Genome-Wide Identification of the BXL Gene Family in Soybean and Expression Analysis Under Salt Stress
Abstract
1. Introduction
2. Results
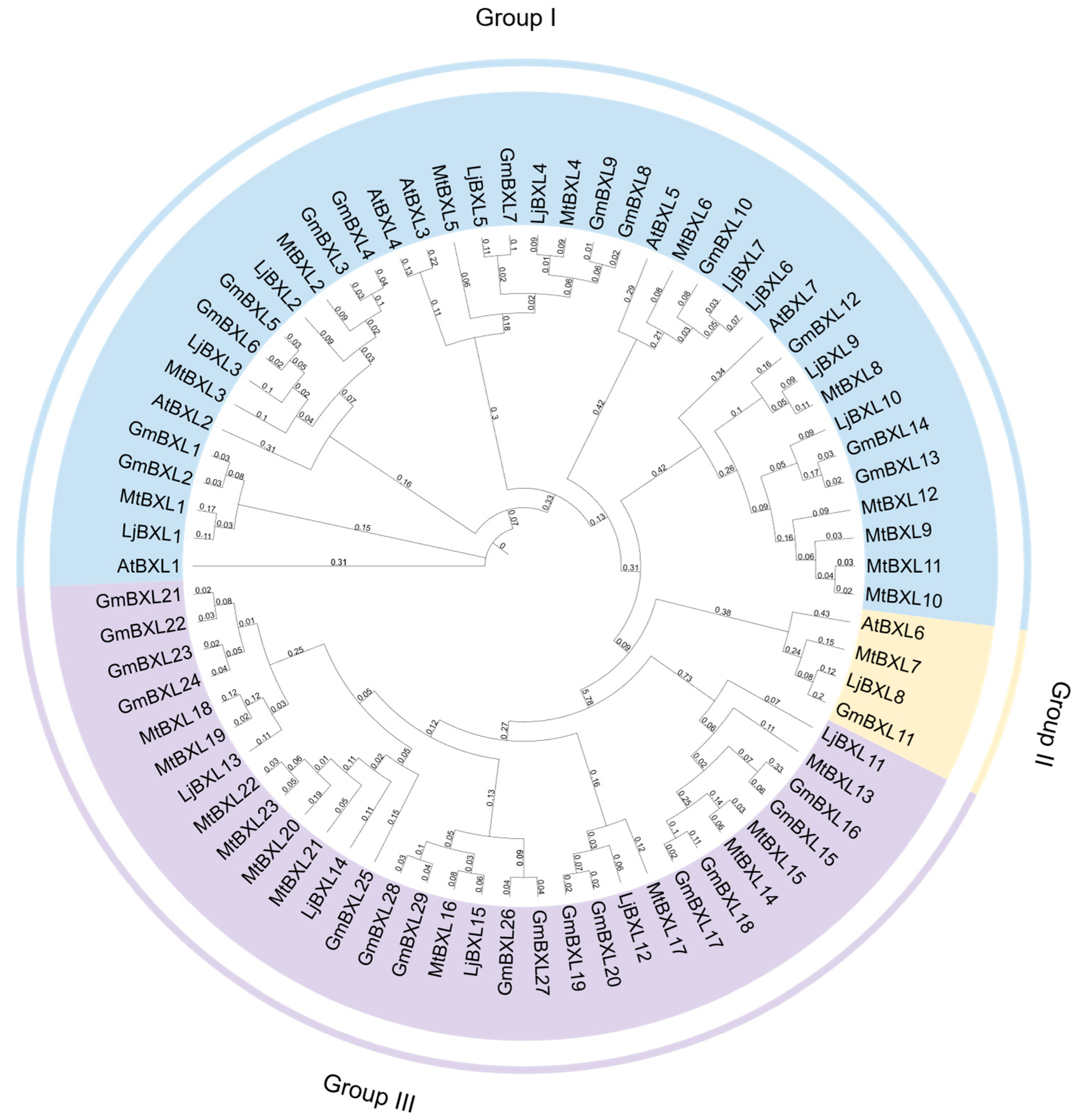
2.1. Identification of the Soybean BXL Genes
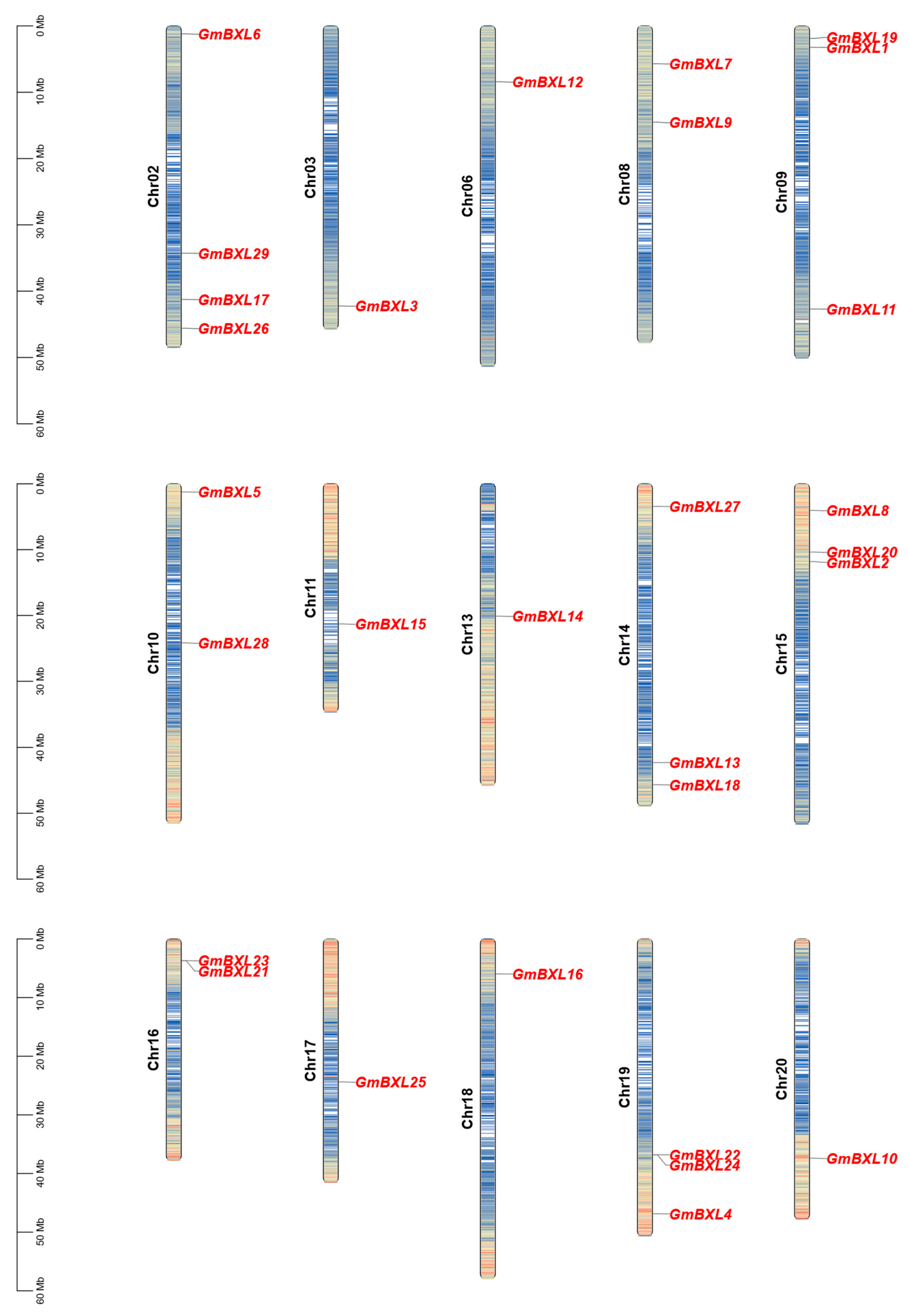
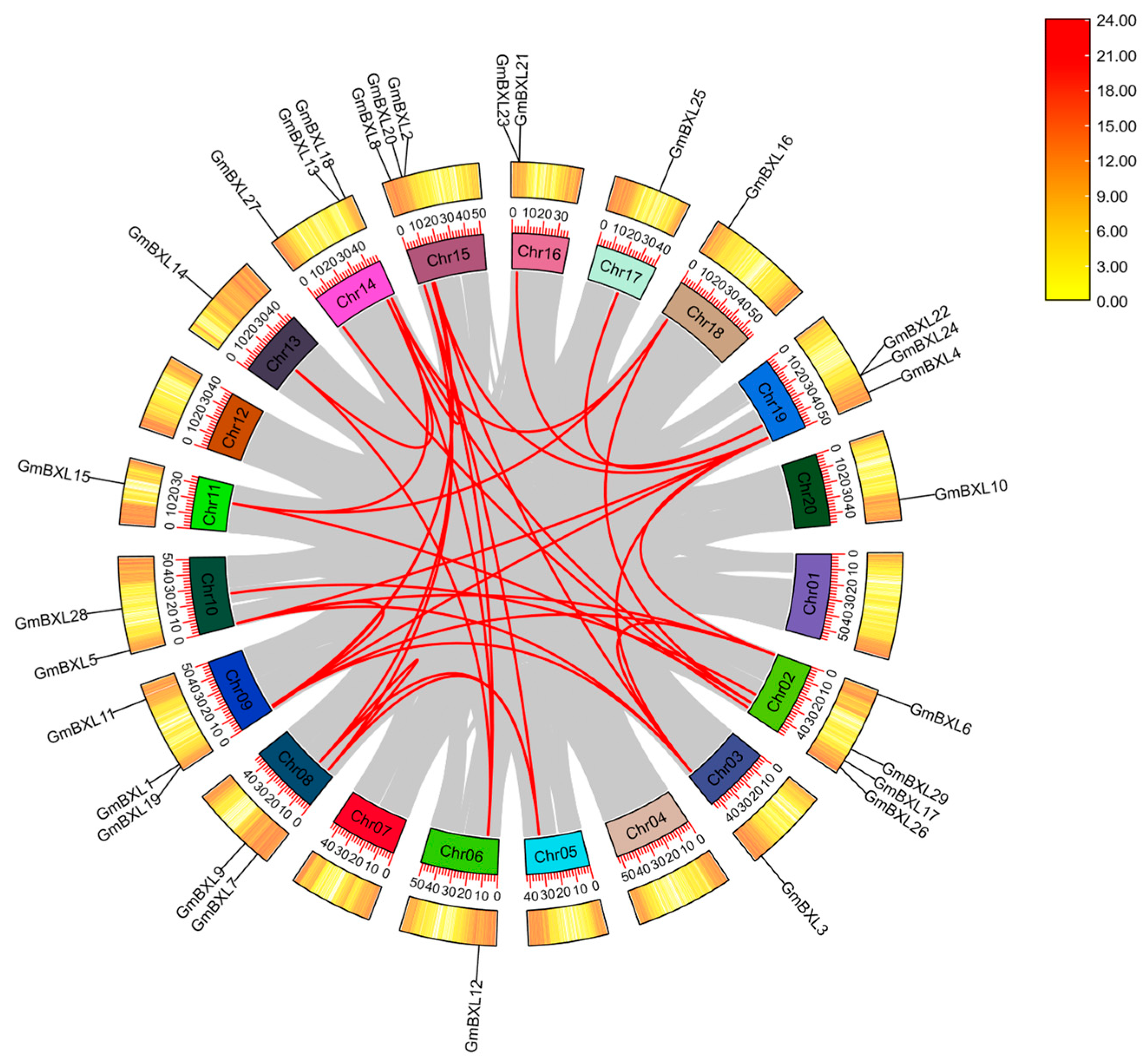
2.2. Chromosomal Distribution and Colinearity Analysis of Soybean BXL Genes
2.3. Analysis of Gene Structures, Conserved Motifs, and Domains
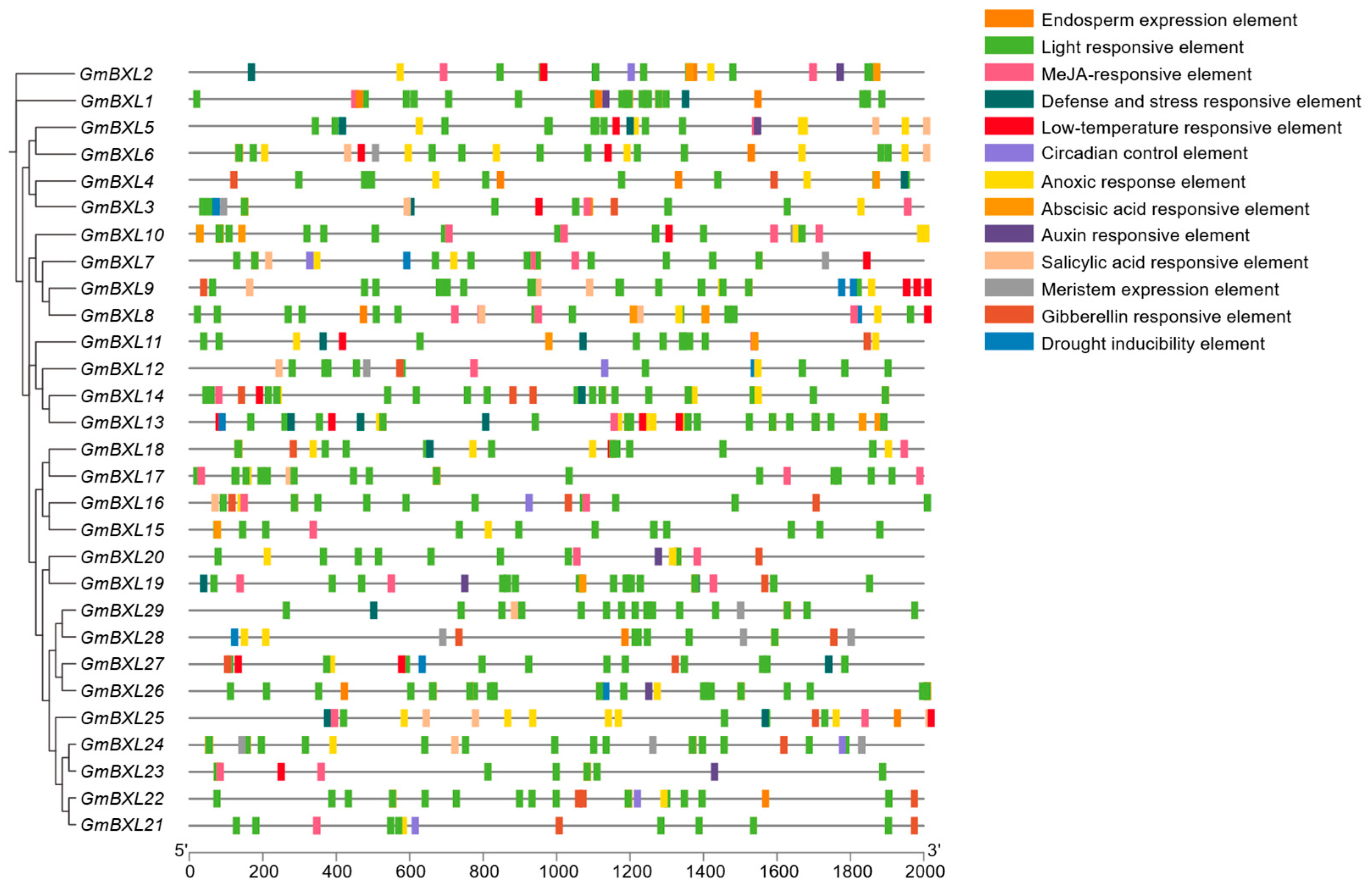
2.4. Analysis of Cis-Elements in BXL Gene Promoters
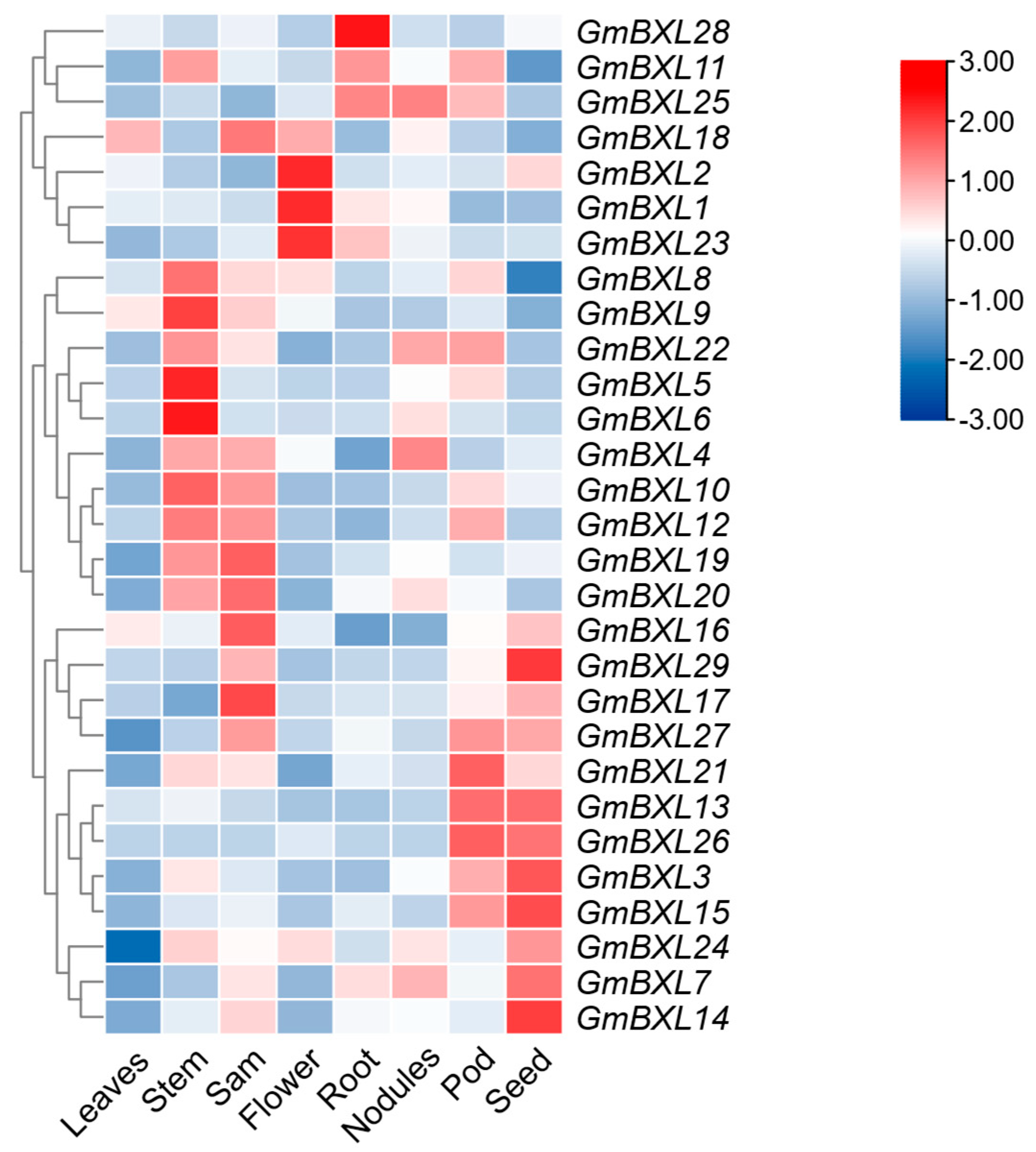
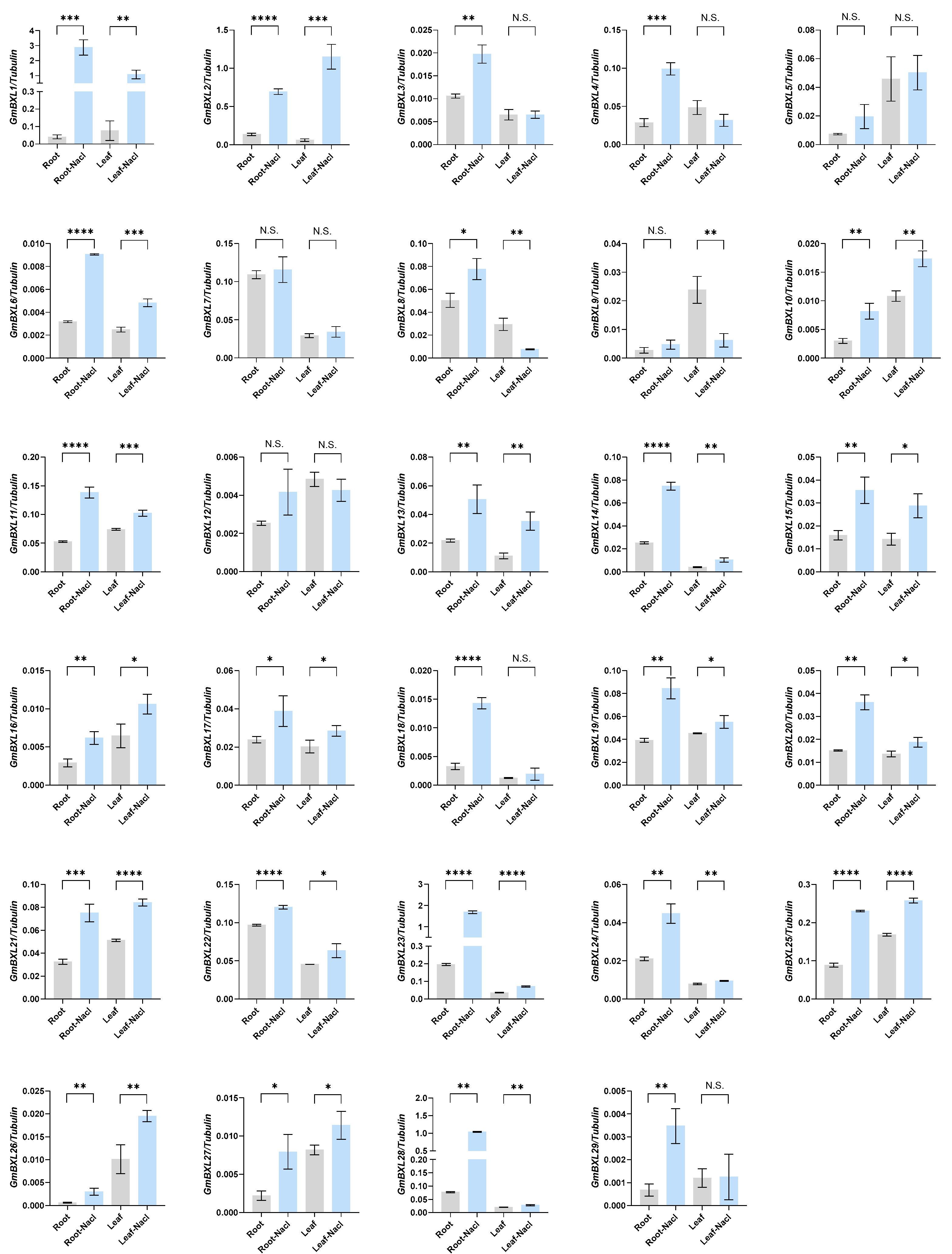
2.5. Response of BXL Genes Under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Annotation of BXL Genes in Soybean
4.3. Prediction of Physicochemical Properties of Soybean BXL Proteins
4.4. Phylogenetic Analysis of the BXL Proteins
4.5. Chromosomal Localization and Collinearity Analysis
4.6. Gene Structure and Conserved Motifs and Domains of BXL Genes
4.7. Analysis of Cis-Acting Elements
4.8. RNA-Seq Data Source and Expression Pattern Analysis of Soybean BXL Genes
4.9. RNA Extraction and Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [PubMed]
- Shtein, I.; Bar-On, B.; Popper, Z. Plant and algal structure: From cell walls to biomechanical function. Plant Physiol. 2018, 164, 56–66. [Google Scholar] [CrossRef]
