Screening for GmRCD1-Interacting Proteins in Glycine Max and Characterization of the GmRCD1-GmNAC058 Interaction
Abstract
1. Introduction
2. Results
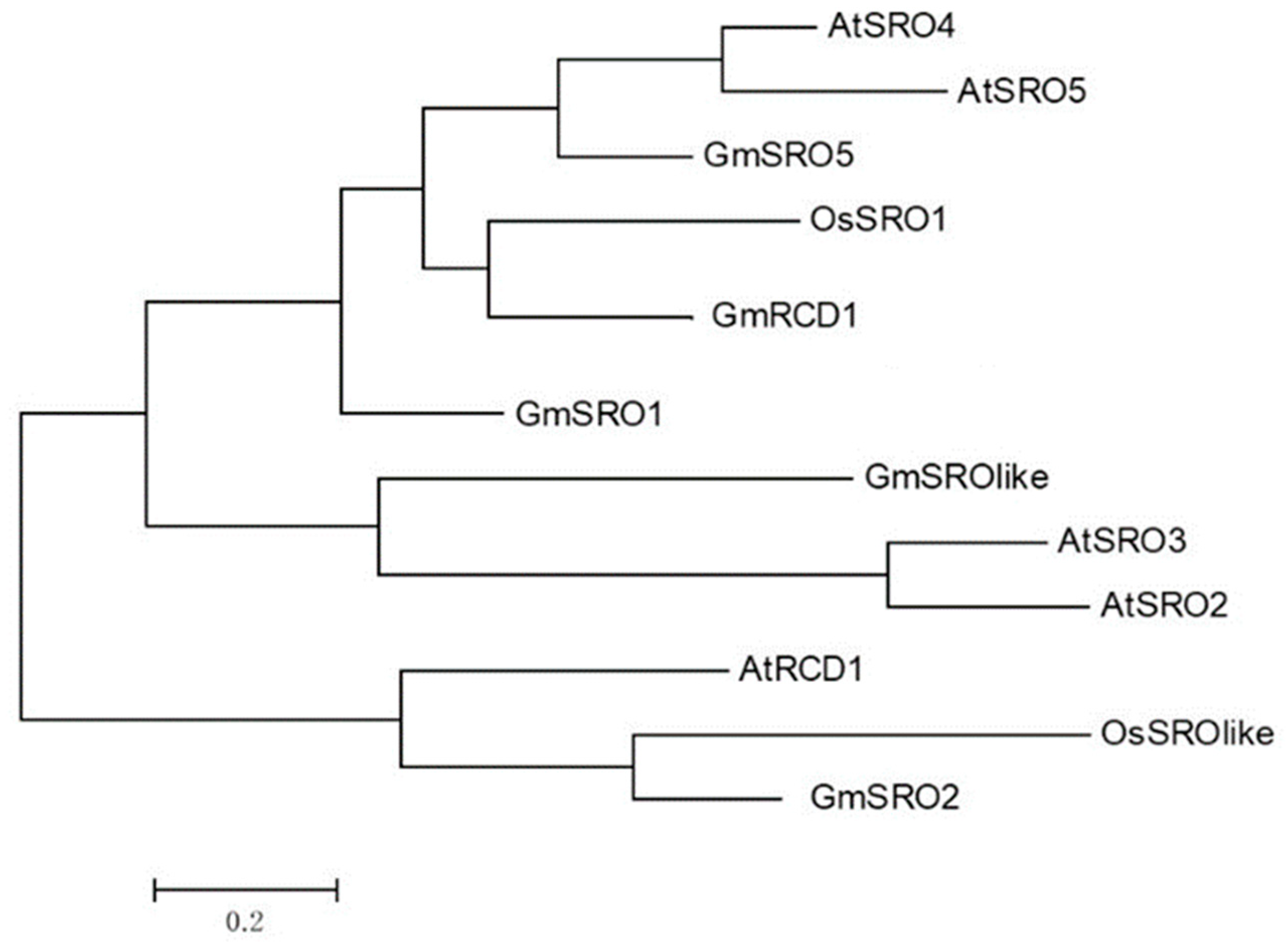
2.1. Genome-Wide Identification and Bioinformatics Analysis of the Soybean SRO Family
2.2. Screening of GmRCD1-Interacting Proteins
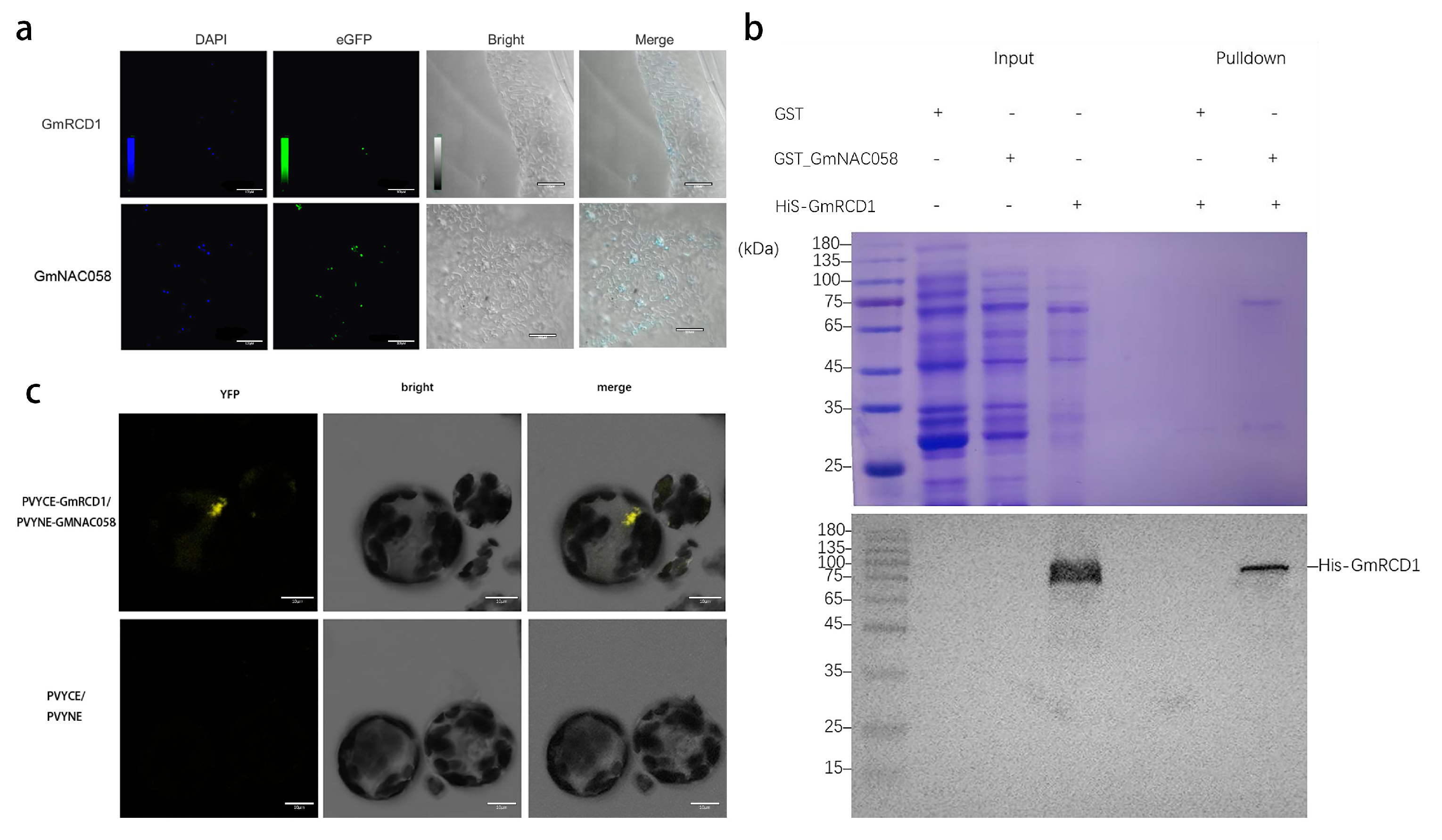
2.3. Verification of Interaction Between GmRCD1 and GmNAC058
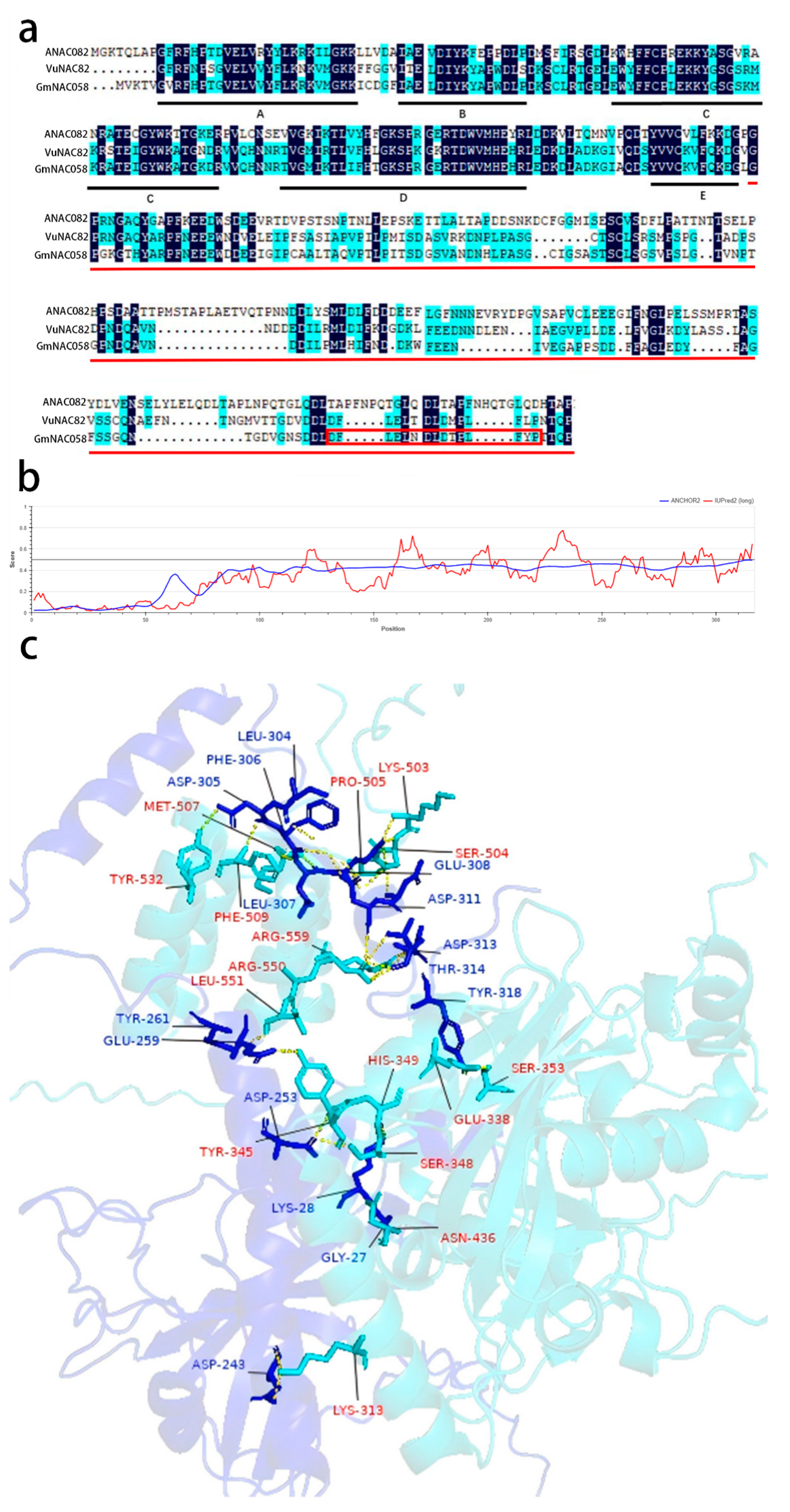
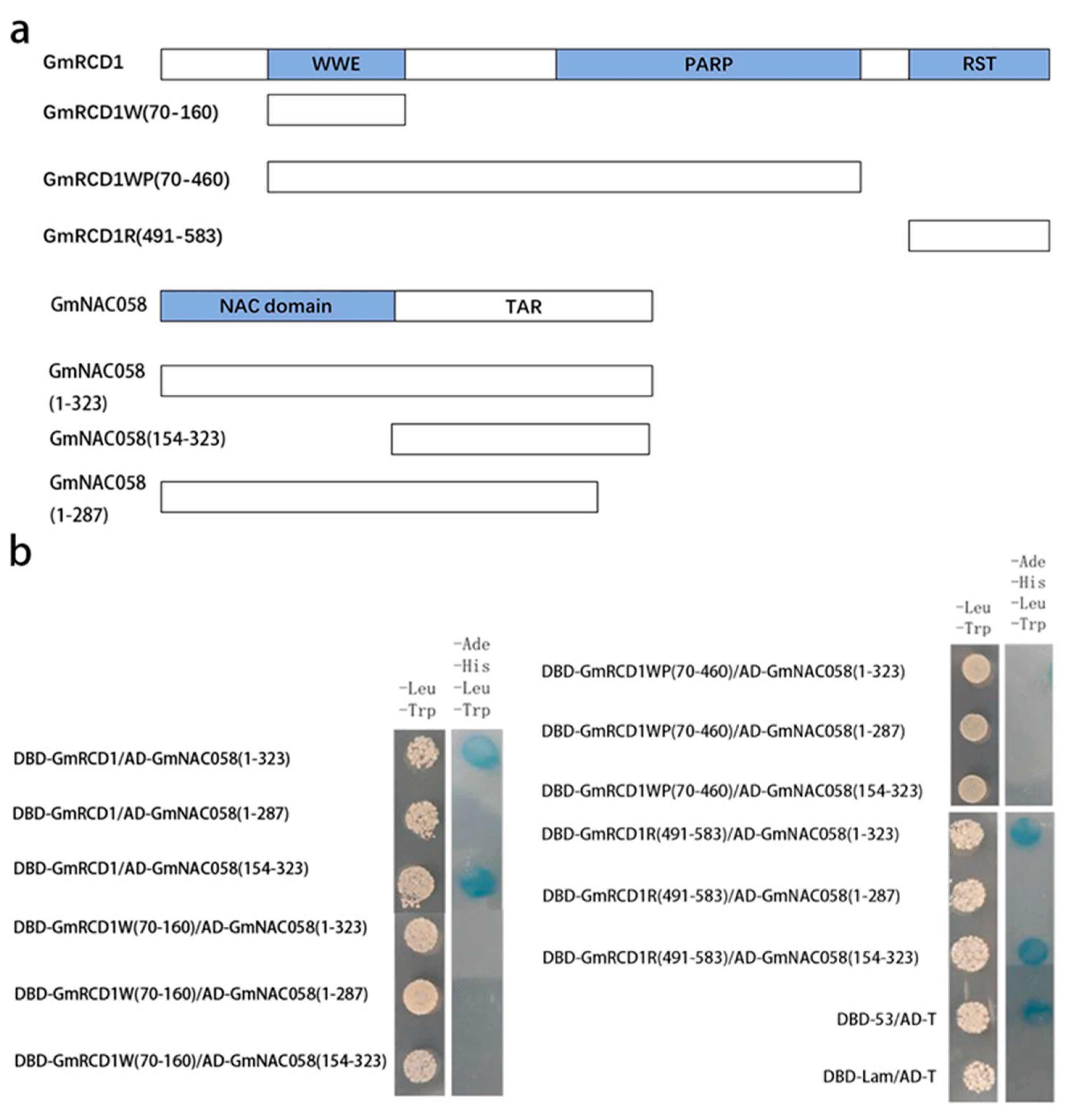
2.4. Identification of Interaction Domains Between GmRCD1 and GmNAC058
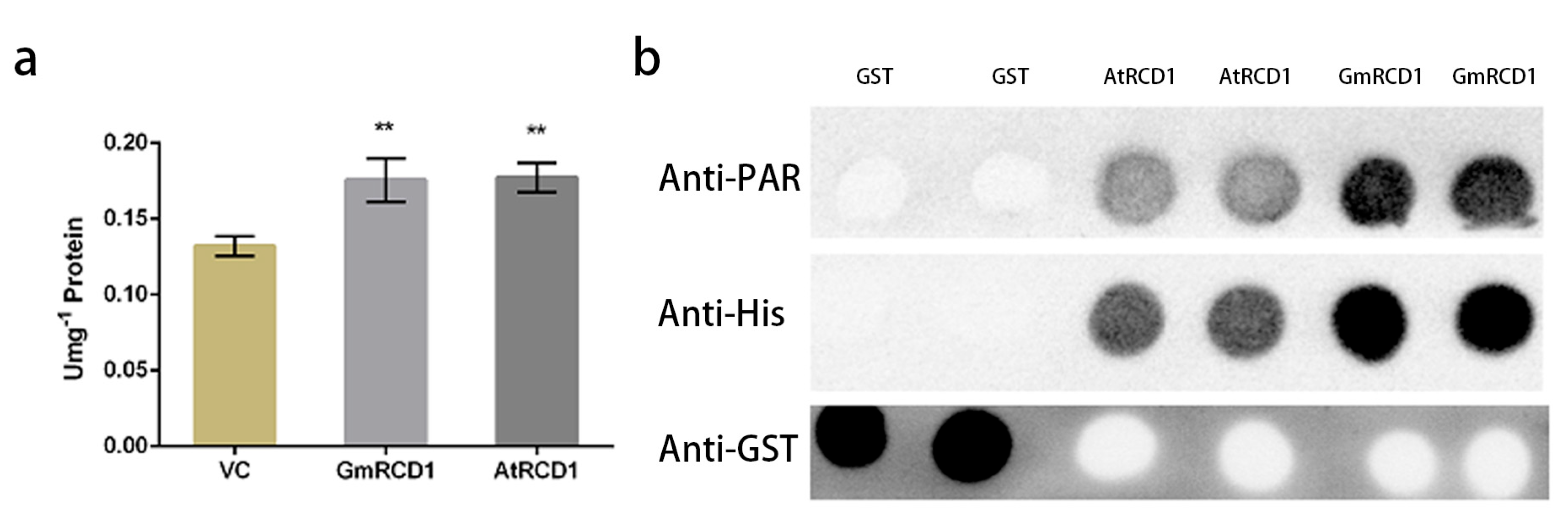
2.5. PARP Activity Assay of GmRCD1
2.6. Assessment of PAR Binding Capacity in RCD1 Protein
3. Discussion
3.1. Functional Investigation of GmRCD1-Interacting Proteins
3.2. Functional Characterization of Protein Domains
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Yeast Two-Hybrid Assay
4.3. Subcellular Localization Analysis
4.4. His-Tagged Proteins Induction and Purification
4.5. GST-Tagged Proteins Induction and Purification
4.6. GST Pull-Down
4.7. BiFC
4.8. PAR Binding Capacity Assay
4.9. PARP Activity Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SRO | Similar To RCD One |
| RCD1 | Radical Cell Death 1 |
| NAC | Nascent Polypeptide-associated Complex |
| MDS | Mitochondrial Dysfunction Stimulon |
| PARP | Poly ADP-ribose Polymerase |
| BiFC | Bimolecular Fluorescence Complementation |
| SPL1 | Squamosa Promoter Binding Protein-like 1 |
| ROS | Reactive Oxygen Species |
| NPR1 | Nonexpresser Of PR Genes 1 |
Appendix A

| Target Gene | Primer Name | Primer Sequence |
|---|---|---|
| Glyma09g34000 | BDGmSRO1F BDGmSRO1R | TCTAGAATGGAAGCAAAAATCGCAAAGGC GGATCCGACTTCCTTCTTGATTGAGC |
| Glyma08g16630 | GmRY3F GmRY3R | GAATTCATGGTTAAAACTGTTGGAGTTC CTCGAGTCAAGGTTGTGTGGTCGGATA |
| Glyma01g36410 | GmRY4F GmRY4R | GGATCCATGGGAGGTGACGCCACTGCCG CTCGAGCTACTGTCCGGAGTTCCCCAAA |
| Glyma15g38830 | GmRY5NF GmRY5NR | GGATCCATGGGTAATAAACTTGGGAGG GAATTCCTACTGCCAAGAAGCACTCTG |
| Glyma08g08430 | GmRY6F GmRY6R | GGATCCATGGTGAAAATCTCACTTGGTGGA GTCGACTTAATGGTGTGACTTTATGCGAAT |
| Glyma05g36490 | GmRY7NF GmRY7NR | GAATTCATGAGTTCGTCTTCGGCAGATGTTTTTCC CTCGAGCTACTGAGATTCATCCATTGGGTCA |
