A High-Throughput Sequencing Strategy for Clinical Repertoire Profiling of T Cell Receptor Beta Chain: Development and Reference Values Across Healthy Adults, Paediatrics, and Cord Blood Units
Abstract
1. Introduction
2. Results
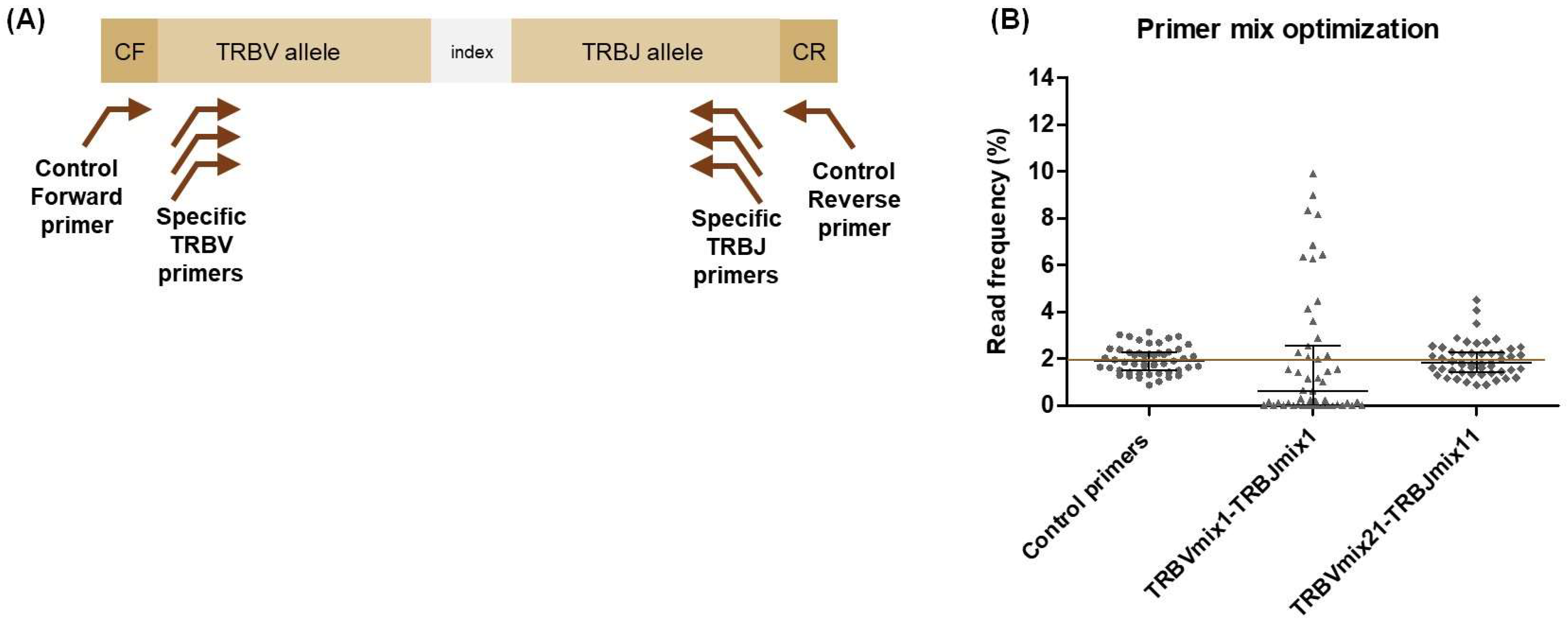
2.1. TCRβ NGS Strategy
2.2. Primer Specificity and Efficiency
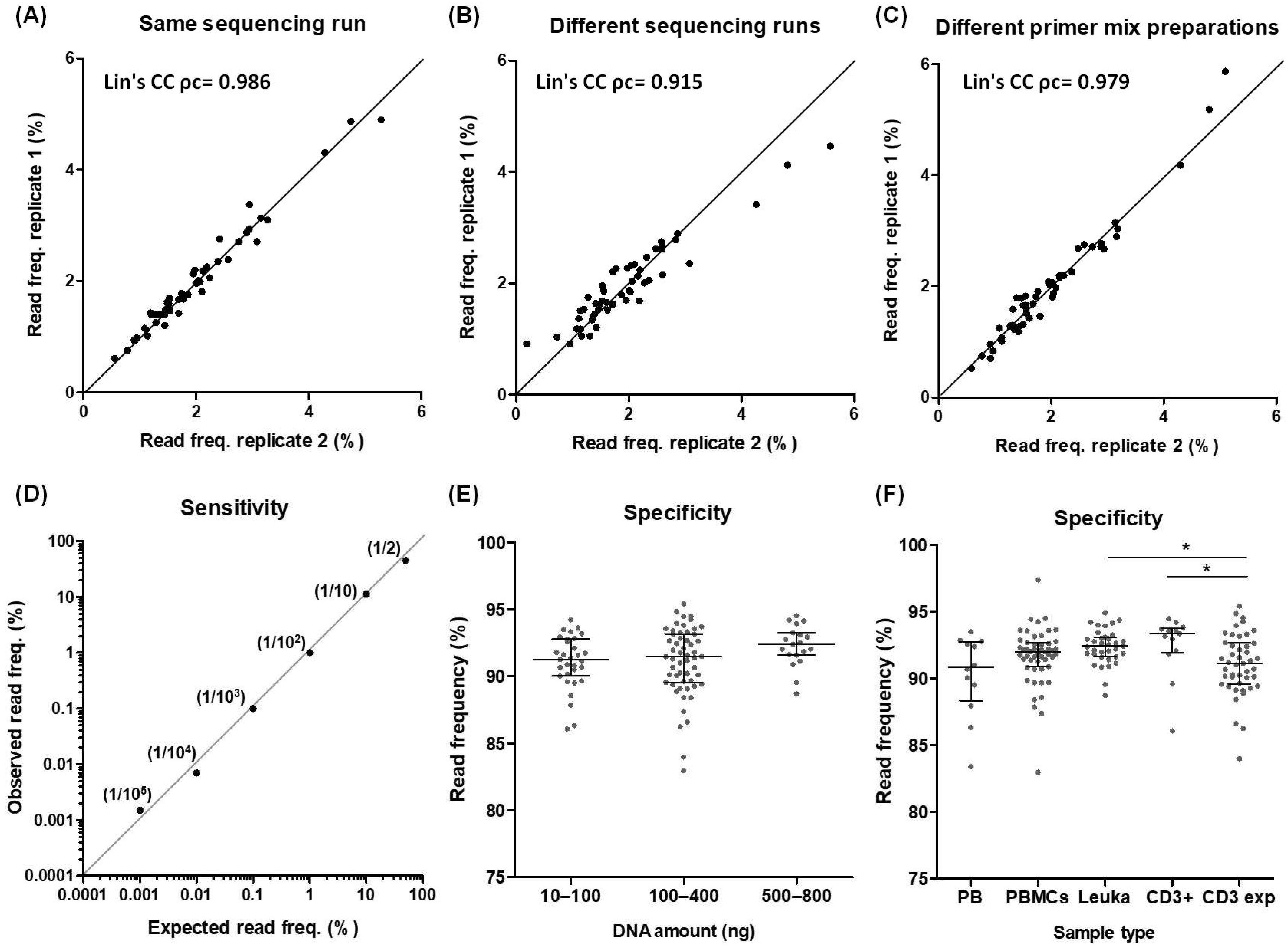
2.3. Reproducibility and Sensitivity
2.4. Establishment of Working Samples
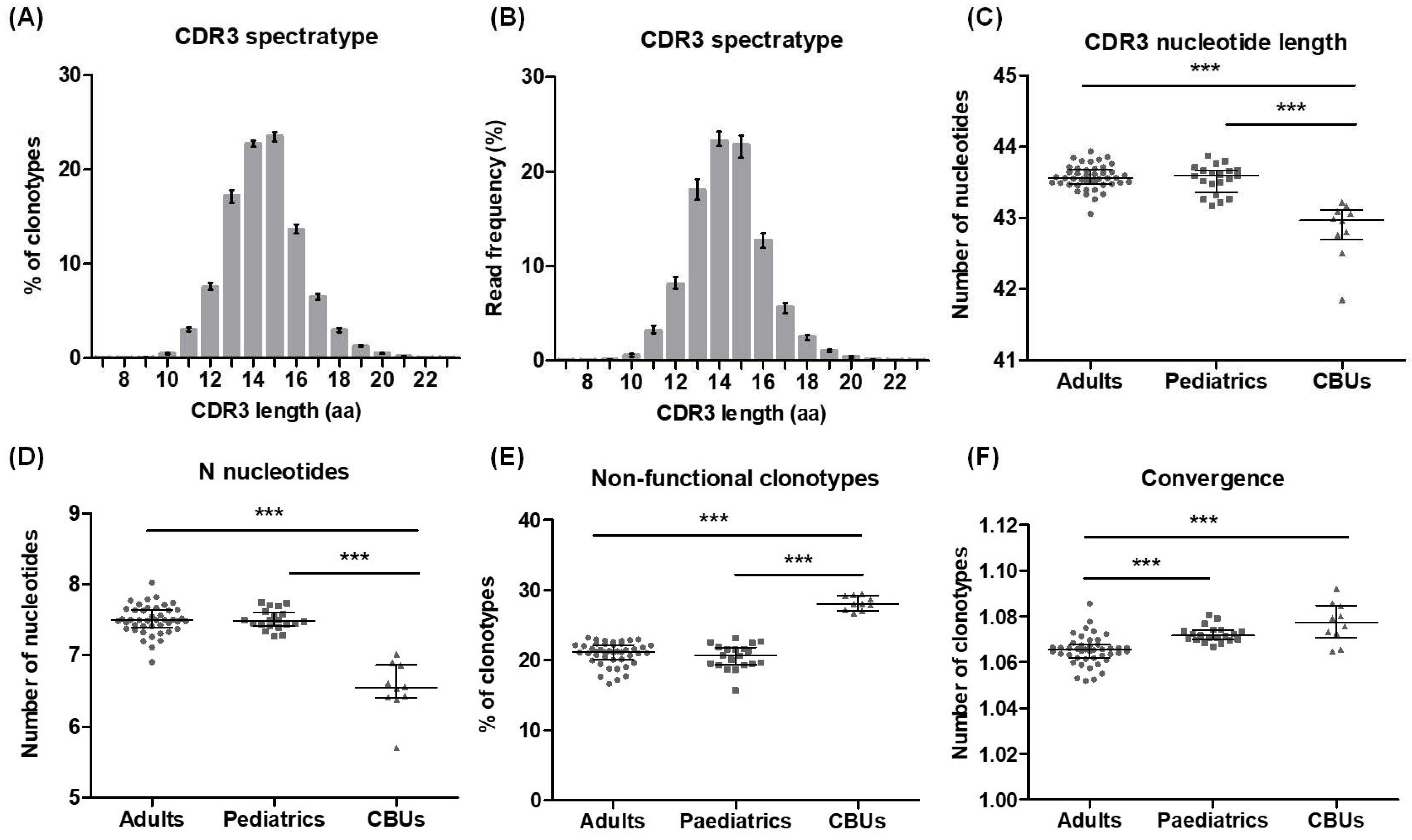
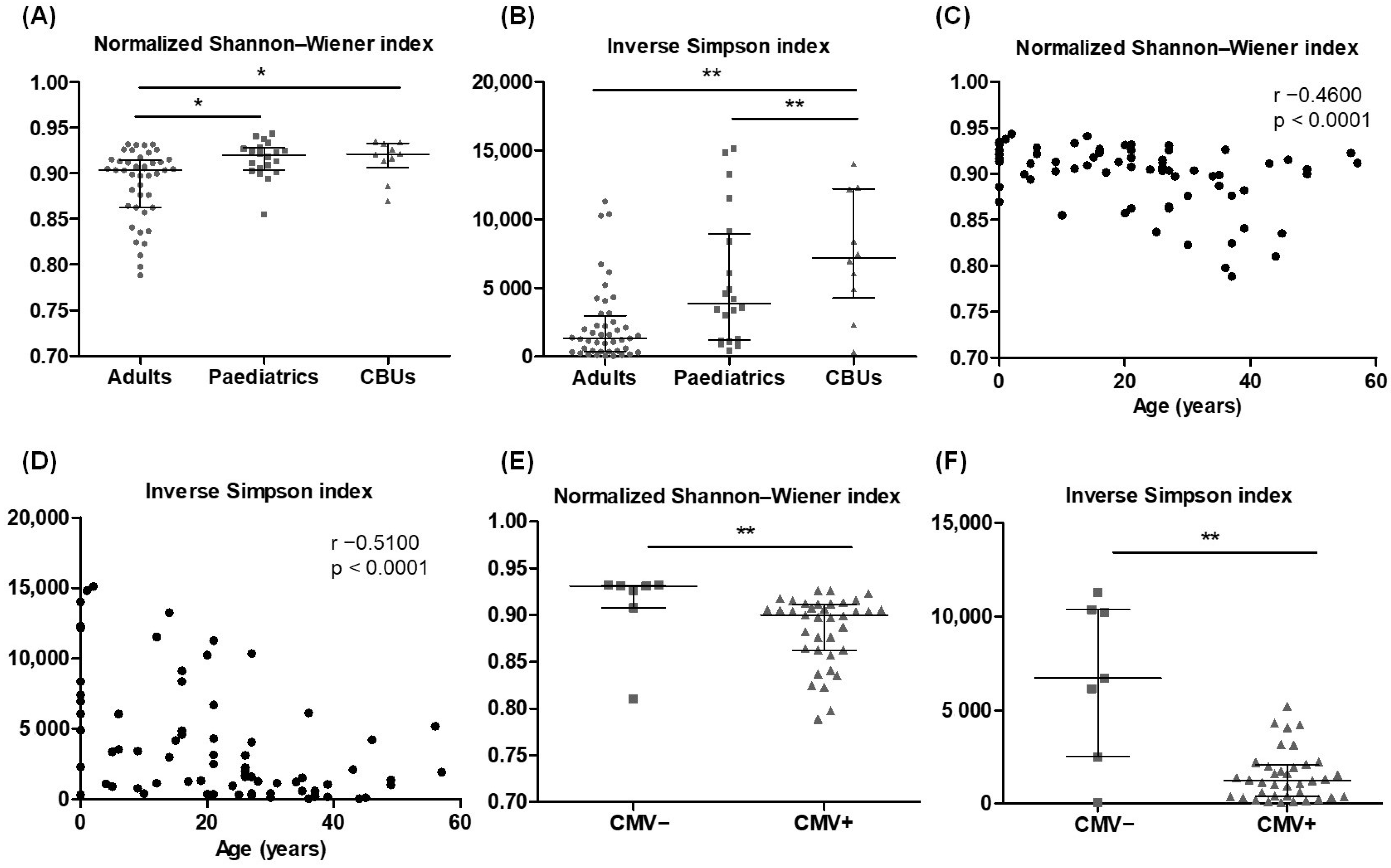
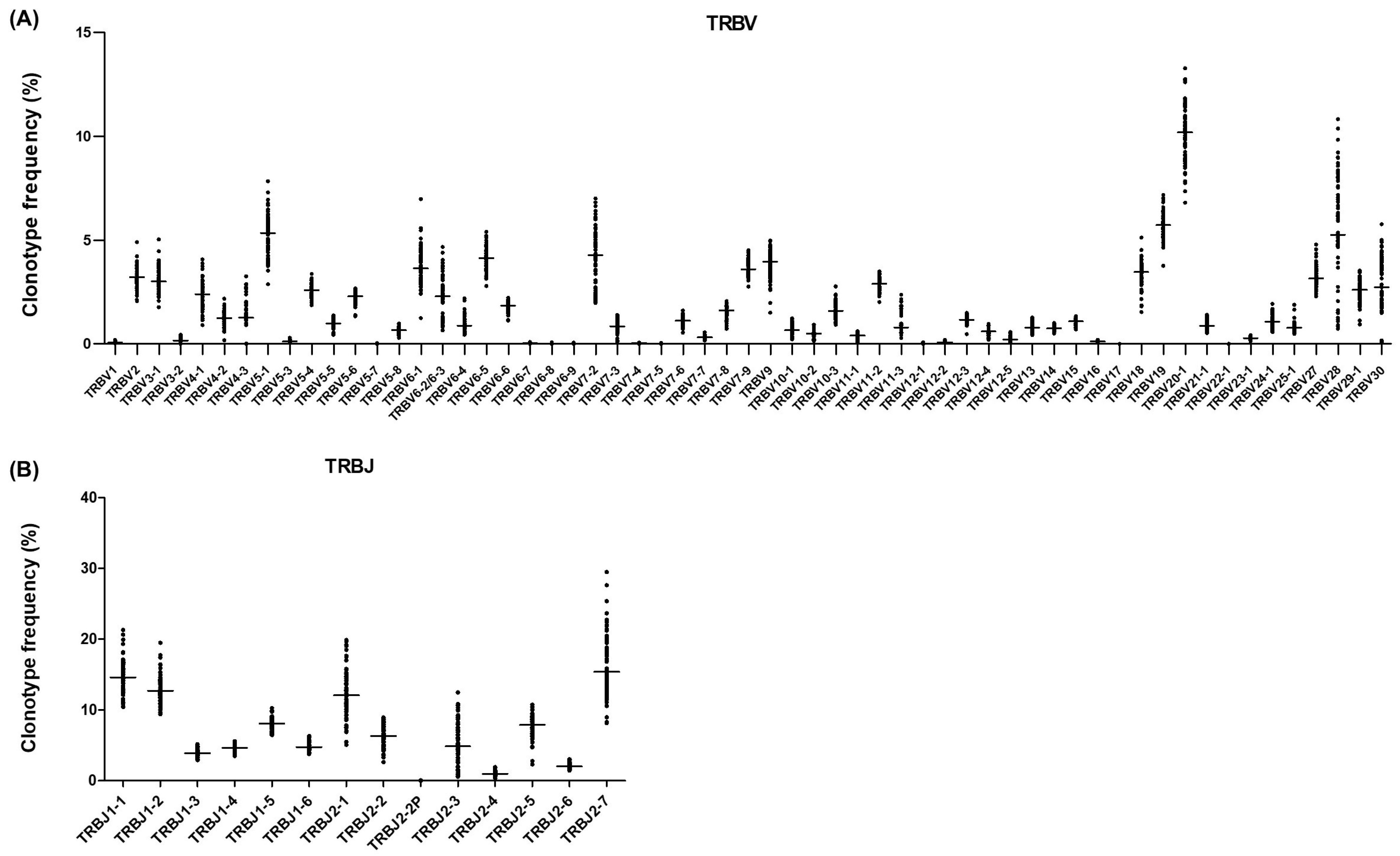
2.5. Reference Values for TCRβ Repertoire Profiling
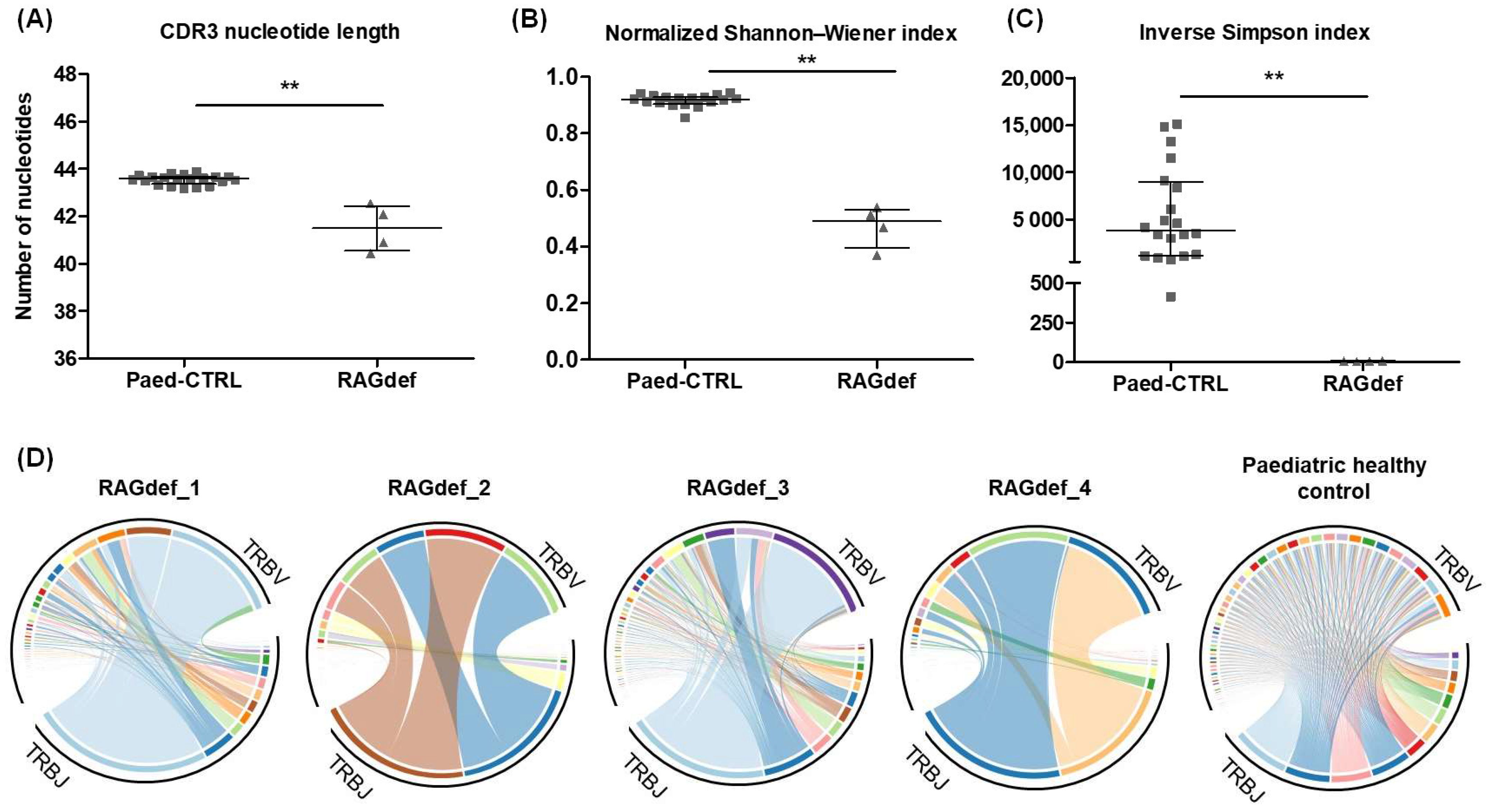
2.6. TCRβ Repertoire Profiling in RAG-SCID/CID Patients
3. Discussion
4. Materials and Methods
4.1. Samples and Ethics Committee
4.2. CD3+ T Cell Quantification Using Flow Cytometry
4.3. Primer Design and Optimisation
4.4. TCRβ Repertoire Profiling in Healthy Donors and Patients with RAG-SCID/CID
4.5. Sequencing Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BST | Blood and Tissue Bank |
| CBU | Umbilical cord blood unit |
| CDR3 | Complementary-determining region 3 |
| CID | Combined immunodeficiency |
| CMV | Cytomegalovirus |
| D | Diversity |
| FR | Framework region |
| gDNA | Genomic DNA |
| HLA | Human Leukocyte Antigen |
| IMGT | ImMunoGeneTics |
| J | Joining |
| MNC | Mononuclear cell |
| mRNA | Messenger RNA |
| NGS | Next-generation sequencing |
| ORF | Open Reading Frame |
| PB | Peripheral blood |
| PBMCs | Peripheral blood mononuclear cells |
| RAG | Recombination activating gene |
| SCID | Severe combined immunodeficiency |
| TCR | T cell receptor |
| TCRβ | T cell receptor β-chain |
| TdT | Terminal deoxynucleotidyl transferase |
| TRAD | T cell receptor alpha diversity gene |
| TRAJ | T cell receptor alpha joining gene |
| TRAV | T cell receptor alpha variable gene |
| TRB | T cell receptor gene |
| TRBD | T cell receptor beta diversity gene |
| TRBJ | T cell receptor beta joining gene |
| TRBV | T cell receptor beta variable gene |
| UTR | Untranslated region |
| V | Variable |
References
- Mazzotti, L.; Gaimari, A.; Bravaccini, S.; Maltoni, R.; Cerchione, C.; Juan, M.; Azucena-Gonzalez, E.; Pasetto, A.; Nascimento, D.; Ancarani, V.; et al. T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer. Int. J. Mol. Sci. 2022, 23, 8590. [Google Scholar] [CrossRef]
