Efficient Hydrolysis of Dichlorvos in Water by Stenotrophomonas acidaminiphila G1 and Methyl Parathion Hydrolase
Abstract
1. Introduction
2. Results
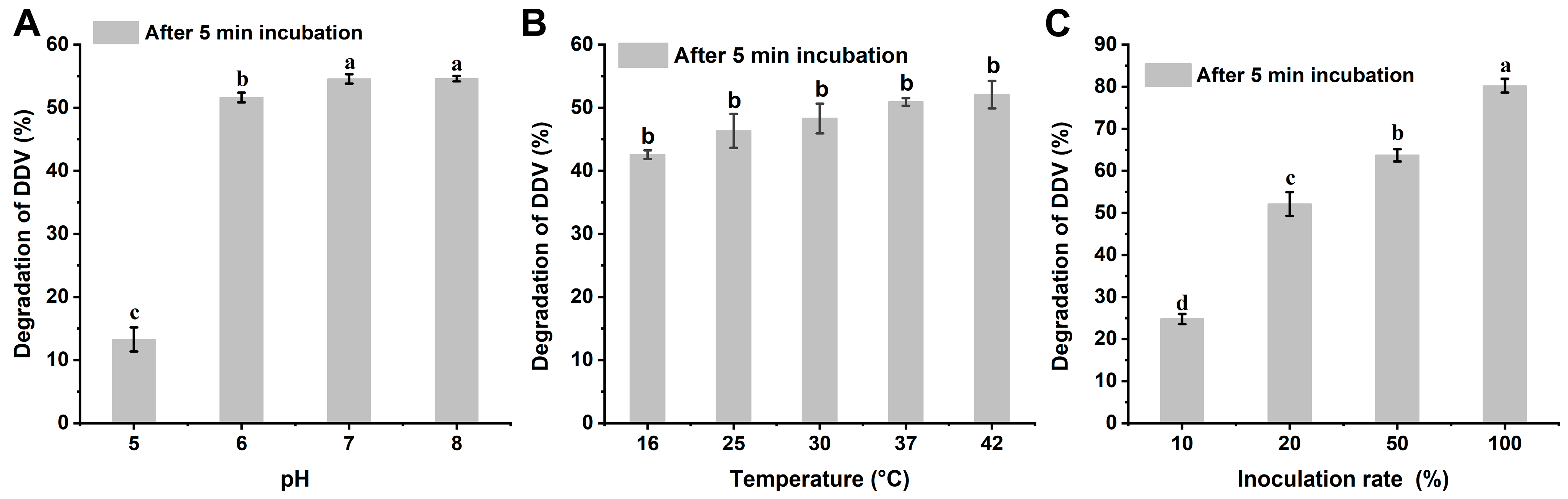
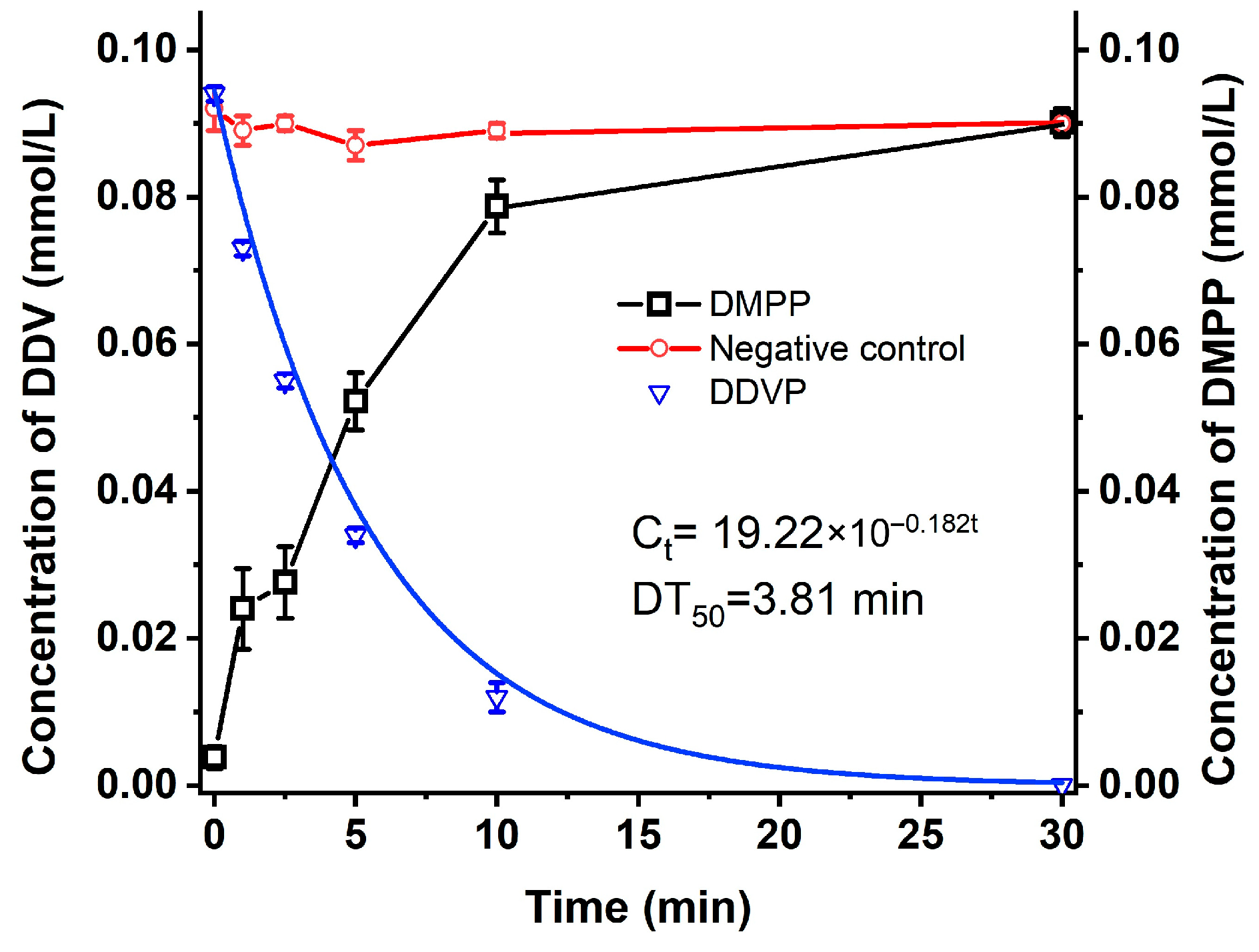
2.1. Metabolism of DDVP by Strain G1
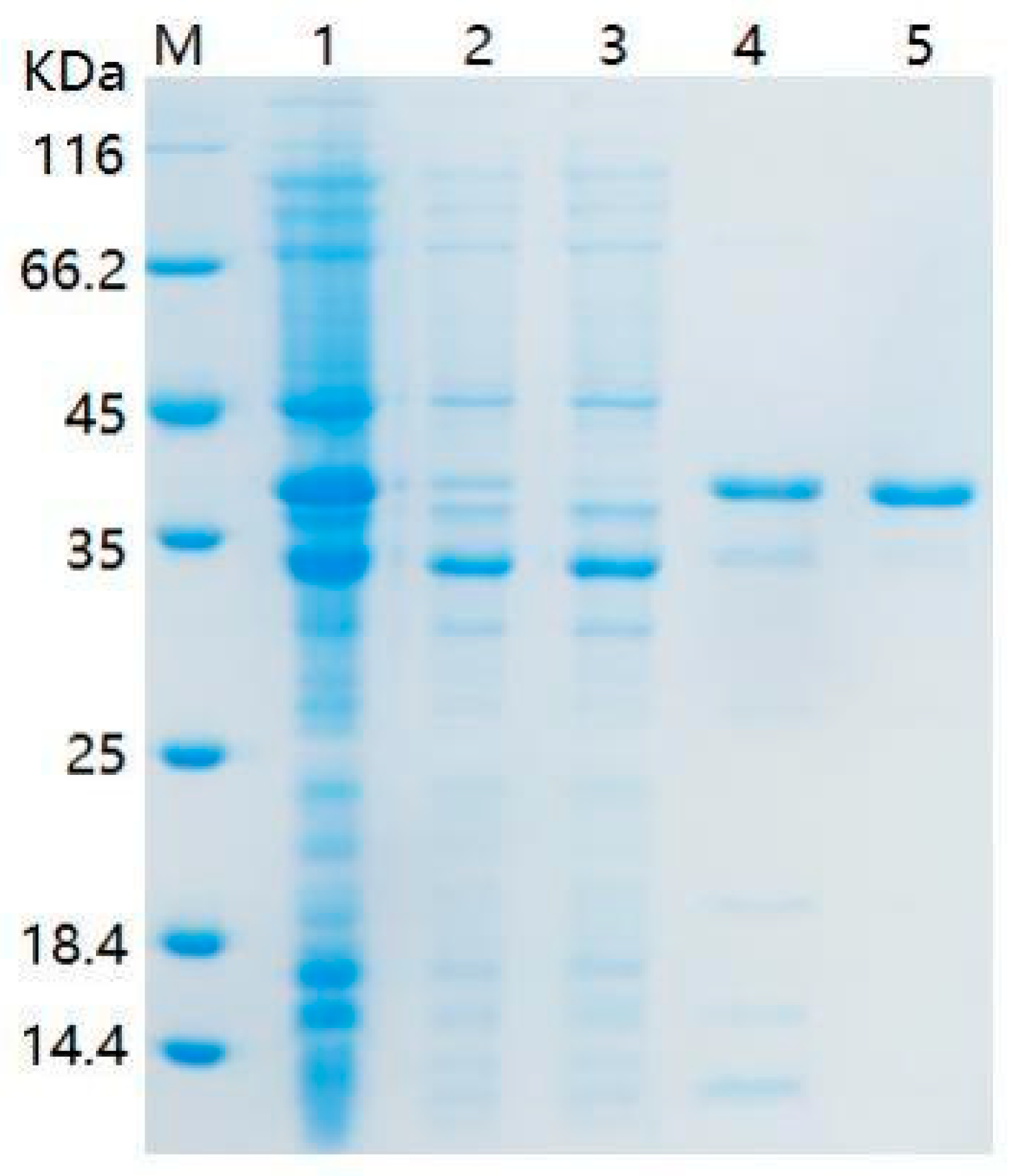
2.2. Genome Features of Strain G1 and Degradation Gene in Strain G1
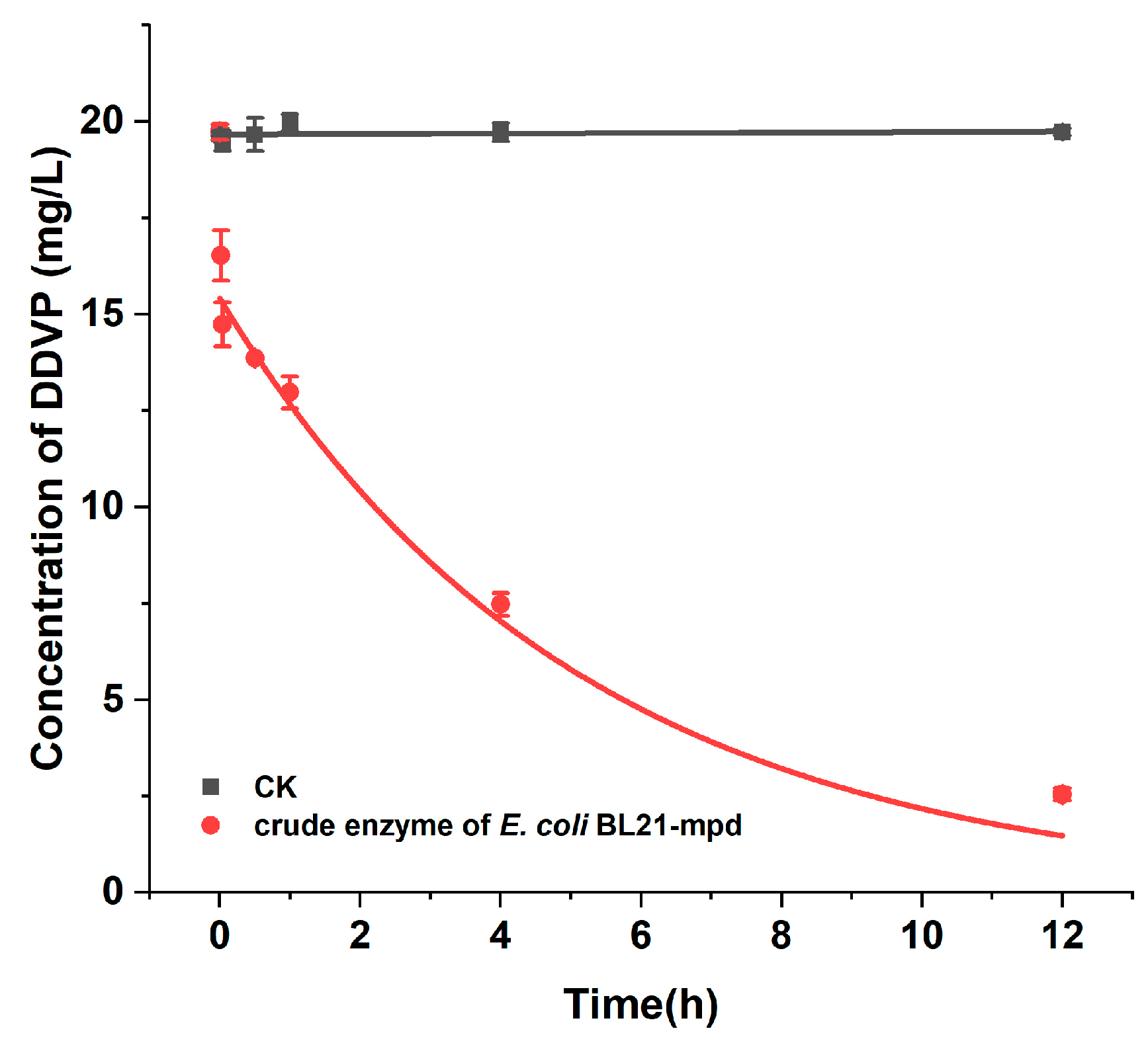
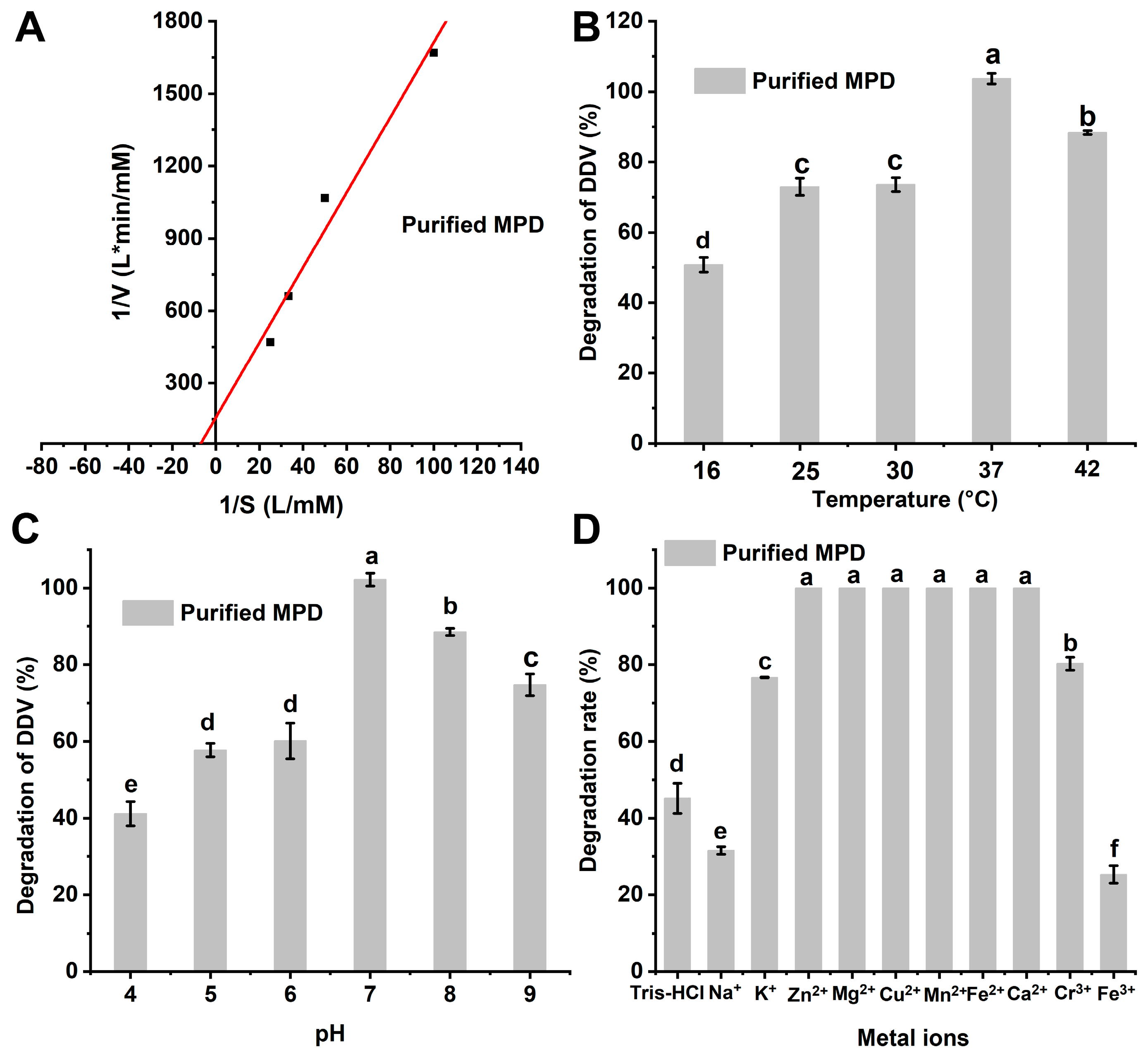
2.3. Degradation Kinetics of Purified Enzyme MPD for DDVP
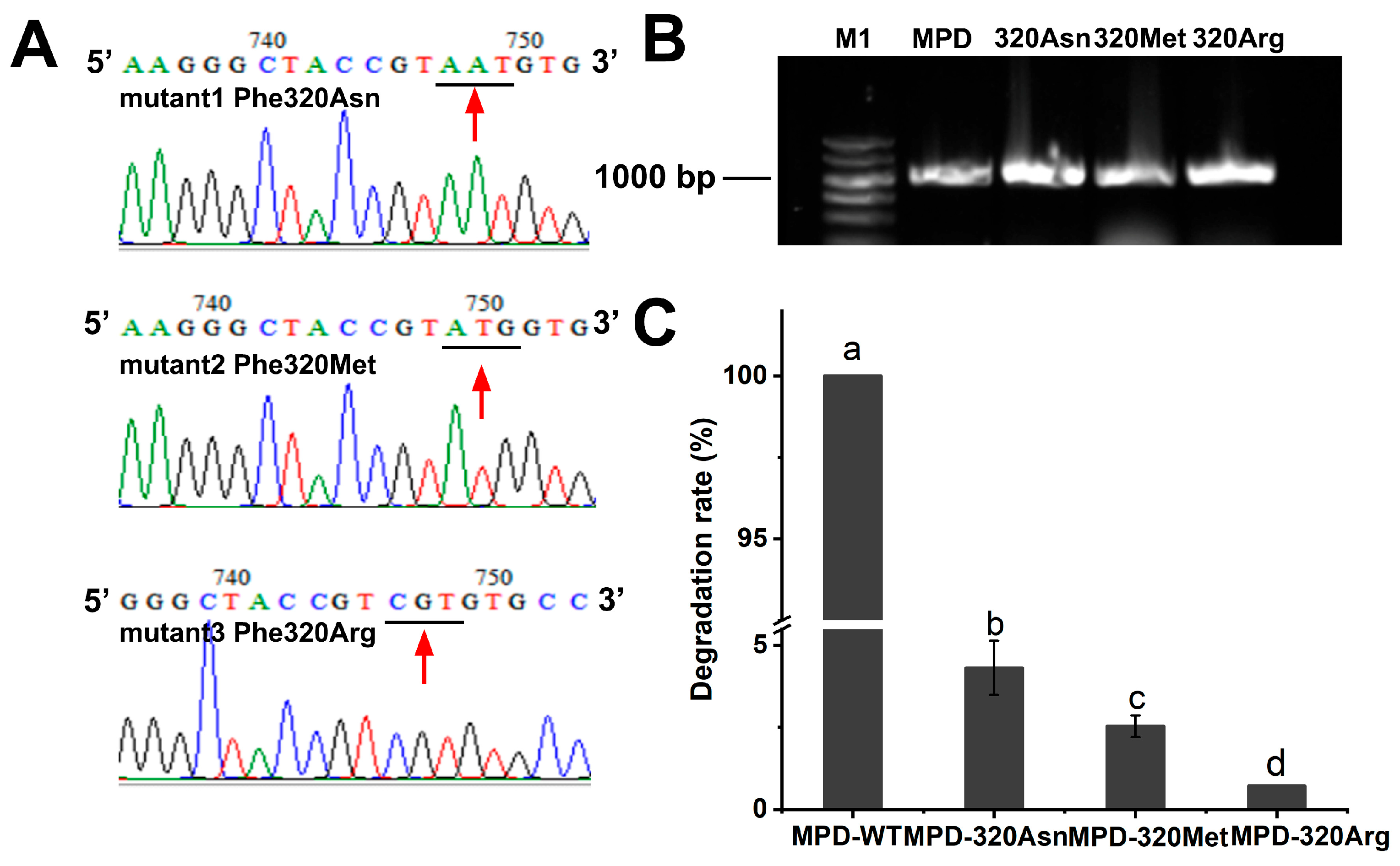
2.4. Catalytic Mechanism of Degrading Enzyme MPD on DDVP
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Strains, Plasmids, and Growth Conditions
4.3. Degradation of DDVP by Strain G1
4.4. Complete Genome Sequencing
4.5. Prediction and Annotation of DDVP Degradation Genes
4.6. Cloning, Expression of the mpd Gene, and Purification of Enzyme MPD
4.7. Determination of Enzyme Activity
4.8. Degradation Characteristics of DDVP by Purified Enzyme
4.9. Analytical Methods
4.10. Homology Modeling and Molecular Docking of DDVP with MPD
4.11. Point Mutation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| DDVP | Dichlorvos |
| CFU | Colony-forming units |
| DMPP | Dimethyl phosphate |
| OPs | Organophosphorus pesticides |
| WHO | World Health Organization |
| EPA | Environmental Protection Agency |
| LB | Luria–Bertani |
| MPS | Massive parallel sequencing |
| PCR | Polymerase chain reaction |
| UPLC | Ultraperformance liquid chromatography |
| MPD | Methyl parathion hydrolase |
References
- Jiang, J.F.; Deng, K.Q.; Duan, R.; An, C.; Dao, F.; Huang, J.L. Iron/manganese-zeolitic imidazolate framework (Fe/Mn-ZIF) nanozyme combined with acetylcholinesterase for colorimetric rapid detection of organophosphorus pesticides. Food Chem. 2025, 473, 143090. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhou, B.H.; Chen, H.L.; Yuan, R.F. Heterogeneous photocatalytic oxidation for the removal of organophosphorus pollutants from aqueous solutions: A review. Sci. Total Environ. 2023, 856, 159048. [Google Scholar]