| GmNAC82(154–323) | GmNAC82(154)F GmNAC82R | GATACATATGGAAGGTCTTGGTCCCGGG GATAGGATCCTCAAGGTTGTGTGGTCGGATA |
| GmNAC82(1–287) | GmNAC82F GmNAC82(287)R | GATACATATGGTTAAAACTGTTGGAGTTC GATAGGATCCAGCAAAGTAGTCTTCCAAGCCC |
| GmRCD1 | BDGmRCD1F BDGmRCD1R | TCTAGAATGGAAGCAAAAATCGCAAAGGC GGATCCGACTTCCTTCTTGATTGAGC |
| GmRCD1W(70–160) | BDGmRCD1WF BDGmRCD1WR | CCATGGGACGGTCCTTAGCTAGGCGT GGATCCTCAGGGGAAAAAACAGCACCCTGC |
| GmRCD1P(260–460) | BDGmRCD1PF BDGmRCD1PR | CCATGGATGCTTATACTGAATCAGT GGATCCTCAACTTCCACAAAAATGACCTT |
| GmRCD1R(491–583) | BDGmRCD1RF BDGmRCD1RR | CCATGGCTCCTAGTATGGTTGCTAGCAC GGATCCTCAGACTTCCTTCTTGATTGAGC |
| Target Gene | Primer Name | Primer Sequence |
|---|---|---|
| GmRCD1 | GmRCD1GF GmRCD1GR | GGATCCATGGAAGCAAAAATCGCAAAGGC CTCGAGTCAGTCTTCTTTCTTGATTGAGTCTT |
| GmNAC82 | GmNAC82-XbaI-F GmNAC82-BamHI-R | TCTAGAATGGTTAAAACTGTTGGAGTTC GGATCCAGGTTGTGTGGTCGGATA |
| Target Gene | Primer Name | Primer Sequence |
|---|---|---|
| GmRCD1 | GmRCD1GF GmRCD1HR | GGATCCATGGAAGCAAAAATCGCAAAGGC GAGCTCTCAGACTTCCTTCTTGATTGAGC |
| GmNAC82 | GSTGmNAC82F GSTGmNAC82R | GAATTCATGGTTAAAACTGTTGGAGTTC CTCGAGTCAAGGTTGTGTGGTCGGATA |
| Target Gene | Primer Name | Primer Sequence |
|---|---|---|
| GmRCD1 | GmRCD1PF GmRCD1PR | GGATCCATGGAAGCAAAAATCGCAAAGGC CTCGAGGACTTCCTTCTTGATTGAGC |
| GmNAC82 | GmNAC82PF GmNAC82PR | TCTAGAATGGTTAAAACTGTTGGAGTTC GGATCCAGGTTGTGTGGTCGGATA |
References
- Oyebamiji, Y.O.; Adigun, B.A.; Shamsudin, N.A.A.; Ikmal, A.M.; Salisu, M.A.; Malike, F.A.; Lateef, A.A. Recent Advancements in Mitigating Abiotic Stresses in Crops. Horticulturae 2024, 10, 156. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.S.; Dietz, K.J. The Significance of Amino Acids and Amino Acid-Derived Molecules in Plant Responses and Adaptation to Heavy Metal Stress. J. Exp. Bot. 2006, 57, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Vandereyken, K.; Van Leene, J.; De Coninck, B.; Cammue, B. Hub Protein Controversy: Taking a Closer Look at Plant Stress Response Hubs. Front. Plant Sci. 2018, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, Y.; Li, J.; Tang, R.; Liang, F.; Tang, R.; Zhou, Y.; Zhang, C. Genome-Wide Identification of SIMILAR TO RCD ONE (SRO) Gene Family in Rapeseed (Brassica napus L.) Reveals Their Role in Drought Stress Response. Plant Signal. Behav. 2024, 19, 2379128. [Google Scholar] [CrossRef]