- Frank, M.L.; Lu, K.; Erdogan, C.; Han, Y.; Hu, J.; Wang, T.; Heymach, J.V.; Zhang, J.; Reuben, A. T-Cell Receptor Repertoire Sequencing in the Era of Cancer Immunotherapy. Clin. Cancer Res. 2023, 29, 994–1008. [Google Scholar] [CrossRef]
- Clambey, E.T.; Davenport, B.; Kappler, J.W.; Marrack, P.; Homann, D. Molecules in medicine mini review: The αβ T cell receptor. J. Mol. Med. 2014, 92, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.J.; Doherty, P.C.; McCluskey, J.; Rossjohn, J. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 2006, 6, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.P.; Giudicelli, V.; Duroux, P.; Jabado-Michaloud, J.; Folch, G.; Aouinti, S.; Carillon, E.; Duvergey, H.; Houles, A.; Paysan-Lafosse, T.; et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015, 43, D413–D422. [Google Scholar] [CrossRef]
- Weng, N.P. Numbers and odds: TCR repertoire size and its age changes impacting on T cell functions. Semin. Immunol. 2023, 69, 101810. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.C.A.; Boyd, S.D. Human adaptive immune receptor repertoire analysis-Past, present, and future. Immunol. Rev. 2018, 284, 9–23. [Google Scholar] [CrossRef]
- Six, A.; Mariotti-Ferrandiz, M.E.; Chaara, W.; Magadan, S.; Pham, H.P.; Lefranc, M.P.; Mora, T.; Thomas-Vaslin, V.; Walczak, A.M.; Boudinot, P. The past, present, and future of immune repertoire biology—The rise of next-generation repertoire analysis. Front. Immunol. 2013, 4, 413. [Google Scholar] [CrossRef]
- Rosati, E.; Dowds, C.M.; Liaskou, E.; Henriksen, E.K.K.; Karlsen, T.H.; Franke, A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017, 17, 61. [Google Scholar] [CrossRef]
- Brüggemann, M.; Kotrová, M.; Knecht, H.; Bartram, J.; Boudjogrha, M.; Bystry, V.; Fazio, G.; Fronková, E.; Giraud, M.; Grioni, A.; et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia 2019, 33, 2241. [Google Scholar] [CrossRef]
- Medina, A.; Puig, N.; Flores-Montero, J.; Jimenez, C.; Sarasquete, M.E.; Garcia-Alvarez, M.; Prieto-Conde, I.; Chillon, C.; Alcoceba, M.; Gutierrez, N.C.; et al. Comparison of next-generation sequencing (NGS) and next-generation flow (NGF) for minimal residual disease (MRD) assessment in multiple myeloma. Blood Cancer J. 2020, 10, 108. [Google Scholar] [CrossRef]
