Transcriptomic Analysis of the Effects of Hydroxysafflor Yellow A on hUC-MSC Senescence via the ECM–Receptor Interaction Pathway
Abstract
1. Introduction
2. Results
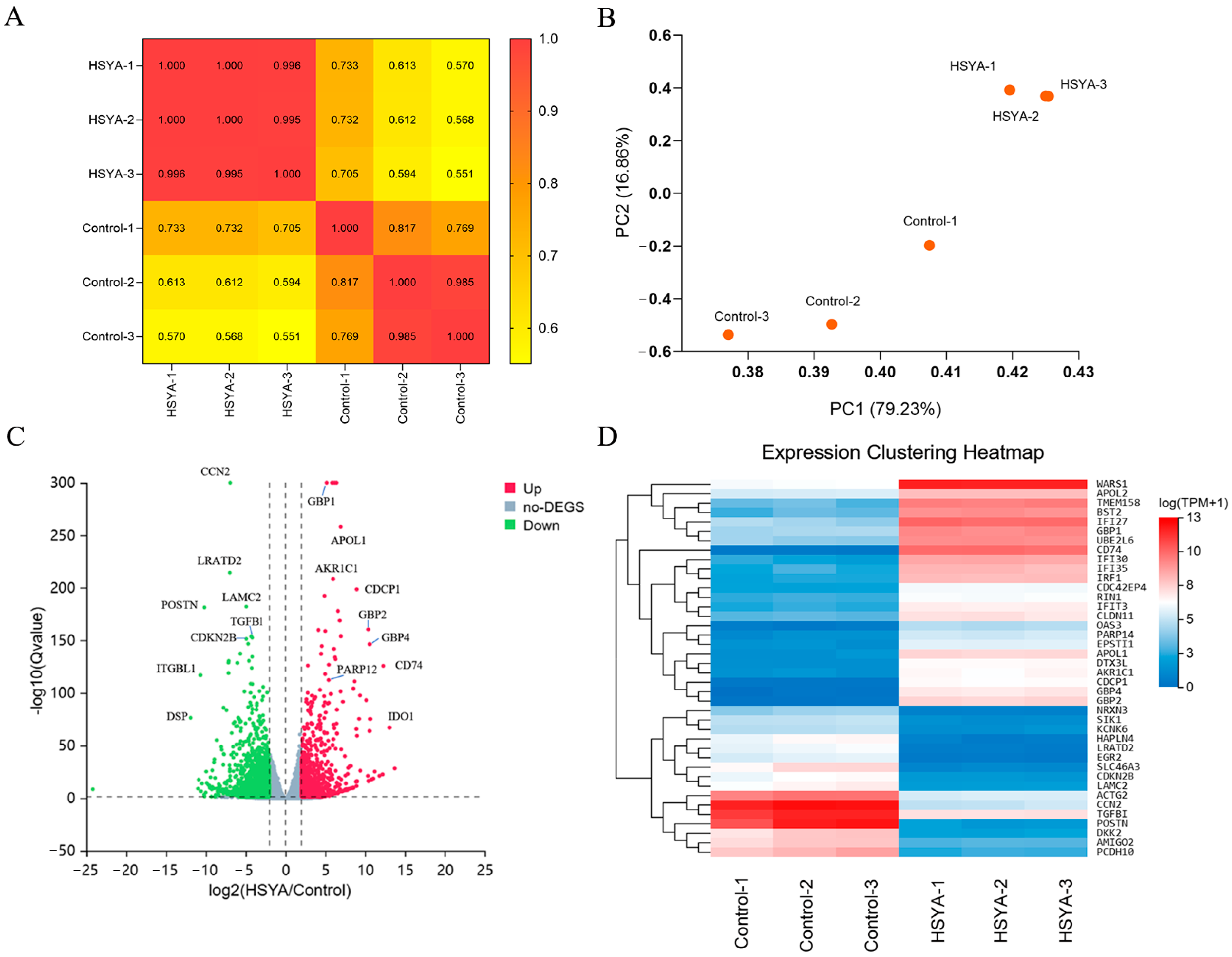
2.1. Differential Gene Expression Analysis
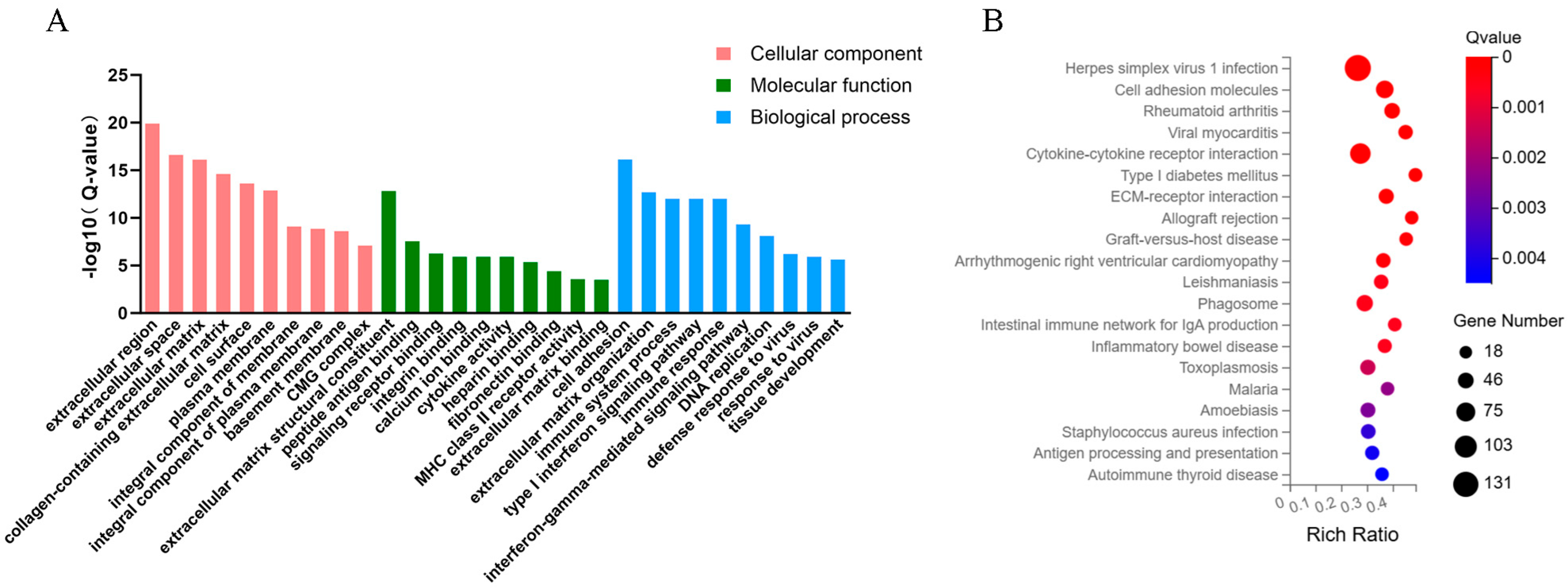
2.2. GO and KEGG Analysis
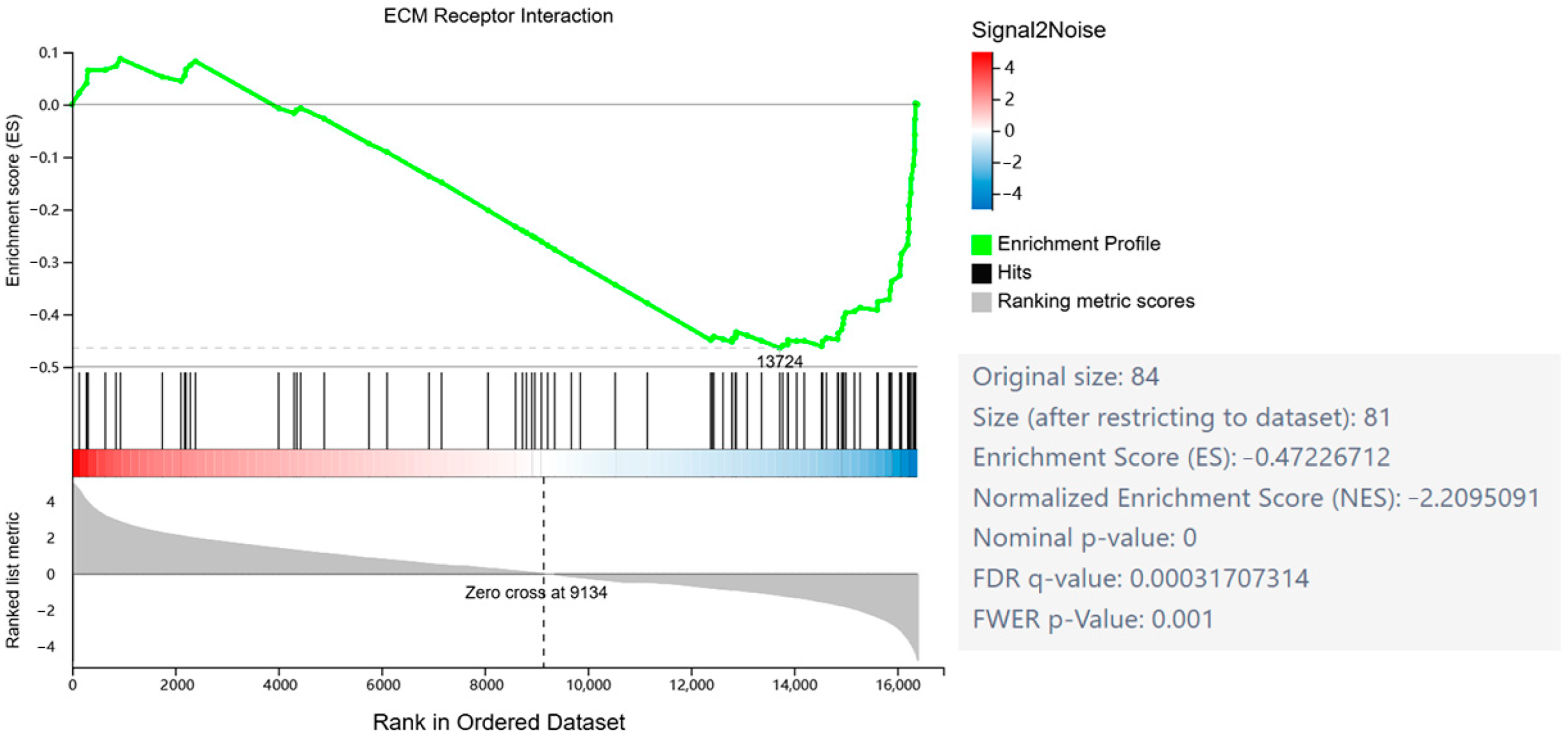
2.3. GSEA
2.4. PPI Network and KDA
2.5. SA-β-Gal Analysis
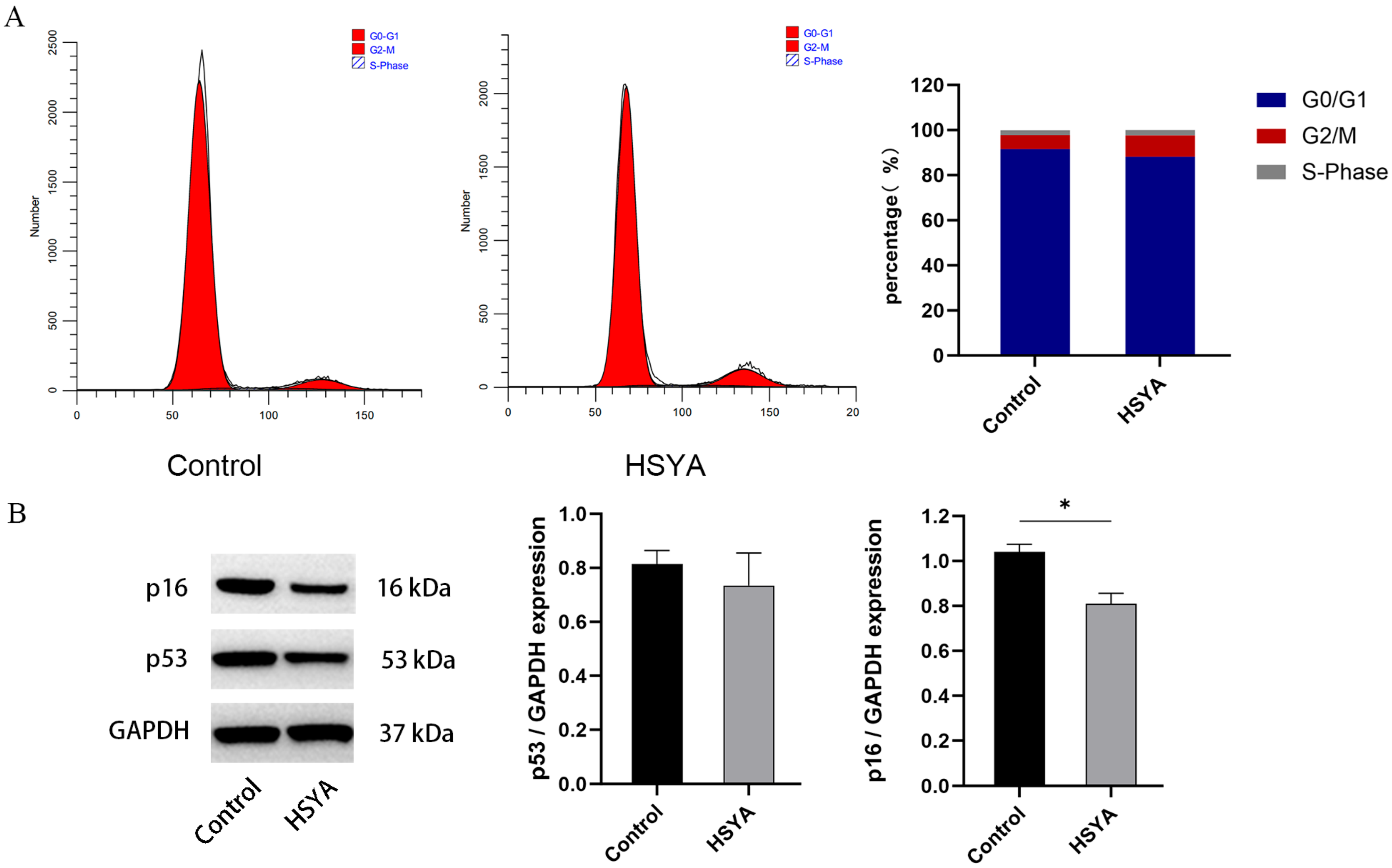
2.6. Cell Cycle and Senescence-Related Protein Expression Analysis
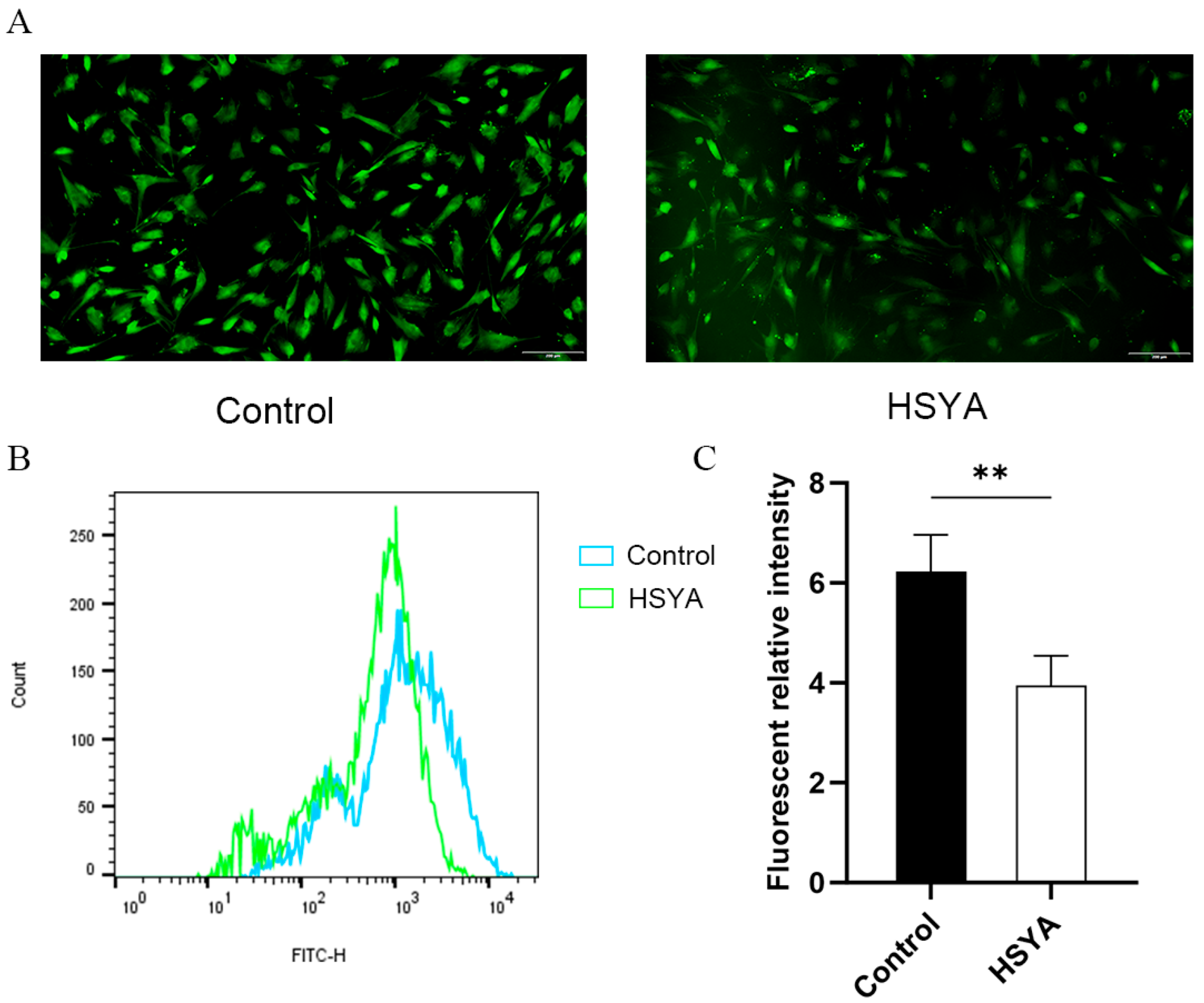
2.7. ROS Levels Analysis
2.8. Gene Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. DEG Analysis
4.3. GO Functional and KEGG Pathway Enrichment Analysis
4.4. GSEA and PPI Network Construction
4.5. SA-β-Gal Measurement
4.6. Cell Cycle Assay
4.7. ROS Measurement
4.8. RT-qPCR Analysis
4.9. Western Blot Analysis
4.10. Data Analysis
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, T.; Wang, B. Application of mesenchymal stem cells for anti-senescence and clinical challenges. Stem Cell Res. Ther. 2023, 14, 260. [Google Scholar] [CrossRef]
- Lee, S.S.; Vũ, T.T.; Weiss, A.S.; Yeo, G.C. Stress-induced senescence in mesenchymal stem cells: Triggers, hallmarks, and current rejuvenation approaches. Eur. J. Cell Biol. 2023, 102, 151331. [Google Scholar] [CrossRef]
- Bruno, A.; Milillo, C.; Anaclerio, F.; Buccolini, C.; Dell’Elice, A.; Angilletta, I.; Gatta, M.; Ballerini, P.; Antonucci, I. Perinatal Tissue-Derived Stem Cells: An Emerging Therapeutic Strategy for Challenging Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 976. [Google Scholar] [CrossRef]
- Reyhani, S.; Abbaspanah, B.; Mousavi, S.H. Umbilical Cord-Derived Mesenchymal Stem Cells in Neurodegenerative Disorders: From Literature to Clinical Practice. Regen. Med. 2020, 15, 1561–1578. [Google Scholar] [CrossRef]