- Mishra, A.; Kumar, J.; Melo, J.S.; Sandaka, B.P. Progressive development in biosensors for detection of dichlorvos pesticide: A review. J. Environ. Chem. Eng. 2021, 9, 105067. [Google Scholar] [CrossRef]
- Quintino-Ottonicar, G.G.; da Silva, L.R.; Maria, V.L.R.d.S.; Pizzo, E.M.; de Santana, A.C.P.; Lenharo, N.R.; Pinho, C.F.; Pereira, S. Exposure to Dichlorvos pesticide alters the morphology of and lipid metabolism in the ventral prostate of rats. Front. Toxicol. 2023, 5, 1207612. [Google Scholar] [CrossRef]
- Fagbohun, I.K.; Idowu, E.T.; Onafuwa, A.O.; Adeneye, A.K.; Adeogun, A.O.; Adetoro, O.O. Knowledge, attitudes and perception of communities on mosquitoes and its control practices in Lagos State, Nigeria. Pan Afr. Med. J. 2021, 38, 44. [Google Scholar] [CrossRef]
- Nteziyaremye, P.; Cherutoi, J.; Makatiani, J.; Muhizi, T. Insecticidal potential of essential oils from Cupressus lusitanica growing in ecological zones of Rwanda against adult housefly, Musca domestica L. Int. J. Trop. Insect Sci. 2023, 43, 895–907. [Google Scholar] [CrossRef]
- Yu, J.X.; Wang, X.G.; Yao, X.L.; Wu, X.M. Safety Evaluation of Heavy Metal Contamination and Pesticide Residues in Coix Seeds in Guizhou Province, China. Foods 2022, 11, 2286. [Google Scholar] [CrossRef]
- Barbieri, M.V.; Monllor-Alcaraz, L.S.; Postigo, C.; de Alda, M.L. Improved fully automated method for the determination of medium to highly polar pesticides in surface and groundwater and application in two distinct agriculture-impacted areas. Sci. Total Environ. 2020, 745, 140650. [Google Scholar] [CrossRef]
- Alhassan, A.B.; Aljahdali, M.O. Behavioural and Biochemical Responses of Freshwater Bivalve Anodonta marginata Exposed to Dichlorvos. Water 2024, 16, 3572. [Google Scholar] [CrossRef]