- Mitchell, A.M.; Michels, A.W. T cell receptor sequencing in autoimmunity. J. Life Sci. 2020, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Michels, A.W. Using the T Cell Receptor as a Biomarker in Type 1 Diabetes. Front. Immunol. 2021, 12, 777788. [Google Scholar] [CrossRef] [PubMed]
- Farmanbar, A.; Kneller, R.; Firouzi, S. RNA sequencing identifies clonal structure of T-cell repertoires in patients with adult T-cell leukemia/lymphoma. NPJ Genomic Med. 2019, 4, 10. [Google Scholar] [CrossRef]
- Lee, Y.N.; Frugoni, F.; Dobbs, K.; Tirosh, I.; Du, L.; Ververs, F.A.; Ru, H.; Ott de Bruin, L.; Adeli, M.; Bleesing, J.H.; et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci. Immunol. 2016, 1, eaah6109. [Google Scholar] [CrossRef]
- Fang, M.; Su, Z.; Abolhassani, H.; Zhang, W.; Jiang, C.; Cheng, B.; Luo, L.; Wu, J.; Wang, S.; Lin, L.; et al. T Cell Repertoire Abnormality in Immunodeficiency Patients with DNA Repair and Methylation Defects. J. Clin. Immunol. 2022, 42, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, S.; Gurnari, C.; Hong, S.; Zhao, R.; Kongkiatkamon, S.; Terkawi, L.; Zawit, M.; Guan, Y.; Awada, H.; Kishtagari, A.; et al. Clinical and basic implications of dynamic T cell receptor clonotyping in hematopoietic cell transplantation. JCI Insight 2021, 6, e149080. [Google Scholar] [CrossRef]
- Schultze-Florey, C.R.; Kuhlmann, L.; Raha, S.; Barros-Martins, J.; Odak, I.; Tan, L.; Xiao, Y.; Ravens, S.; Hambach, L.; Venturini, L.; et al. Clonal expansion of CD8+ T cells reflects graft-versus-leukemia activity and precedes durable remission following DLI. Blood Adv. 2021, 5, 4485–4499. [Google Scholar] [CrossRef]
- Goel, M.; Eugster, A.; Schetelig, J.; Bonifacio, E.; Bornhäuser, M.; Link-Rachner, C.S. Potential of TCR sequencing in graft-versus-host disease. Bone Marrow Transplant. 2023, 58, 239–246. [Google Scholar] [CrossRef]
- Leick, M.; Gittelman, R.M.; Yusko, E.; Sanders, C.; Robins, H.; DeFilipp, Z.; Nikiforow, S.; Ritz, J.; Chen, Y. T Cell Clonal Dynamics Determined by High-Resolution TCR-β Sequencing in Recipients after Allogeneic Hematopoietic Cell Transplantation. Biol. Blood Marrow Transplant. 2020, 26, 1567–1574. [Google Scholar] [CrossRef]
- Kuzich, J.A.; Kankanige, Y.; Guinto, J.; Ryland, G.; McBean, M.; Wong, E.; Koldej, R.; Collins, J.; Westerman, D.; Ritchie, D.; et al. T cell receptor beta locus sequencing early post-allogeneic stem cell transplant identifies patients at risk of initial and recurrent cytomegalovirus infection. Bone Marrow Transplant. 2021, 56, 2582–2590. [Google Scholar] [CrossRef]