- Jaspers, P.; Overmyer, K.; Wrzaczek, M.; Vainonen, J.P.; Blomster, T.; Salojarvi, J.; Reddy, R.A.; Kangasjarvi, J. The RST and PARP-Like Domain Containing SRO Protein Family: Analysis of Protein Structure, Function and Conservation in Land Plants. BMC Genom. 2010, 11, 170. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, L.; Ren, J.; Pan, F. Arabidopsis SIMILAR TO RCD-ONE Genes Are Ubiquitous and Respond to Multiple Abiotic Stresses Through Diverse Signaling Pathways. J. Biosci. 2019, 44, 129. [Google Scholar] [CrossRef]
- Teotia, S.; Lamb, R.S. The Paralogous Genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 Have Partially Redundant Functions During Arabidopsis Development. Plant Physiol. 2009, 151, 180–198. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Huang, H.; Liu, W.; Huang, Y.; Xu, J.; Huang, S.; Wang, W. Genome-Wide Characterization of a SRO Gene Family Involved in Response to Biotic and Abiotic Stresses in Banana (Musa spp.). BMC Plant Biol. 2019, 19, 211. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, X.; Liu, Z.; Wu, B. Comprehensive Analysis of StSRO Gene Family and Its Expression in Response to Different Abiotic Stresses in Potato. Int. J. Mol. Sci. 2022, 23, 13518. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi Blue, Y.; Kusumi, J.; Satake, A. Copy Number Analyses of DNA Repair Genes Reveal the Role of Poly (ADP-Ribose) Polymerase (PARP) in Tree Longevity. iScience 2021, 24, 102779. [Google Scholar] [CrossRef]
- Jiang, W.; Geng, Y.; Liu, Y.; Chen, S.; Cao, S.; Li, W.; Wang, J.; Wang, Q.; Zhang, X.; Yin, Y.; et al. Genome-Wide Identification and Characterization of SRO Gene Family in Wheat: Molecular Evolution and Expression Profiles During Different Stresses. Plant Physiol. Biochem. 2020, 154, 590–611. [Google Scholar] [CrossRef] [PubMed]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inze, D.; Kushnir, S. CEO1, a New Protein from Arabidopsis thaliana, Protects Yeast Against Oxidative Damage. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.; Tuominen, H.; Kettunen, R.; Betz, C.; Langebartels, C.; Sandermann, H.J.; Kangasjarvi, J. Ozone-Sensitive Arabidopsis rcd1 Mutant Reveals Opposite Roles for Ethylene and Jasmonate Signaling Pathways in Regulating Superoxide-Dependent Cell Death. Plant Cell 2000, 12, 1849–1862. [Google Scholar] [CrossRef]
- Jaspers, P.; Blomster, T.; Brosche, M.; Salojarvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F. Unequally Redundant RCD1 and SRO1 Mediate Stress and Developmental Responses and Interact with Transcription Factors in Arabidopsis. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Watahiki, M.K.; Yamamoto, K.T. Overexpression of the RADICAL-INDUCED CELL DEATH1 (RCD1) Gene of Arabidopsis Causes Weak rcd1 Phenotype with Compromised Oxidative-Stress Responses. Biosci. Biotechnol. Biochem. 2006, 70, 1827–1831. [Google Scholar] [CrossRef][Green Version]
- Katiyar-Agarwal, S.; Zhu, J.; Kim, K.; Agarwal, M.; Fu, X.; Huang, A.; Zhu, J. The Plasma Membrane Na+/H+ Antiporter SOS1 Interacts with RCD1 and Functions in Oxidative Stress Tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 18816–18821. [Google Scholar] [CrossRef]
- Vainonen, J.P.; Jaspers, P.; Wrzaczek, M.; Lamminmaki, A.; Reddy, R.A.; Vaahtera, L.; Brosche, M.; Kangasjarvi, J. RCD1-DREB2A Interaction in Leaf Senescence and Stress Responses in Arabidopsis thaliana. Biochem. J. 2012, 442, 573–581. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A Methyl Viologen-Resistant Mutant of Arabidopsis, Which Is Allelic to Ozone-Sensitive rcd1, Is Tolerant to Supplemental Ultraviolet-B Irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef]
- De Clercq, I.; Vermeirssen, V.; Van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inze, A.; Ng, S.; Ivanova, A.; Rombaut, D. The Membrane-Bound NAC Transcription Factor ANAC013 Functions in Mitochondrial Retrograde Regulation of the Oxidative Stress Response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A Membrane-Bound NAC Transcription Factor, ANAC017, Mediates Mitochondrial Retrograde Signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef]