- Carolina Di Santo, M.; Ilina, N.; Pagano, E.; Sozzi, G. A Japanese plum α-l-arabinofuranosidase/β-D-xylosidase gene is developmentally regulated by alternative splicing. Plant Sci. 2015, 231, 173–183. [Google Scholar] [CrossRef]
- Cleemuput, G.; Hessing, M.; Van Oort, M.; Deconynck, M.; Delcour, J. Purification and characterization of a [bata]-D-xylosidase and an endo-xylanase from wheat flour. Plant Physiol. 1997, 113, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Knob, A.; Terrasan, C.; Carmona, E. β-xylosidases from filamentous fungi: An overview. World J. Microbiol. Biotechnol. 2010, 26, 389–407. [Google Scholar] [CrossRef]
- Sinnott, M. Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 1990, 90, 1171–1202. [Google Scholar] [CrossRef]
- Minic, Z.; Rihouey, C.; Trung Do, C.; Lerouge, P.; Jouanin, L. Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol. 2004, 135, 867–878. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Goujon, T.; Minic, Z.; El Amrani, A.; Lerouxel, O.; Aletti, E.; Lapierre, C.; Joseleau, J.; Jouanin, L. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J. 2003, 33, 677–690. [Google Scholar] [CrossRef]
- Lee, E.; Koizumi, N.; Sano, H. Identification of genes that are up-regulated in concert during sugar depletion in Arabidopsis. Plant Cell Environ. 2004, 27, 337–345. [Google Scholar] [CrossRef]
- Lee, E.; Matsumura, Y.; Soga, K.; Hoson, T.; Koizumi, N. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 2007, 48, 405–413. [Google Scholar] [CrossRef]