- Milano, F.; Emerson, R.O.; Salit, R.; Guthrie, K.A.; Thur, L.A.; Dahlberg, A.; Robins, H.S.; Delaney, C. Impact of T Cell Repertoire Diversity on Mortality Following Cord Blood Transplantation. Front. Oncol. 2020, 10, 583349. [Google Scholar] [CrossRef] [PubMed]
- Buhler, S.; Bettens, F.; Dantin, C.; Ferrari-Lacraz, S.; Ansari, M.; Mamez, A.C.; Masouridi-Levrat, S.; Chalandon, Y.; Villard, J. Genetic T-cell receptor diversity at 1 year following allogeneic hematopoietic stem cell transplantation. Leukemia 2020, 34, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Huisman, W.; Roex, M.C.J.; Hageman, L.; Koster, E.A.S.; Veld, S.A.J.; Hoogstraten, C.; van Balen, P.; van Egmond, H.M.; van Bergen, C.A.M.; Einsele, H.; et al. Tracking the progeny of adoptively transferred virus-specific T cells in patients posttransplant using TCR sequencing. Blood Adv. 2023, 7, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.D.; Darko, S.; Lang, H.; Ransier, A.; Lazarski, C.A.; Wang, Y.; Hanley, P.J.; Davila, B.J.; Heimall, J.R.; Ambinder, R.F.; et al. T-cell receptor sequencing demonstrates persistence of virus-specific T cells after antiviral immunotherapy. Br. J. Haematol. 2019, 187, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Mora-Buch, R.; Tomás-Marín, M.; Enrich, E.; Antón-Iborra, M.; Martorell, L.; Valdivia, E.; Lara-de-León, A.G.; Aran, G.; Piron, M.; Querol, S.; et al. Virus-Specific T Cells From Cryopreserved Blood During an Emergent Virus Outbreak for a Potential Off-the-Shelf Therapy. Transplant. Cell Ther. 2023, 29, 572.e1–572.e13. [Google Scholar] [CrossRef]
- Delmonte, O.M.; Villa, A.; Notarangelo, L.D. Immune dysregulation in patients with RAG deficiency and other forms of combined immune deficiency. Blood 2020, 135, 610–619. [Google Scholar] [CrossRef]
- Carlson, C.S.; Emerson, R.O.; Sherwood, A.M.; Desmarais, C.; Chung, M.W.; Parsons, J.M.; Steen, M.S.; LaMadrid-Herrmannsfeldt, M.A.; Williamson, D.W.; Livingston, R.J.; et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 2013, 4, 2680. [Google Scholar] [CrossRef]