- Tao, J.; Wu, F.; Wen, H.; Liu, X.; Luo, W.; Gao, L.; Jiang, Z.; Mo, B.; Chen, X.; Kong, W. RCD1 Promotes Salt Stress Tolerance in Arabidopsis by Repressing ANAC017 Activity. Int. J. Mol. Sci. 2023, 24, 9793. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Vainonen, J.P.; Hunter, K.; Tossavainen, H.; Tiwari, A.; Jarvi, S.; Hellman, M.; Aarabi, F.; Alseekh, S.; Wybouw, B. Arabidopsis RCD1 Coordinates Chloroplast and Mitochondrial Functions Through Interaction with ANAC Transcription Factors in Arabidopsis. eLife 2019, 8, e43284. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of an Arabidopsis Transcription Factor, DREB2A, Involved in Drought-Responsive Gene Expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Theisen, F.F.; Prestel, A.; Elkjaer, S.; Leurs, Y.H.A.; Morffy, N.; Strader, L.C.; O’Shea, C.; Teilum, K.; Kragelund, B.B.; Skriver, K. Molecular Switching in Transcription Through Splicing and Proline-Isomerization Regulates Stress Responses in Plants. Nat. Commun. 2024, 15, 592. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xi, M.; Liu, T.; Wu, X.; Ju, L.; Wang, D. The Central Role of Transcription Factors in Bridging Biotic and Abiotic Stress Responses for Plants’ Resilience. New Crops 2024, 1, 100005. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.; et al. Transcriptomic Reprogramming in Soybean Seedlings Under Salt Stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Hu, J.; Zhuang, Y.; Li, X.; Jing, X.; Zhang, R.; Zhang, L.; Wang, Y.; Liu, Y.; Huang, J.; Wang, P.; et al. Time-Series Transcriptome Comparison Reveals the Gene Regulation Network Under Salt Stress in Soybean (Glycine max) Roots. BMC Plant Biol. 2022, 22, 157. [Google Scholar] [CrossRef]
- Melo, B.P.; Fraga, O.T.; Silva, J.C.F.; Ferreira, D.O.; Brustolini, O.J.B.; Carpinetti, P.A.; Machado, J.P.B.; Reis, P.A.B.; Fontes, E.P.B. Revisiting the Soybean GmNAC Superfamily. Front. Plant Sci. 2018, 9, 1864. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean During Development and Dehydration Stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Sipari, N.; Lihavainen, J.; Keinanen, M. Metabolite Profiling of Paraquat Tolerant Arabidopsis thaliana Radical-Induced Cell Death1 (rcd1)—A Mediator of Antioxidant Defence Mechanisms. Antioxidants 2022, 11, 2034. [Google Scholar] [CrossRef]