- Bauer, K.; Nayem, S.; Lehmann, M.; Wenig, M.; Shu, L.; Ranf, S.; Geigenberger, P.; Vlotr, A. β-D-XYLOSIDASE 4 modulates systemic immune signaling in Arabidopsis thaliana. Front. Plant Sci. 2023, 13, 1096800. [Google Scholar] [CrossRef]
- Itai, A.; Yoshida, K.; Tanabe, K.; Tamura, F. A β-D-xylosidase-like gene is expressed during fruit ripening in Japanese pear (Pyrus pyrifolia Nakai). J. Exp. Bot. 1999, 50, 877–878. [Google Scholar] [CrossRef]
- Genero, M.; Gismondi, M.; Monti, L.; Gabilondo, J.; Budde, C.; Andreo, C.; Lara, M.; Drincovich, M.; Bustamante, C. Cell wall-related genes studies on peach cultivars with differential susceptibility to woolliness: Looking for candidates as indicators of chilling tolerance. Plant Cell Rep. 2016, 35, 1235–1246. [Google Scholar] [CrossRef]
- Bustamante, C.; Civello, P.; Martinez, G. Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit. Plant Sci. 2009, 177, 49–56. [Google Scholar] [CrossRef]
- Lai, B.; Chen, C.; Liu, C.; Zhu, C.; Du, L. Genome-wide identification of BXL family genes in Brassica juncea var. tumida and their expression during swelling of stem. J. Huazhong Agric. Univ. 2020, 39, 155–163. [Google Scholar]
- Chen, J.; Qu, C.; Chang, R.; Suo, J.; Yu, J.; Sun, X.; Liu, G.; Xu, Z. Genome-wide identification of BXL genes in Populus trichocarpa and their expression under different nitrogen treatments. Biotech 2020, 10, 57. [Google Scholar] [CrossRef]