- Tan, K.T.; Ding, L.W.; Sun, Q.Y.; Lao, Z.T.; Chien, W.; Ren, X.; Xiao, J.F.; Loh, X.Y.; Xu, L.; Lill, M.; et al. Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines. BMC Cancer 2018, 18, 940. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Davydov, A.N.; Frenkel, F.E.; Fanchi, L.; Zolotareva, O.I.; Hemmers, S.; Putintseva, E.V.; Obraztsova, A.S.; Shugay, M.; et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017, 35, 908–911. [Google Scholar] [CrossRef]
- Britanova, O.V.; Shugay, M.; Merzlyak, E.M.; Staroverov, D.B.; Putintseva, E.V.; Turchaninova, M.A.; Mamedov, I.Z.; Pogorelyy, M.V.; Bolotin, D.A.; Izraelson, M.; et al. Dynamics of Individual T Cell Repertoires: From Cord Blood to Centenarians. J. Immunol. 2016, 196, 5005–5013. [Google Scholar] [CrossRef]
- Yang, L.; Jin, R.; Lu, D.; Ge, Q. T cell Tolerance in Early Life. Front. Immunol. 2020, 11, 576261. [Google Scholar] [CrossRef]
- George, J.F.; Schroeder, H.W. Developmental regulation of D beta reading frame and junctional diversity in T cell receptor-beta transcripts from human thymus. J. Immunol. 1992, 148, 1230–1239. [Google Scholar] [CrossRef]
- Zemlin, M.; Schelonka, R.L.; Bauer, K.; Schroeder, H.W. Regulation and chance in the ontogeny of B and T cell antigen receptor repertoires. Immunol. Res. 2002, 26, 265–278. [Google Scholar] [CrossRef]
- Rudd, B.D.; Venturi, V.; Li, G.; Samadder, P.; Ertelt, J.M.; Way, S.S.; Davenport, M.P.; Nikolich-Zugich, J. Nonrandom attrition of the naive CD8+ T-cell pool with aging governed by T-cell receptor:pMHC interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 13694–13699. [Google Scholar] [CrossRef] [PubMed]
- Britanova, O.V.; Putintseva, E.V.; Shugay, M.; Merzlyak, E.M.; Turchaninova, M.A.; Staroverov, D.B.; Bolotin, D.A.; Lukyanov, S.; Bogdanova, E.A.; Mamedov, I.Z.; et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014, 192, 2689–2698. [Google Scholar] [CrossRef]
- Krishna, C.; Chowell, D.; Gönen, M.; Elhanati, Y.; Chan, T.A. Genetic and environmental determinants of human TCR repertoire diversity. Immun. Ageing 2020, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Nguyen, T.; Achour, A.; Ko, A.; Cifello, J.; Ling, C.; Sharma, J.; Hiroi, T.; Zhang, Y.; Chia, C.W.; et al. Longitudinal analysis reveals age-related changes in the T cell receptor repertoire of human T cell subsets. J. Clin. Investig. 2022, 132, e158122. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Y.; Yang, X.; Shuai, P.; Yang, L.; Wen, X.; Zhong, X.; Yang, S.; Xu, S.; Liu, Y.; Zhang, Z. Evaluation and comparison of adaptive immunity through analyzing the diversities and clonalities of T-cell receptor repertoires in the peripheral blood. Front. Immunol. 2022, 13, 916430. [Google Scholar] [CrossRef]