- Zhang, P.; Dong, B.; Yuan, P.; Li, X. Human Umbilical Cord Mesenchymal Stem Cells Promoting Knee Joint Chondrogenesis for the Treatment of Knee Osteoarthritis: A Systematic Review. J. Orthop. Surg. Res. 2023, 18, 639. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, W.; Ge, L.; Lu, M. Mesenchymal Stem Cell Senescence during Aging: From Mechanisms to Rejuvenation Strategies. Aging Dis. 2023, 14, 1651–1676. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lin, J.; Tang, J.; Chen, Z.; Qian, X.; Gao, W.Q.; Xu, H. Decreased Immunomodulatory and Secretory Capability of Aging Human Umbilical Cord Mesenchymal Stem Cells In Vitro. Biochem. Biophys. Res. Commun. 2020, 525, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Li, D.; Fu, J.; Shi, Q.; Lu, Y.; Ju, X. Comparison of Biological Properties of Umbilical Cord-Derived Mesenchymal Stem Cells from Early and Late Passages: Immunomodulatory Ability Is Enhanced in Aged Cells. Mol. Med. Rep. 2015, 11, 166–174. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Matveeva, D.K.; Ezdakova, M.I.; Ratushnyy, A.Y. Modification of the Properties of Extracellular Matrix of Senescent Mesenchymal Stem Cells. Bull. Exp. Biol. Med. 2023, 175, 569–575. [Google Scholar] [CrossRef]
- Vidović, T.; Ewald, C.Y. Longevity-Promoting Pathways and Transcription Factors Respond to and Control Extracellular Matrix Dynamics During Aging and Disease. Front. Aging. 2022, 3, 935220. [Google Scholar] [CrossRef]
- Voronkina, I.V.; Smagina, L.V.; Bildyug, N.B.; Musorina, A.S.; Poljanskaya, G.G. Dynamics of Matrix Metalloproteinase Activity and Extracellular Matrix Proteins Content in the Process of Replicative Senescence of Human Mesenchymal Stem Cells. Cell Tiss. Biol. 2020, 14, 349–357. [Google Scholar] [CrossRef]