- Ben Salem, I.; Boussabbeh, M.; Kantaoui, H.; Bacha, H.; Abid-Essefi, S. Crocin, the main active saffron constituent, mitigates dichlorvos-induced oxidative stress and apoptosis in HCT-116 cells. Biomed. Pharmacother. 2016, 82, 65–71. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Park, S.; Vijayasarathy, S.; Swaminathan, A.; Sivasantosh, S.; Kim, Y.; Yoo, G.; Madhumitha, H.; Mubarakali, D.; Cho, N.M. Cellular metabolism and health impacts of dichlorvos: Occurrence, detection, prevention, and remedial strategies—A review. Environ. Res. 2024, 242, 117600. [Google Scholar] [CrossRef]
- Ben Salem, I.; Boussabbeh, M.; Bacha, H.; Abid, S. Dichlorvos-induced toxicity in HCT116 cells: Involvement of oxidative stress and apoptosis. Pestic. Biochem. Physiol. 2015, 119, 62–66. [Google Scholar] [CrossRef]
- Zhang, P.C.; Zhou, Z.X.; Yao, J.Q.; Jiang, Y.H.; Lei, H.; Xie, Z.J.; Li, J.; Zhao, X.L.; Zhu, L.; Wan, M.H.; et al. Effects of pesticide dichlorvos on liver injury in rats and related toxicity mechanisms. Ecotoxicol. Environ. Saf. 2025, 290, 117747. [Google Scholar] [CrossRef]
- Saka, W.A.; Adeogun, A.E.; Adisa, V.I.; Olayioye, A.; Igbayilola, Y.D.; Akhigbe, R.E. L-arginine attenuates dichlorvos-induced testicular toxicity in male Wistar rats by suppressing oxidative stress-dependent activation of caspase 3-mediated apoptosis. Biomed. Pharmacother. 2024, 178, 117136. [Google Scholar] [CrossRef]
- Li, D.Y.; Ma, X.Y.; Zhang, S.Y.; Wang, Y.K.; Han, Y.N.; Chen, R.; Wang, X.C.; Ngo, H.H. Aquatic photolysis of high-risk chemicals of emerging concern from secondary effluent mediated by sunlight irradiation for ecological safety and the enhanced methods. Water Res. 2023, 238, 120002. [Google Scholar] [CrossRef]
- Kumar, R.; George, L.; Jun, Z.; Mukherji, S. Photocatalytic activity of graphene oxide-TiO2 nanocomposite on dichlorvos and malathion and assessment of toxicity changes due to photodegradation. Chemosphere 2022, 308, 136402. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Jonathan, S.G.; Ahmad, R. Degradation of 2, 2-Dichlorovinyl dimethyl phosphate (dichlorvos) through the rhizosphere interaction between Panicum maximum Jacq and some selected fungi. Chemosphere 2019, 221, 403–411. [Google Scholar] [CrossRef]