- Gong, M.; Li, X.; Zheng, A.; Xu, H.; Xie, S.; Yan, R.; Wu, H.; Wang, Z. Age-related changes in the TRB and IGH repertoires in healthy adult males and females. Immunol. Lett. 2021, 240, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, L.; Shi, B.; He, X.; Peng, A.; Li, Y.; Zhang, T.; Sun, S.; Ma, R.; Yao, X. Analyzing the CDR3 Repertoire with respect to TCR-Beta Chain V-D-J and V-J Rearrangements in Peripheral T Cells using HTS. Sci. Rep. 2016, 6, 29544. [Google Scholar] [CrossRef]
- Hou, X.; Wang, M.; Lu, C.; Xie, Q.; Cui, G.; Chen, J.; Du, Y.; Dai, Y.; Diao, H. Analysis of the Repertoire Features of TCR Beta Chain CDR3 in Human by High-Throughput Sequencing. Cell Physiol. Biochem. 2016, 39, 651–667. [Google Scholar] [CrossRef] [PubMed]
- Rudilla, F.; Carrasco-Benso, M.P.; Pasamar, H.; López-Montañés, M.; Andrés-Rozas, M.; Tomás-Marín, M.; Company, D.; Moya, C.; Larrea, L.; Guerreiro, M.; et al. Development and characterization of a cell donor registry for virus-specific T cell manufacture in a blood bank. HLA 2024, 103, e15419. [Google Scholar] [CrossRef] [PubMed]

| (A) First PCR—TRB Specific Amplification | |||||
| PCR Reaction (Final Volume: 25 µL) | Thermocycler Program | ||||
| Reagent | Stock Concentration | Volume/Reaction | Cycles | Temperature | Time |
| PCR buffer without MgCl2 | 10X | 2.5 µL | 1 | 50 °C | 2 min |
| MgCl2 | 50 mM | 0.75 µL | 70 °C | 20 min | |
| dNTPs | 10 mM | 0.5 µL | 95 °C | 10 min | |
| TRBV primer mix | 3 µM | 5 µL | 35 | 95 °C | 15 s |
| TRBJ primer mix | 3.4 µM | 5 µL | 62 °C | 15 s | |
| Platinum Taq DNA Polymerase | 0.1 µL | 72 °C | 30 s | ||
| H2O | 6.15 µL | 1 | 72 °C | 3 min | |
| DNA | 5 µL | ||||
| (B) Second PCR—Library Preparation | |||||
| PCR Reaction (Final Volume: 20 µL) | Thermocycler Program | ||||
| Reagent | Stock Concentration | Volume/Reaction | Cycles | Temperature | Time |
| FastStart HF Buffer without MgCl2 | 10X | 2 µL | 1 | 95 °C | 10 min |
| MgCl2 | 25 mM | 3.6 µL | 15 | 95 °C | 15 s |
| DMSO | 1 µL | 60 °C | 30 s | ||
| PCR Nucleotide Mix | 10 mM | 0.4 µL | 72 °C | 1 min | |