- Matveeva, D.; Kashirina, D.; Ezdakova, M.; Larina, I.; Buravkova, L.; Ratushnyy, A. Senescence-Associated Alterations in Matrisome of Mesenchymal Stem Cells. Int. J. Mol. Sci. 2024, 25, 5332. [Google Scholar] [CrossRef]
- Liu, J.; Ding, Y.; Liu, Z.; Liang, X. Senescence in Mesenchymal Stem Cells: Functional Alterations, Molecular Mechanisms, and Rejuvenation Strategies. Front. Cell Dev. Biol. 2020, 8, 258. [Google Scholar] [CrossRef]
- Weng, Z.; Wang, Y.; Ouchi, T.; Liu, H.; Qiao, X.; Wu, C.; Zhao, Z.; Li, L.; Li, B. Mesenchymal Stem/Stromal Cell Senescence: Hallmarks, Mechanisms, and Combating Strategies. Stem Cells Transl. Med. 2022, 11, 356–371. [Google Scholar] [CrossRef]
- Al-Azab, M.; Safi, M.; Idiiatullina, E.; Al-Shaebi, F.; Zaky, M.Y. Aging of Mesenchymal Stem Cell: Machinery, Markers, and Strategies of Fighting. Cell. Mol. Biol. Lett. 2022, 27, 69. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, O.; Chen, S.; Zhou, Y. Aging and Mesenchymal Stem Cells: Therapeutic Opportunities and Challenges in the Older Group. Gerontology 2022, 68, 339–352. [Google Scholar] [CrossRef]
- Saliev, T.; Singh, P.B. From Bench to Bedside: Translating Cellular Rejuvenation Therapies into Clinical Applications. Cells 2024, 13, 2052. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Gianoncelli, A. An Updated Overview on the Role of Small Molecules and Natural Compounds in the “Young Science” of Rejuvenation. Antioxidants 2023, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.H.; Dong, J.C.; Bian, Q. Small molecules for mesenchymal stem cell fate determination. World J. Stem Cells 2019, 11, 1084–1103. [Google Scholar] [CrossRef]