- Parte, S.G.; Mohekar, A.D.; Kharat, A.S. Aerobic dichlorvos degradation by Pseudomonas stutzeri smk: Complete pathway and implications for toxicity in Mus musculus. Iran. J. Microbiol. 2020, 12, 138–147. [Google Scholar] [CrossRef]
- Sun, J.N.; Si, G.Y.; Liu, H.Y.; Li, Y.Q.; Wang, X.H.; Chen, J. Degradation effects on dichlorvos by a biocontrol strain, Trichoderma atroviride T23. J. Integr. Agric. 2023, 22, 2746–2758. [Google Scholar] [CrossRef]
- Ma, W.W.; Zhao, Y.Y.; Sun, H.; Zhang, Z.W.; Huang, L.L. Oral Administration of Lactiplantibacillus plantarum CCFM8661 Alleviates Dichlorvos-Induced Toxicity in Mice. Foods 2024, 13, 3211. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, W.P.; Li, J.Y.; Pang, S.M.; Mishra, S.; Bhatt, P.; Zeng, D.X.; Chen, S.H. Emerging Technologies for Degradation of Dichlorvos: A Review. Int. J. Environ. Res. Public Health 2021, 18, 5789. [Google Scholar]
- Malakootian, M.; Shahesmaeili, A.; Faraji, M.; Amiri, H.; Silva Martinez, S. Advanced oxidation processes for the removal of organophosphorus pesticides in aqueous matrices: A systematic review and meta-analysis. Process Saf. Environ. Prot. 2020, 134, 292–307. [Google Scholar] [CrossRef]
- Youfeng, Z.; Dongdong, Z.; Ling, H. Enantioselective biodegradation and enantiomerization of dichlorprop in soils. Chemosphere 2020, 258, 127322. [Google Scholar] [CrossRef]
- Ning, J.; Gang, G.; Bai, Z.; Hu, Q.; Qi, H.; Ma, A.; Zhuan, X.; Zhuang, G. In situ enhanced bioremediation of dichlorvos by a phyllosphere Flavobacterium strain. Front. Environ. Sci. Eng. 2012, 6, 231–237. [Google Scholar] [CrossRef]