- Brosche, M.; Blomster, T.; Salojarvi, J.; Cui, F.; Sipari, N.; Leppala, J.; Lamminmaki, A.; Tomai, G.; Narayanasamy, S.; Reddy, R.A.; et al. Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 2014, 10, e1004112. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Petrov, V.; Yun, D.J.; Gechev, T. Revisiting Plant Salt Tolerance: Novel Components of the SOS Pathway. Trends Plant Sci. 2023, 28, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, X.; Zhao, H.; Wang, Y.; Wei, X.; Wang, P.; Zhan, J.; Liu, L.; Li, F.; Ge, X. GhRCD1 Regulates Cotton (Gossypium hirsutum) Somatic Embryogenesis by Modulating the GhMYC3-GhMYB44-GhLBD18 Transcriptional Cascade. New Phytol. 2023, 240, 207–223. [Google Scholar] [CrossRef]
- Yi, S.Y.; Min, S.R.; Kwon, S.Y. NPR1 Is Instrumental in Priming for the Enhanced flg22-Induced MPK3 and MPK6 Activation. Plant Pathol. J. 2015, 31, 192–194. [Google Scholar] [CrossRef]
- Moon, J.; Parry, G.; Estelle, M. The Ubiquitin-Proteasome Pathway and Plant Development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef]
- Lu, X.; Nowicka, U.; Sridharan, V.; Liu, F.; Randles, L.; Hymel, D.; Dyba, M.; Tarasov, S.G.; Tarasova, N.I.; Zhao, X.Z.; et al. Structure of the Rpn13-Rpn2 Complex Provides Insights for Rpn13 and Uch37 as Anticancer Targets. Nat. Commun. 2017, 8, 15540. [Google Scholar] [CrossRef]
- Luo, G.; Gu, H.; Liu, J.; Qu, L.J. Four Closely-Related RING-Type E3 Ligases, APD1-4, Are Involved in Pollen Mitosis II Regulation in Arabidopsis. J. Integr. Plant Biol. 2012, 54, 814–827. [Google Scholar] [CrossRef]
- Cho, S.K.; Ryu, M.Y.; Kim, J.H.; Hong, J.S.; Oh, T.R.; Kim, W.T.; Yang, S.W. RING E3 Ligases: Key Regulatory Elements Are Involved in Abiotic Stress Responses in Plants. BMB Rep. 2017, 50, 393–400. [Google Scholar] [CrossRef]
- Du, Q.L.; Cui, W.Z.; Zhang, C.H.; Yu, D.Y. GmRFP1 Encodes a Previously Unknown RING-Type E3 Ubiquitin Ligase in Soybean (Glycine max). Mol. Biol. Rep. 2010, 37, 685–693. [Google Scholar] [CrossRef]
