygenase of the Brassinosteroid Biosynthetic Pathway, Correlates with Brassinosteroid Deficiency in Planta. J. Biol. Chem. 2001, 276, 25687–25691. [Google Scholar] [CrossRef]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.-C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef]
- Min, Y.K.; Asami, T.; Fujioka, S.; Murofushi, N.; Yamaguchi, I.; Yoshida, S. New lead compounds for brassinosteroid biosynthesis inhibitors. Bioorg. Med. Chem. Lett. 1999, 9, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Okamoto, M.; Shinoda, S.; Kushiro, T.; Koshiba, T.; Kamiya, Y.; Hirai, N.; Todoroki, Y.; Sakata, K.; Nambara, E.; et al. A Plant Growth Retardant, Uniconazole, Is a Potent Inhibitor of ABA Catabolism in Arabidopsis. Biosci. Biotechnol. Biochem. 2006, 70, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-Localized BZR1 Mediates Brassinosteroid-Induced Growth and Feedback Suppression of Brassinosteroid Biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Ma, J.; Zhang, M.; Li, X.; Sun, Y.; Zhang, M.; Ding, Z. Auxin promotes hypocotyl elongation by enhancing BZR1 nuclear accumulation in Arabidopsis. Sci. Adv. 2023, 9, eade2493. [Google Scholar] [CrossRef]
- Sun, L.; Feraru, E.; Feraru, M.I.; Waidmann, S.; Wang, W.; Passaia, G.; Wang, Z.-Y.; Wabnik, K.; Kleine-Vehn, J. PIN-LIKES Coordinate Brassinosteroid Signaling with Nuclear Auxin Input in Arabidopsis thaliana. Curr. Biol. 2020, 30, 1579–1588.e6. [Google Scholar] [CrossRef]
- Mateo-Bonmatí, E.; Casanova-Sáez, R.; Šimura, J.; Ljung, K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021, 232, 642–654. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Leung, D.W.M.; He, Z.; Zhang, J.; Peng, X.; Liu, E. Overexpression of OsIAAGLU reveals a role for IAA–glucose conjugation in modulating rice plant architecture. Plant Cell Rep. 2019, 38, 731–739. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Prerostova, S.; Černý, M.; Dobrev, P.I.; Motyka, V.; Hluskova, L.; Zupkova, B.; Gaudinova, A.; Knirsch, V.; Janda, T.; Brzobohatý, B.; et al. Light Regulates the Cytokinin-Dependent Cold Stress Responses in Arabidopsis. Front. Plant Sci. 2021, 11, 608711. [Google Scholar] [CrossRef]
- Nguyen, H.N.; Nguyen, T.Q.; Kisiala, A.B.; Emery, R.J.N. Beyond transport: Cytokinin ribosides are translocated and active in regulating the development and environmental responses of plants. Planta 2021, 254, 45. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Moonlighting PPKL1 reveals a role of cytokinin in regulating rice grain size. Mol. Plant 2022, 15, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Záveská Drábková, L.; Honys, D.; Motyka, V. Evolutionary diversification of cytokinin-specific glucosyltransferases in angiosperms and enigma of missing cis-zeatin O-glucosyltransferase gene in Brassicaceae. Sci. Rep. 2021, 11, 7885. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Harlina, P.W.; Ewas, M.; Zhenyuan, P.; Nie, X.; Gallego, P.P.; Ullah Khan, S.; Nishawy, E.; Khan, A.H.; Jia, H. Foliar applied 24-epibrassinolide alleviates salt stress in rice (Oryza sativa L.) by suppression of ABA levels and upregulation of secondary metabolites. J. Plant Interact. 2021, 16, 533–549. [Google Scholar] [CrossRef]