- Yadav, S.; Verma, S.K.; Chaudhary, H.S. Isolation and Characterization of Organophosphate Pesticides Degrading Bacteria from Contaminated Agricultural Soil. OnLine J. Biol. Sci. 2015, 15, 113–125. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Zhang, G.-S.; Zhang, Z.-H.; Xu, J.-H.; Li, S.-P. Isolation and Characterization of a Dichlorvos-Degrading Strain DDV-1 of Ochrobactrum sp. Pedosphere 2006, 16, 64–71. [Google Scholar] [CrossRef]
- Oncescu, T.; Oancea, P.; Enache, M.; Popescu, G.; Dumitru, L.; Kamekura, M. Halophilic bacteria are able to decontaminate dichlorvos, a pesticide, from saline environments. Open Life Sci. 2007, 2, 563–573. [Google Scholar] [CrossRef]
- Yuan, S.; Li, C.; Yu, H.; Xie, Y.; Guo, Y.; Yao, W. Selective uptake determines the variation in degradation of organophosphorus pesticides by Lactobacillus plantarum. Food Chem. 2021, 360, 130106. [Google Scholar] [CrossRef]
- Zhao, K.; Yu, Y.; Jiang, D.; Wang, D.; Li, Z.-M.; Huang, G.-Z.; Bai, Z.-H.; Bai, Z.H. Degradation of Dichlorvos by Rhodobacter sphaeroides. Environ. Sci. 2009, 4, 1200–1204. [Google Scholar]
- Sukirtha, T.; Usharani, M. Production and Qualitative Analysis of Biosurfactant and Biodegradation of the Organophosphate by Nocardia mediterranie. J. Bioremediat. Biodegrad. 2013, 4, 198–205. [Google Scholar]
- Sun, J.; Karuppiah, V.; Li, Y.; Pandian, S.; Kumaran, S.; Chen, J. Role of cytochrome P450 genes of Trichoderma atroviride T23 on the resistance and degradation of dichlorvos. Chemosphere 2022, 290, 133173. [Google Scholar] [CrossRef]
- Cruz-Alcalde, A.; Sans, C.; Esplugas, S. Priority pesticide dichlorvos removal from water by ozonation process: Reactivity, transformation products and associated toxicity. Sep. Purif. Technol. 2018, 192, 123–129. [Google Scholar] [CrossRef]
- Richardson, D.D.; Caruso, J.A. Screening organophosphorus nerve agent degradation products in pesticide mixtures by GC-ICPMS. Anal. Bioanal. Chem. 2007, 389, 679–682. [Google Scholar] [CrossRef]