- Wang, J.; Qin, H.; Zhou, S.; Wei, P.; Zhang, H.; Zhou, Y.; Miao, Y.; Huang, R. The Ubiquitin-Binding Protein OsDSK2a Mediates Seedling Growth and Salt Responses by Regulating Gibberellin Metabolism in Rice. Plant Cell 2020, 32, 414–428. [Google Scholar] [CrossRef]
- Chao, L.; Liu, Y.; Chen, D.; Xue, X.; Mao, Y.; Chen, X. Arabidopsis Transcription Factors SPL1 and SPL12 Confer Plant Thermotolerance at Reproductive Stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.; Geng, Z.; Zhang, Y.; Xue, W.; Ma, L.; Yang, J.; Jin, Y.; Wang, S.; Zhuo, S.; Zhang, Y. Genome-Wide Characterization of NmrA-Like Proteins and the Regulatory Function of Soybean GmNmrA6 in Response to Salt and Oxidative Stresses. Environ. Exp. Bot. 2023, 213, 105447. [Google Scholar] [CrossRef]
- Sipari, N.; Lihavainen, J.; Shapiguzov, A.; Kangasjarvi, J.; Keinanen, M. Primary Metabolite Responses to Oxidative Stress in Early-Senescing and Paraquat Resistant Arabidopsis thaliana rcd1 (Radical-Induced Cell Death1). Front. Plant Sci. 2020, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, Generalist in Plant Life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. The SNAC1-Targeted Gene OsSRO1c Modulates Stomatal Closure and Oxidative Stress Tolerance by Regulating Hydrogen Peroxide in Rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef]
- Ohbayashi, I.; Lin, C.Y.; Shinohara, N.; Matsumura, Y.; Machida, Y.; Horiguchi, G.; Tsukaya, H.; Sugiyama, M. Evidence for a Role of ANAC082 as a Ribosomal Stress Response Mediator Leading to Growth Defects and Developmental Alterations in Arabidopsis. Plant Cell 2017, 29, 2644–2660. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Kragelund, B.B. The Molecular Basis for Cellular Function of Intrinsically Disordered Protein Regions. Nat. Rev. Mol. Cell Biol. 2024, 25, 187–211. [Google Scholar] [CrossRef] [PubMed]
- Kjaersgaard, T.; Jensen, M.K.; Christiansen, M.W.; Gregersen, P.; Skriver, K. Senescence-Associated BarleyNAC (NAM, ATAF1,2, CUC) Transcription Factor Interacts with Radical-Induced Cell Death 1 Through a Disordered Regulatory Domain. J. Biol. Chem. 2011, 286, 35418–35429. [Google Scholar] [CrossRef]