- Malaga, S.; Janeczko, A.; Janowiak, F.; Waligórski, P.; Oklestkova, J.; Dubas, E.; Krzewska, M.; Nowicka, A.; Surówka, E.; Rapacz, M.; et al. Involvement of homocastasterone, salicylic and abscisic acids in the regulation of drought and freezing tolerance in doubled haploid lines of winter barley. Plant Growth Regul. 2020, 90, 173–188. [Google Scholar] [CrossRef]
- Choudhary, A.; Senthil-Kumar, M. Drought attenuates plant defence against bacterial pathogens by suppressing the expression of CBP60g/SARD1 during combined stress. Plant Cell Environ. 2022, 45, 1127–1145, Erratum in Plant Cell Environ. 2022, 45, 1962. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Yuan, S.; Jiang, H.; Wang, Y.; Zhang, L.; Shi, Z.; Jiao, L.; Meng, D. A 3R-MYB transcription factor is involved in Methyl Jasmonate-Induced disease resistance in Agaricus bisporus and has implications for disease resistance in Arabidopsis. J. Adv. Res. 2025, 73, 117–131. [Google Scholar] [CrossRef]
- Ren, C.; Han, C.; Peng, W.; Huang, Y.; Peng, Z.; Xiong, X.; Zhu, Q.; Gao, B.; Xie, D. A Leaky Mutation in DWARF4 Reveals an Antagonistic Role of Brassinosteroid in the Inhibition of Root Growth by Jasmonate in Arabidopsis. Plant Physiol. 2009, 151, 1412–1420. [Google Scholar] [CrossRef]
- Hilker, M.; Schmülling, T. Stress priming, memory, and signalling in plants. Plant Cell Environ. 2019, 42, 753–761. [Google Scholar] [CrossRef]
- Kalachova, T.; Janda, M.; Šašek, V.; Ortmannová, J.; Nováková, P.; Dobrev, I.P.; Kravets, V.; Guivarc’h, A.; Moura, D.; Burketová, L.; et al. Identification of salicylic acid-independent responses in an Arabidopsis phosphatidylinositol 4-kinase beta double mutant. Ann. Bot. 2020, 125, 775–784. [Google Scholar] [CrossRef]
- Derevyanchuk, M.; Litvinovskaya, R.; Khripach, V.; Martinec, J.; Kravets, V. Effect of 24-epibrassinolide on Arabidopsis thaliana alternative respiratory pathway under salt stress. Acta Physiol. Plant. 2015, 37, 215. [Google Scholar] [CrossRef]
- Dobrev, P.I.; Vankova, R. Quantification of Abscisic Acid, Cytokinin, and Auxin Content in Salt-Stressed Plant Tissues. In Plant Salt Tolerance; Shabala, S., Cuin, T.A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 913, pp. 251–261. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bukhonska, Y.; Derevyanchuk, M.; Filepova, R.; Martinec, J.; Dobrev, P.; Ruelland, E.; Kravets, V. Brassinosteroid Synthesis and Perception Differently Regulate Phytohormone Networks in Arabidopsis thaliana. Int. J. Mol. Sci. 2025, 26, 9644. https://doi.org/10.3390/ijms26199644
Bukhonska Y, Derevyanchuk M, Filepova R, Martinec J, Dobrev P, Ruelland E, Kravets V. Brassinosteroid Synthesis and Perception Differently Regulate Phytohormone Networks in Arabidopsis thaliana. International Journal of Molecular Sciences. 2025; 26(19):9644. https://doi.org/10.3390/ijms26199644
Chicago/Turabian StyleBukhonska, Yaroslava, Michael Derevyanchuk, Roberta Filepova, Jan Martinec, Petre Dobrev, Eric Ruelland, and Volodymyr Kravets. 2025. "Brassinosteroid Synthesis and Perception Differently Regulate Phytohormone Networks in Arabidopsis thaliana" International Journal of Molecular Sciences 26, no. 19: 9644. https://doi.org/10.3390/ijms26199644
APA StyleBukhonska, Y., Derevyanchuk, M., Filepova, R., Martinec, J., Dobrev, P., Ruelland, E., & Kravets, V. (2025). Brassinosteroid Synthesis and Perception Differently Regulate Phytohormone Networks in Arabidopsis thaliana. International Journal of Molecular Sciences, 26(19), 9644. https://doi.org/10.3390/ijms26199644