- Magny, R.; Lejeune, M.; Mégarbane, B.; Houzé, P.; Labat, L. Cas d’intoxication au dichlorvos: Identification de métabolites et de produits de dégradation par une approche utilisant les réseaux moléculaires. Toxicol. Anal. Clin. 2022, 34, 207–208. [Google Scholar] [CrossRef]
- Abe, K.; Yoshida, S.; Suzuki, Y.; Mori, J.; Doi, Y.; Takahashi, S.; Kera, Y. Haloalkylphosphorus Hydrolases Purified from Sphingomonas sp. Strain TDK1 and Sphingobium sp. Strain TCM1. Appl. Environ. Microbiol. 2014, 80, 5866–5873. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yuan, X.; Li, Y.; Wang, X.; Chen, J. The pathway of 2,2-dichlorovinyl dimethyl phosphate (DDVP) degradation by Trichoderma atroviride strain T23 and characterization of a paraoxonase-like enzyme. Appl. Microbiol. Biotechnol. 2019, 103, 8947–8962. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, Z.; Wang, D.; Guan, S.; Han, W. Molecular Dynamics Simulations of Acylpeptide Hydrolase Bound to Chlorpyrifosmethyl Oxon and Dichlorvos. Int. J. Mol. Sci. 2015, 16, 6217–6234. [Google Scholar] [CrossRef]
- Okoroiwu, H.U.; Iwara, I.A. Dichlorvos toxicity: A public health perspective. Interdiscip. Toxicol. 2019, 11, 129–137. [Google Scholar] [CrossRef]
- Dong, Y.; Bartlam, M.; Sun, L.; Zhou, Y.; Zhang, Z.-P.; Zhang, C.-G.; Rao, Z.; Zhang, X.E. Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3. J. Mol. Biol. 2005, 353, 655–663. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, J.-J.; Wang, S.-J.; Zhang, X.-E.; Zhou, N.-Y. Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. strain WBC-3. Biochem. Biophys. Res. Commun. 2005, 334, 1107–1114. [Google Scholar] [CrossRef]
- Schenk, G.; Mateen, I.; Ng, T.-K.; Pedroso, M.M.; Mitić, N.; Jafelicci, M.; Marques, R.F.; Gahan, L.R.; Ollis, D.L. Organophosphate-degrading metallohydrolases: Structure and function of potent catalysts for applications in bioremediation. Coord. Chem. Rev. 2016, 317, 122–131. [Google Scholar] [CrossRef]
- Li, Y.; Yang, H.; Xu, F. Identifying and engineering a critical amino acid residue to enhance the catalytic efficiency of Pseudomonas sp. methyl parathion hydrolase. Appl. Microbiol. Biotechnol. 2018, 102, 6537–6545. [Google Scholar] [CrossRef]