| FastStart HF Enzyme | 0.2 µL | 1 | 72 °C | 3 min | |
| H2O | 7.8 µL | ||||
| Barcode | 2 µM | 4 µL | |||
| First PCR product | 1 µL | ||||
| T Cells Analysed | Replicate | Total Reads | TRB Alignment | Observed Diversity | D50 | Normalised Shannon–Wiener Index | Inverse Simpson Index | |
|---|---|---|---|---|---|---|---|---|
| Diverse T cell repertoire | 10,000 | 1 | 108,297 | 88% | 4718 | 527 | 0.9074 | 1094.18 |
| 2 | 160,836 | 90% | 5696 | 519 | 0.8900 | 933.81 | ||
| 50,000 | 1 | 833,190 | 92% | 33192 | 2981 | 0.8936 | 760.16 | |
| 2 | 704,224 | 91% | 33686 | 3086 | 0.8972 | 1048.24 | ||
| 100,000 | 1 | 1,085,346 | 93% | 55812 | 4900 | 0.8958 | 1028.91 | |
| 2 | 1,079,824 | 93% | 59041 | 5057 | 0.8942 | 951.34 | ||
| Clonal T cell repertoire | 10,000 | 1 | 134,485 | 90% | 224 | 6 | 0.6245 | 17.25 |
| 2 | 138,687 | 89% | 226 | 6 | 0.6233 | 15.05 | ||
| 50,000 | 1 | 763,219 | 92% | 562 | 7 | 0.5506 | 17.81 | |
| 2 | 767,594 | 92% | 569 | 6 | 0.5269 | 14.75 | ||
| 100,000 | 1 | 1,186,708 | 91% | 958 | 7 | 0.4996 | 17.15 | |
| 2 | 1,080,508 | 91% | 902 | 7 | 0.5075 | 18.02 |
| Adults | Paediatrics | CBUs | ||
|---|---|---|---|---|
| Type of sample | PBMCs | Leukapheresis | PBMCs | MNCs |
| N | 9 | 35 | 20 | 10 |
| Age (years) | 28 (19–57) | 11 (1–17) | 0 | |
| Sex | 65.91% F, 34.09% M | 50% F, 50% M | ND | |
| IgG CMV | 15.91% Neg, 84.09% Pos | ND | ND | |
| HLA class I (HLA-A, -B, -C) | 23% ≥ 1 HM loci, 77% HT loci | ND | ND | |
| HLA class II (HLA-DRB1, -DQB1) | 25% ≥ 1 HM loci, 75% HT loci | ND | ND | |
| Adults | Paediatrics | CBUs | p-Value | |
|---|---|---|---|---|
| CDR3 nucleotide length | 43.56 (43.48–43.68) | 43.59 (43.36–43.67) | 42.97 (42.70–43.11) | 0.0001 |
| N-nucleotides added to CDR3 | 7.50 (7.39–7.64) | 7.48 (7.42–7.61) | 6.55 (6.41–6.87) | <0.0001 |
| Non-functional clonotypes (%) | 21.16 (20.02–22.14) | 20.64 (19.32–21.78) | 28.01 (27.07–29.15) | <0.0001 |
| Convergence | 1.065 (1.062–1.068) | 1.072 (1.070–1.074) | 1.077 (1.071–1.085) | <0.0001 |
| Observed diversity | 43,639 (37,303–47,255) | 45,242 (39,992–52,400) | 28,250 (22,106–40,594) | ND |
| D50 | 3811 (3081–4903) | 4483 (3803–6053) | 2861 (2015–4827) | ND |
| Norm. Shannon–Wiener index | 0.9037 (0.8631–0.9147) | 0.9199 (0.9036–0.9282) | 0.9214 (0.9064–0.9331) | 0.0021 |