- Wu, H.; Dong, W.; He, Z.; Li, Y.; Xie, H.; Sun, M.; Shen, Y.; Sun, X. Genome-wide identification and expression analysis of the rice BXL gene family. Biotechnol. Bull. 2025, 41, 87–98. [Google Scholar]
- Papiernik, S.; Grieve, C.; Lesch, S.; Yates, S. Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil Sci. Plant Anal. 2005, 36, 951–967. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.; Chan, L.; Law, C.; Li, M.; Lam, H. Twenty years of mining salt tolerance genes in soybean. Mol. Breed. 2023, 43, 45. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Qu, Y.; Guo, Y.; Yu, L.; Liu, Y.; Jiang, J.; Chen, J.; Ren, Y.; Liu, G.; Tian, L.; et al. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014, 80, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Yim, A.; Tao, Y.; Wong, F.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef]
- Qu, Y.; Guan, R.; Bose, J.; Henderson, S.; Wege, S.; Qiu, L.; Gilliham, M. Soybean CHX-type ion transport protein GmSALT3 confers leaf Na+ exclusion via a root derived mechanism, and Cl-exclusion via a shoot derived process. Plant Cell Environ. 2021, 44, 856–869. [Google Scholar] [CrossRef]
- Yung, W.; Wang, Q.; Huang, M.; Wong, F.; Liu, A.; Ng, M.; Li, K.; Sze, C.; Li, M.; Lam, H. Priminginduced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 2022, 109, 1575–1590. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef]