- Moradi, S.; Khani, S.; Ansari, M.; Shahlaei, M. Atomistic details on the mechanism of organophosphates resistance in insects: Insights from homology modeling, docking and molecular dynamic simulation. J. Mol. Liq. 2019, 276, 59–66. [Google Scholar] [CrossRef]

| Gene Number | Locus | Annotation |
|---|---|---|
| G1_GM000376 | Chr1:451815:452579:+ | c-di-GMP-specific phosphodiesterase class I |
| G1_GM000403 | Chr1:479899:482832:+ | Sensor domain-containing phosphodiesterase |
| G1_GM000617 | Chr1:725113:726333:− | 2′3′-cyclic phosphodiesterase |
| G1_GM000878 | Chr1:1012855:1014471:− | Phosphodiesterase |
| G1_GM001006 | Chr1:1174990:1175985:− | Methyl-parathion hydrolase mpd |
| G1_GM001037 | Chr1:1214103:1214729:− | c-di-GMP phosphodiesterase class II |
| G1_GM001072 | Chr1:1257629:1259686:− | CdpA cyclic di-GMP phosphodiesterase |
| G1_GM001177 | Chr1:1376461:1377048:+ | RNA2′,3′-cyclic phosphodiesterase |
| G1_GM001721 | Chr1:1922141:1923490:+ | Phosphodiesterase |
| G1_GM001738 | Chr1:1942976:1945048:− | c-di-GMP-specific phosphodiesterase class I |
| G1_GM001887 | Chr1:2112072:2113784:+ | c-di-GMP phosphodiesterase |
| G1_GM001963 | Chr1:2197887:2200340:+ | c-di-GMP phosphodiesterase A |
| G1_GM002029 | Chr1:2274043:2275569:− | 3′,5′-cyclic AMP phosphodiesterase CpdA T |
| G1_GM002209 | Chr1:2470901:2473153:+ | Phosphodiesterase |
| G1_GM002415 | Chr1:2714768:2716021:+ | Phosphodiesterase |
| G1_GM003033 | Chr1:3405300:3405869:+ | Type I phosphodiesterase |
| G1_GM003125 | Chr1:3504090:3505742:+ | Type I phosphodiesterase |
| G1_GM003196 | Chr1:3576332:3577663:− | Diguanylate phosphodiesterase |
| G1_GM003204 | Chr1:3587564:3588784:+ | c-di-GMP-specific phosphodiesterase class I |
| G1_GM003579 | Chr1:4007203:4008258:− | Glycerophosphodiester phosphodiesterase |
| G1_GM003053 | Chr1:3427595:3428083:+ | Phosphotriesterase family |