- Chen, S.; Hilcove, S.; Ding, S. Exploring stem cell biology with small molecules. Mol. Biosyst. 2006, 2, 18–24. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, C.; Gu, L.; Chen, H.; Huo, Y.; Wang, S.; Tao, J.; Xu, C.; Zhang, Q.; Ma, M.; et al. Hydroxysafflor Yellow A and Tenuigenin Exhibit Neuroprotection Effects against Focal Cerebral Ischemia via Differential Regulation of JAK2/STAT3 and SOCS3 Signaling Interaction. Mol. Neurobiol. 2024, 61, 5584–5600. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Feng, Y.; Li, Y.; Liao, H. Pharmacological Actions, Molecular Mechanisms, Pharmacokinetic Progressions, and Clinical Applications of Hydroxysafflor Yellow A in Antidiabetic Research. J. Immunol. Res. 2021, 2021, 4560012. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Duan, J.; Li, H.; Wang, Y.; Wang, F.; Chu, J.; Sun, J.; Liu, M.; Wang, C.; Lu, C.; et al. Hydroxysafflor Yellow A Ameliorates Renal Fibrosis by Suppressing TGF-β1-Induced Epithelial-to-Mesenchymal Transition. PLoS ONE 2016, 11, e0153409. [Google Scholar] [CrossRef]
- Hu, T.; Wei, G.; Xi, M.; Yan, J.; Wu, X.; Wang, Y.; Zhu, Y.; Wang, C.; Wen, A. Corrigendum: Synergistic Cardioprotective Effects of Danshensu and Hydroxysafflor Yellow A Against Myocardial Ischemia-Reperfusion Injury Are Mediated Through the Akt/Nrf2/HO-1 Pathway. Int. J. Mol. Med. 2021, 48, 212. [Google Scholar] [CrossRef]
- Liu, L.; Cui, Q.; Song, J.; Yang, Y.; Zhang, Y.; Qi, J.; Zhao, J. Hydroxysafflower Yellow A Inhibits Vascular Adventitial Fibroblast Migration via NLRP3 Inflammasome Inhibition through Autophagy Activation. Int. J. Mol. Sci. 2022, 24, 172. [Google Scholar] [CrossRef]
- Huang, Y.Z.; Zhao, L.; Zhu, Y.; Tian, S.J.; Zhang, W.; Liu, S.; Ge, J.F. Interrupting TGF-β1/CCN2/Integrin-α5β1 Signaling Alleviates High Mechanical-Stress Caused Chondrocyte Fibrosis. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1233–1241. [Google Scholar]