| Inverse Simpson index | 1350 (375–2976) | 3860 (1175–8939) | 7190 (4257–12,196) | 0.0002 |
| RAGdef _1 | RAGdef_2 | RAGdef_3 | RAGdef_4 | |
|---|---|---|---|---|
| Diagnostic (gene affected) | CID (RAG1) | SCID (RAG2) | CID (RAG1) | SCID (RAG2) |
| Age (months) | 24 | 2.5 | 48 | 2 |
| Sex | M | M | M | M |
| Lymphocytes (×109/L) | 13.90 | 0.60 | 1.70 | 1.10 |
| CD3+ cells (×109/L) | 13.73 | 0.02 | 0.87 | 0.52 |
| CD4+ cells (×109/L) | 0.94 | 0 | 0.17 | 0.47 |
| CD8+ cells (×109/L) | 7.20 | 0.04 | 0.25 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enrich, E.; Antón-Iborra, M.; Hobeich, C.; Mora-Buch, R.; Lara-de-León, A.G.; Parra-Martínez, A.; Sánchez, B.; Vidal, F.; Soler-Palacin, P.; Rudilla, F. A High-Throughput Sequencing Strategy for Clinical Repertoire Profiling of T Cell Receptor Beta Chain: Development and Reference Values Across Healthy Adults, Paediatrics, and Cord Blood Units. Int. J. Mol. Sci. 2025, 26, 9590. https://doi.org/10.3390/ijms26199590
Enrich E, Antón-Iborra M, Hobeich C, Mora-Buch R, Lara-de-León AG, Parra-Martínez A, Sánchez B, Vidal F, Soler-Palacin P, Rudilla F. A High-Throughput Sequencing Strategy for Clinical Repertoire Profiling of T Cell Receptor Beta Chain: Development and Reference Values Across Healthy Adults, Paediatrics, and Cord Blood Units. International Journal of Molecular Sciences. 2025; 26(19):9590. https://doi.org/10.3390/ijms26199590
Chicago/Turabian StyleEnrich, Emma, Mireia Antón-Iborra, Carlos Hobeich, Rut Mora-Buch, Ana Gabriela Lara-de-León, Alba Parra-Martínez, Belén Sánchez, Francisco Vidal, Pere Soler-Palacin, and Francesc Rudilla. 2025. "A High-Throughput Sequencing Strategy for Clinical Repertoire Profiling of T Cell Receptor Beta Chain: Development and Reference Values Across Healthy Adults, Paediatrics, and Cord Blood Units" International Journal of Molecular Sciences 26, no. 19: 9590. https://doi.org/10.3390/ijms26199590
APA StyleEnrich, E., Antón-Iborra, M., Hobeich, C., Mora-Buch, R., Lara-de-León, A. G., Parra-Martínez, A., Sánchez, B., Vidal, F., Soler-Palacin, P., & Rudilla, F. (2025). A High-Throughput Sequencing Strategy for Clinical Repertoire Profiling of T Cell Receptor Beta Chain: Development and Reference Values Across Healthy Adults, Paediatrics, and Cord Blood Units. International Journal of Molecular Sciences, 26(19), 9590. https://doi.org/10.3390/ijms26199590