- Chen, H.; Chen, X.; Gu, H.; Wu, B.; Zhang, H.; Yuan, X.; Cui, X. GmHKT1;4, a novel soybean gene regulating Na+/K+ ratio in roots enhances salt tolerance in transgenic plants. Plant Growth Regul. 2014, 73, 299–308. [Google Scholar] [CrossRef]
- Chen, H.; He, H.; Yu, D. Overexpression of a novel soybean gene modulating Na+ and K+ transport enhances salt tolerance in transgenic tobacco plants. Physiol. Plant 2011, 141, 11–18. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Q.; Shao, Y.; Zhang, B.; Feng, A.; Meng, F.; Li, W. GmLEA2–1, a late embryogenesis abundant protein gene isolated from soybean (Glycine Max (L.) Merr.), confers tolerance to abiotic stress. Acta Biol. Hung. 2018, 69, 270–282. [Google Scholar] [CrossRef]
- Liao, H.; Wong, F.; Phang, T.; Cheung, M.; Li, W.; Shao, G.; Yan, X.; Lam, H. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 2003, 318, 103–111. [Google Scholar] [CrossRef]
- Li, W.; Shao, G.; Lam, H. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 2008, 178, 80–91. [Google Scholar] [CrossRef]
- Golldack, D.; Luking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Khan, S.; Li, M.; Wang, S.; Yin, H. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.; Wong, F.; Yung, W.; Ku, Y.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Long, S.; Zhao, C. Maintenance of cell wall integrity under high salinity. Int. J. Mol. Sci. 2021, 22, 3260. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Hafiz, M.; Muhammda, I.; Faheem, S.; Jeong, D.; Seung, H.; Soo, I.; Gyuhwa, C. Systems identification and characterization of cell wall reassembly and degradation related geges in Glycine max (L.) Merill, a bioenergy legume. Sci. Rep. 2017, 7, 10862. [Google Scholar]
- Jin, T.; Shan, Z.; Zhou, S.; Yang, Q.; Gai, J.; Li, Y. GmDNAJC7 from soybean is involved in plant tolerance to alkaline-salt, salt, and drought stresses. Agronomy 2022, 12, 1419. [Google Scholar] [CrossRef]
- Goodstein, D.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11, Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2, New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2022, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.; Mitra, A.; Baumgarten, A.; Young, N.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gen families in Arabidopsis thaliana . BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Resil. Stab. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Hruba, P.; Honys, D.; Twell, D.; Capkova, V.; Tupy, J. Expression of beta-galactosidase and beta-xylosidase genes during microspore and pollen development. Planta 2005, 220, 931–940. [Google Scholar] [CrossRef]
- Xu, C.; Shan, J.; Liu, T.; Wang, Q.; Ji, Y.; Zhang, Y.; Wang, M.; Xia, N.; Zhao, L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M.; et al. Functional Evolution of Phosphatidylethanolamine Binding Proteins in Soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef]