| Protein | Substrate | Bond | Length (Å) | Kd (μM) | ΔG (kcal/mol) | Molecular-Interaction Energy (kcal/mol) | Electrostatic Potential Energy (kcal/mol) |
|---|---|---|---|---|---|---|---|
| MPD | DDVP | Hydrogen bond | 2.60 | 144.15 | −5.24 | −6.25 | −1.01 |
| Strains | Concentration (mg/L) | Time (h) | Degradation Rate (%) | DT50 (h) | Reference |
|---|---|---|---|---|---|
| Flavobacterium YD-4 | 400 | 48 | 60.89 | 15.60 | [24] |
| Lactobacillus plantarum LAB | 50 | 24 | 100 | / | [28] |
| Pseudomonas stutzeri SMK | 0.05 | 144 | 80 | / | [18] |
| Pseudomonas AUG12 | 100 | 140 | / | / | [25] |
| Rhodobacter sphaeroides EBL0706 | 400 | 12 | 98 | / | [29] |
| Ochrobactrum sp. dichlorvos-1 | 100 | 24 | 100 | / | [26] |
| Halophilic bacteria TL4 | 32.99 | / | / | 3.10 | [27] |
| Halophilic bacteria T10/1 | 32.99 | / | / | 4.47 | |
| Nocardia mediterranei | 10 | 72 | 100 | / | [30] |
| S. acidaminiphila G1 | 20 | 0.5 | 100 | 0.056 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, Q.; Hua, R. Efficient Hydrolysis of Dichlorvos in Water by Stenotrophomonas acidaminiphila G1 and Methyl Parathion Hydrolase. Int. J. Mol. Sci. 2025, 26, 9572. https://doi.org/10.3390/ijms26199572
Mei Q, Hua R. Efficient Hydrolysis of Dichlorvos in Water by Stenotrophomonas acidaminiphila G1 and Methyl Parathion Hydrolase. International Journal of Molecular Sciences. 2025; 26(19):9572. https://doi.org/10.3390/ijms26199572
Chicago/Turabian StyleMei, Quyang, and Rimao Hua. 2025. "Efficient Hydrolysis of Dichlorvos in Water by Stenotrophomonas acidaminiphila G1 and Methyl Parathion Hydrolase" International Journal of Molecular Sciences 26, no. 19: 9572. https://doi.org/10.3390/ijms26199572
APA StyleMei, Q., & Hua, R. (2025). Efficient Hydrolysis of Dichlorvos in Water by Stenotrophomonas acidaminiphila G1 and Methyl Parathion Hydrolase. International Journal of Molecular Sciences, 26(19), 9572. https://doi.org/10.3390/ijms26199572