- Tejera-Muñoz, A.; Marquez-Exposito, L.; Tejedor-Santamaría, L.; Rayego-Mateos, S.; Orejudo, M.; Suarez-Álvarez, B.; López-Larrea, C.; Ruíz-Ortega, M.; Rodrigues-Díez, R.R. CCN2 Increases TGF-β Receptor Type II Expression in Vascular Smooth Muscle Cells: Essential Role of CCN2 in the TGF-β Pathway Regulation. Int. J. Mol. Sci. 2021, 23, 375. [Google Scholar] [CrossRef] [PubMed]
- Zaykov, V.; Chaqour, B. The CCN2/CTGF Interactome: An Approach to Understanding the Versatility of CCN2/CTGF Molecular Activities. J. Cell Commun. Signal. 2021, 15, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.; Fisher, G.J. Role of Age-Associated Alterations of the Dermal Extracellular Matrix Microenvironment in Human Skin Aging: A Mini-Review. Gerontology 2015, 61, 427–434. [Google Scholar] [CrossRef]
- Mavrogonatou, E.; Papadopoulou, A.; Pratsinis, H.; Kletsas, D. Senescence-Associated Alterations in the Extracellular Matrix: Deciphering Their Role in the Regulation of Cellular Function. Am. J. Physiol. Cell Physiol. 2023, 325, C633–C647. [Google Scholar] [CrossRef] [PubMed]
- Levi, N.; Papismadov, N.; Solomonov, I.; Sagi, I.; Krizhanovsky, V. The ECM Path of Senescence in Aging: Components and Modifiers. FEBS J. 2020, 287, 2636–2646. [Google Scholar] [CrossRef]
- Yang, S.; Liao, W. Hydroxysafflor Yellow A Attenuates Oxidative Stress Injury-Induced Apoptosis in the Nucleus Pulposus Cell Line and Regulates Extracellular Matrix Balance via CA XII. Exp. Ther. Med. 2022, 23, 182. [Google Scholar] [CrossRef]