- Wendel, J. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef]
- Kondrashov, F.; Rogozin, I.; Wolf, Y.; Koonin, E. Selection in the evolution of gene duplications. Genome Biol. 2002, 3, RESEARCH0008. [Google Scholar] [CrossRef]
- Conant, G.; Wolfe, K. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Hou, Z.; Li, H.; Li, Z.; Fang, C.; Kong, L.; Li, Y.; Du, H.; Li, T.; Wang, L.; et al. Agronomical selection on loss-of-function of GIGANTEA simultaneously facilitates soybean salt tolerance and early maturity. J. Integr. Plant Biol. 2022, 64, 1866–1882. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Zhuang, Y.; Sun, W.; Cao, H.; Xu, R.; Kong, F.; Zhang, D. A novel miR160a-GmARF16-GmMYC2 module determines soybean salt tolerance and adaptation. New Phytol. 2024, 241, 2176–2192. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.; Gasteiger, E.; Bairoch, A.; Sanchez, J.; Williams, K.; Appel, R.; Hochstrasser, D. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–532. [Google Scholar]
- Pei, X.; Wang, F.; Du, H.; He, M.; Li, L.; Gou, C.; Chen, Z.; Wang, Y.; Kong, F.; Zhao, L. Genome-wide identification and functional prediction of BYPASS1-related (BPS1) homologs in soybean. Mol. Breed. 2023, 43, 59. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Bailey, T.; Boden, M.; Buske, F.; Frith, M.; Grant, C.; Clementi, L.; Ren, J.; Li, W.; Noble, W. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Kelley, L.; Mezulis, S.; Yates, C.; Wass, M.; Sternberg, M. The Phyre2 web portal for protein modeling, prediction and analysis. Nature 2015, 10, 845–858. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Lai, B.; Hu, W.; You, M.; Wang, L.; Su, T. Genome-Wide Identification of the BXL Gene Family in Soybean and Expression Analysis Under Salt Stress. Int. J. Mol. Sci. 2025, 26, 9534. https://doi.org/10.3390/ijms26199534
Wen Y, Lai B, Hu W, You M, Wang L, Su T. Genome-Wide Identification of the BXL Gene Family in Soybean and Expression Analysis Under Salt Stress. International Journal of Molecular Sciences. 2025; 26(19):9534. https://doi.org/10.3390/ijms26199534
Chicago/Turabian StyleWen, Yimin, Biwei Lai, Weijie Hu, Mengyang You, Lingshuang Wang, and Tong Su. 2025. "Genome-Wide Identification of the BXL Gene Family in Soybean and Expression Analysis Under Salt Stress" International Journal of Molecular Sciences 26, no. 19: 9534. https://doi.org/10.3390/ijms26199534
APA StyleWen, Y., Lai, B., Hu, W., You, M., Wang, L., & Su, T. (2025). Genome-Wide Identification of the BXL Gene Family in Soybean and Expression Analysis Under Salt Stress. International Journal of Molecular Sciences, 26(19), 9534. https://doi.org/10.3390/ijms26199534