- Pan, R.; Zhang, Y.; Zang, B.; Tan, L.; Jin, M. Hydroxysafflor Yellow A Inhibits TGF-β1-Induced Activation of Human Fetal Lung Fibroblasts In Vitro. J. Pharm. Pharmacol. 2016, 68, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Yang, D.; Sun, X.; Liu, W.; Wang, L.; Li, X.; Man, X.; Fu, Q. Effects of Hydroxysafflor Yellow A on Proliferation and Collagen Synthesis of Rat Vascular Adventitial Fibroblasts Induced by Angiotensin II. Int. J. Clin. Exp. Pathol. 2014, 7, 5772–5781. [Google Scholar] [PubMed]
- Qian, Y.; Chen, X. ID1, Inhibitor of Differentiation/DNA Binding, Is an Effector of the p53-Dependent DNA Damage Response Pathway. J. Biol. Chem. 2008, 283, 22410–22416. [Google Scholar] [CrossRef]
- Liu, H.; Jia, D.; Li, A.; Chau, J.; He, D.; Ruan, X.; Liu, F.; Li, J.; He, L.; Li, B. p53 Regulates Neural Stem Cell Proliferation and Differentiation via BMP-Smad1 Signaling and Id1. Stem Cells Dev. 2013, 22, 913–927. [Google Scholar] [CrossRef]
- Je, Y.J.; Choi, D.K.; Sohn, K.C.; Kim, H.R.; Im, M.; Lee, Y.; Lee, J.H.; Kim, C.D.; Seo, Y.J. Inhibitory Role of Id1 on TGF-β-Induced Collagen Expression in Human Dermal Fibroblasts. Biochem. Biophys. Res. Commun. 2014, 444, 81–85. [Google Scholar] [CrossRef]
- Chen, X.; Zankl, A.; Niroomand, F.; Liu, Z.; Katus, H.A.; Jahn, L.; Tiefenbacher, C. Upregulation of ID Protein by Growth and Differentiation Factor 5 (GDF5) Through a Smad-Dependent and MAPK-Independent Pathway in HUVSMC. J. Mol. Cell. Cardiol. 2006, 41, 26–33. [Google Scholar] [CrossRef]
- Liang, Y.Y.; Brunicardi, F.C.; Lin, X. Smad3 Mediates Immediate Early Induction of Id1 by TGF-β. Cell Res. 2009, 19, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Dituri, F.; Cossu, C.; Mancarella, S.; Giannelli, G. The Interactivity between TGFβ and BMP Signaling in Organogenesis, Fibrosis, and Cancer. Cells 2019, 8, 1130. [Google Scholar] [CrossRef]
- Cabrita, M.A.; Bose, R.; Vanzyl, E.J.; Pastic, A.; Marcellus, K.A.; Pan, E.; Hamill, J.D.; McKay, B.C. The p53 Protein Induces Stable miRNAs That Have the Potential to Modify Subsequent p53 Responses. Gene 2017, 608, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.A.; Espinosa, J.M. The Impact of Post-Transcriptional Regulation in the p53 Network. Brief. Funct. Genom. 2013, 12, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Jones, M.C. Cell Cycle Control by Cell-Matrix Interactions. Curr. Opin. Cell Biol. 2024, 86, 102288. [Google Scholar] [CrossRef]
- Narasimha, A.M.; Kaulich, M.; Shapiro, G.S.; Choi, Y.J.; Sicinski, P.; Dowdy, S.F. Cyclin D Activates the Rb Tumor Suppressor by Mono-Phosphorylation. eLife 2014, 3, e02872. [Google Scholar] [CrossRef]
- Matson, J.P.; Cook, J.G. Cell Cycle Proliferation Decisions: The Impact of Single Cell Analyses. FEBS J. 2017, 284, 362–375. [Google Scholar] [CrossRef]
- Rex, N.; Melk, A.; Schmitt, R. Cellular Senescence and Kidney Aging. Clin. Sci. 2023, 137, 1805–1821. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; de Jong, T.V.; Melov, S.; Guryev, V.; Campisi, J.; Demaria, M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr. Biol. 2017, 27, 2652–2660.e4. [Google Scholar] [CrossRef]


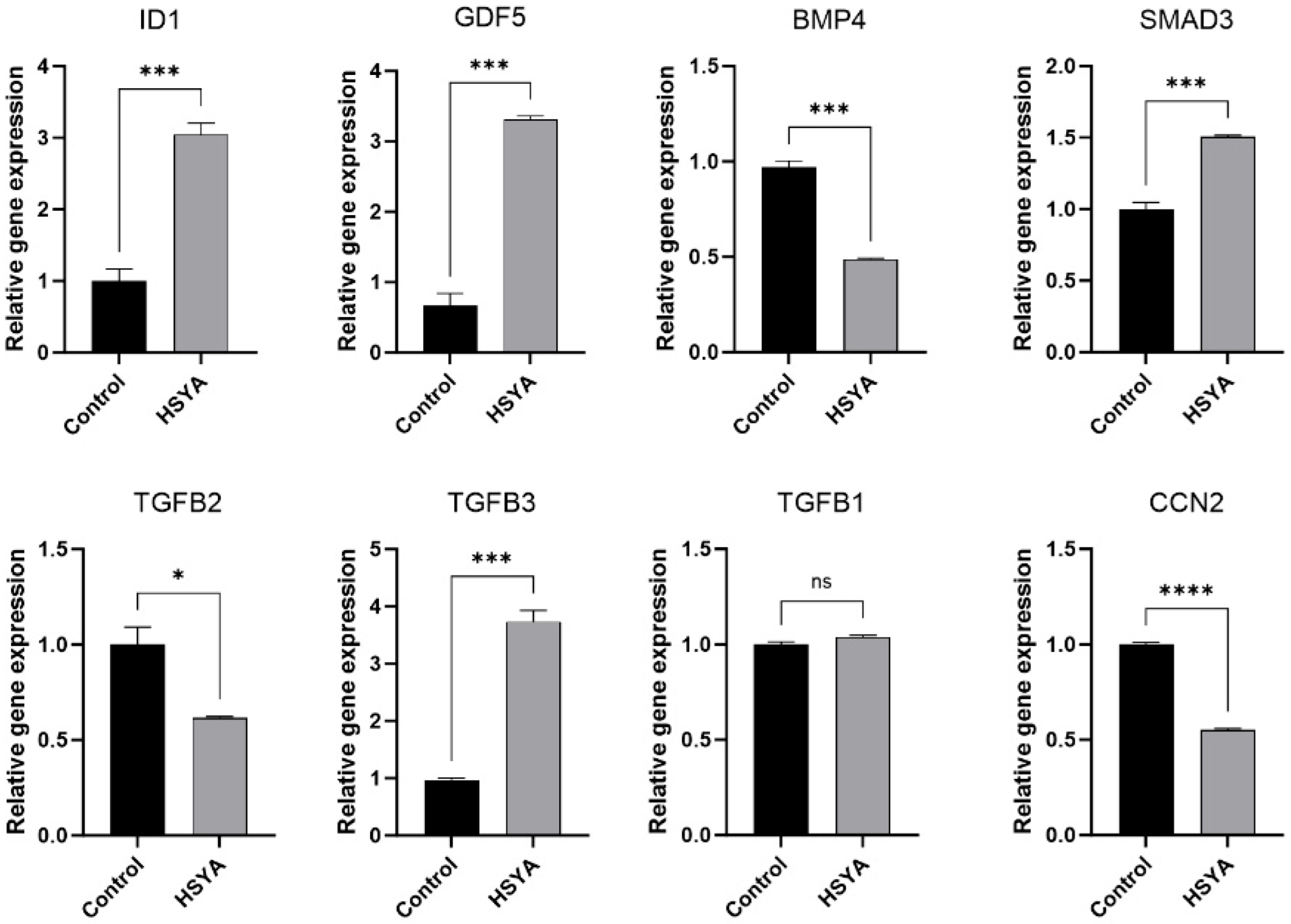
| Gene Symbol | Control Average FPKM | HSYA Average FPKM | log2 (HSYA/Control) | Q-Value |
|---|---|---|---|---|
| ID1 | 4.35 | 144.62 | 4.79 | 1.71 × 10−88 |
| GDF5 | 0.29 | 12.31 | 5.21 | 6.58 × 10−46 |
| SMAD3 | 3.21 | 13.31 | 1.89 | 4.33 × 10−37 |
| BMP4 | 14.04 | 3.01 | −2.37 | 2.24 × 10−25 |
| TGFB2 | 19.54 | 2.86 | −3.03 | 4.52 × 10−17 |
| ZFYVE16 | 6.75 | 1.95 | −1.91 | 1.40 × 10−13 |
| BMPR2 | 18.71 | 4.75 | −2.22 | 5.02 × 10−13 |
| ACVRL1 | 0.71 | 4.34 | 2.47 | 6.16 × 10−13 |
| TGFB3 | 0.21 | 1.77 | 2.87 | 9.66 × 10−13 |
| DCN | 129.94 | 39.06 | −1.95 | 1.31 × 10−12 |
| Gene Symbol | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| ID1 | GTTGGAGCTGAACTCGGAATCC | ACACAAGATGCGATCGTCCGCA |
| GDF5 | AACAGCAGCGTGAAGTTGGAGG | ACACGTACCTCTGCTTCCTGAC |
| SMAD3 | TGAGGCTGTCTACCAGTTGACC | GTGAGGACCTTGTCAAGCCACT |
| BMP4 | CTGGTCTTGAGTATCCTGAGCG | TCACCTCGTTCTCAGGGATGCT |
| TGFB2 | AAGAAGCGTGCTTTGGATGCGG | ATGCTCCAGCACAGAAGTTGGC |
| TGFB3 | CTAAGCGGAATGAGCAGAGGATC | TCTCAACAGCCACTCACGCACA |
| TGFB1 | TACCTGAACCCGTGTTGCTCTC | GTTGCTGAGGTATCGCCAGGAA |
| CCN2 | CTTGCGAAGCTGACCTGGAAGA | CCGTCGGTACATACTCCACAGA |
| β-actin | GGGAAATCGTGCGTGACATT | AGGTAGTTTCGTGGATGCCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Zhu, Q.; Feng, X.; Chou, X.; Lu, T. Transcriptomic Analysis of the Effects of Hydroxysafflor Yellow A on hUC-MSC Senescence via the ECM–Receptor Interaction Pathway. Int. J. Mol. Sci. 2025, 26, 9579. https://doi.org/10.3390/ijms26199579
Wang S, Zhu Q, Feng X, Chou X, Lu T. Transcriptomic Analysis of the Effects of Hydroxysafflor Yellow A on hUC-MSC Senescence via the ECM–Receptor Interaction Pathway. International Journal of Molecular Sciences. 2025; 26(19):9579. https://doi.org/10.3390/ijms26199579
Chicago/Turabian StyleWang, Siyun, Qi Zhu, Xueer Feng, Xinghua Chou, and Tao Lu. 2025. "Transcriptomic Analysis of the Effects of Hydroxysafflor Yellow A on hUC-MSC Senescence via the ECM–Receptor Interaction Pathway" International Journal of Molecular Sciences 26, no. 19: 9579. https://doi.org/10.3390/ijms26199579
APA StyleWang, S., Zhu, Q., Feng, X., Chou, X., & Lu, T. (2025). Transcriptomic Analysis of the Effects of Hydroxysafflor Yellow A on hUC-MSC Senescence via the ECM–Receptor Interaction Pathway. International Journal of Molecular Sciences, 26(19), 9579. https://doi.org/10.3390/ijms26199579