- Tossavainen, H.; Hellman, M.; Vainonen, J.P.; Kangasjarvi, J.; Permi, P. 1H, 13C and 15N NMR Chemical Shift Assignments of A. thaliana RCD1 RST. Biomol. NMR Assign. 2017, 11, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Bugge, K.; Staby, L.; Salladini, E.; Falbe-Hansen, R.G.; Kragelund, B.B.; Skriver, K. αα-Hub Domains and Intrinsically Disordered Proteins: A Decisive Combo. J. Biol. Chem. 2021, 296, 100226. [Google Scholar] [CrossRef] [PubMed]
- DaRosa, P.A.; Wang, Z.; Jiang, X.; Pruneda, J.N.; Cong, F.; Klevit, R.E.; Xu, W. Allosteric Activation of the RNF146 Ubiquitin Ligase by a Poly(ADP-Ribosyl)ation Signal. Nature 2015, 517, 223–226. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, H.; Wang, Q.; Si, C.; Yang, Y.; Yu, Y.; Zhou, L.; Ding, L.; Song, A.; Xu, D.; et al. CmRCD1 Represses Flowering by Directly Interacting with CmBBX8 in Summer Chrysanthemum. Hortic. Res. 2021, 8, 79. [Google Scholar] [CrossRef]
- Menke, F.L. Plants Get on PAR with Poly(ADP-Ribosyl)ation. EMBO Rep. 2016, 17, 1677–1678. [Google Scholar] [CrossRef]
- Liu, S.; Liu, S.; Wang, M.; Wei, T.; Meng, C.; Wang, M.; Xia, G. A Wheat SIMILAR TO RCD-ONE Gene Enhances Seedling Growth and Abiotic Stress Resistance by Modulating Redox Homeostasis and Maintaining Genomic Integrity. Plant Cell 2014, 26, 164–180. [Google Scholar] [CrossRef]
- Vogt, S.; Feijs, K.; Hosch, S.; De Masi, R.; Lintermann, R.; Loll, B.; Wirthmueller, L. The Superior Salinity Tolerance of Bread Wheat Cultivar Suzhou No. 3 Is Unlikely to Be Caused by Elevated Ta-sro1 Poly-(ADP-Ribose) Polymerase Activity. Plant Cell 2022, 34, 4130–4137. [Google Scholar] [CrossRef]
- Vainonen, J.P.; Gossens, R.; Krasensky-Wrzaczek, J.; De Masi, R.; Danciu, I.; Puukko, T.; Battchikova, N.; Jonak, C.; Wirthmueller, L.; Wrzaczek, M.; et al. Poly(ADP-ribose)-binding Protein RCD1 Is a Plant PARylation Reader Regulated by Photoregulatory Protein Kinases. Commun. Biol. 2023, 6, 395. [Google Scholar] [CrossRef]
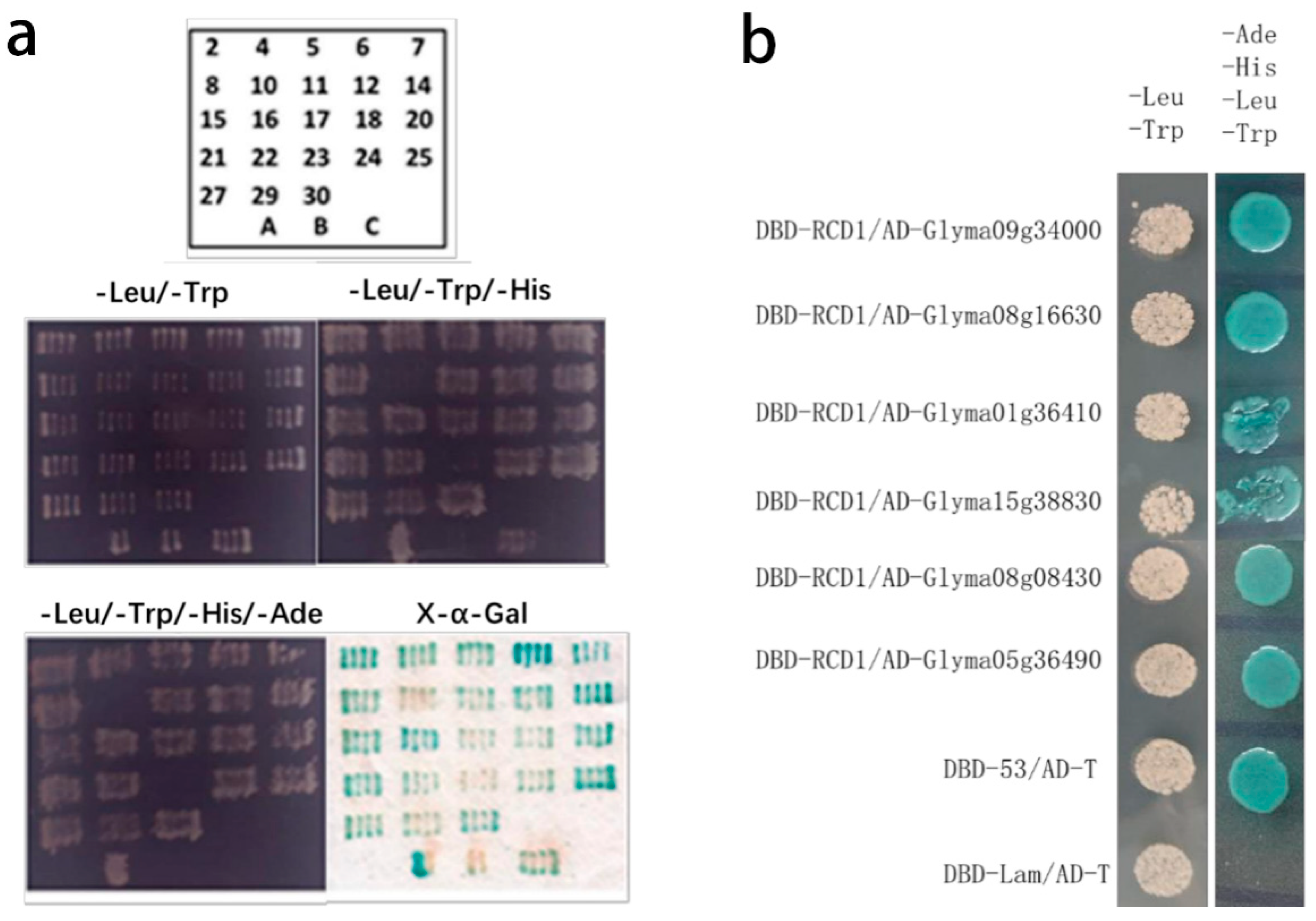
| No. | Gene ID | Classification |
|---|---|---|
| 11 | Glyma09g34000 | GmSRO1 |
| 8 | Glyma04g39140 | NAC transcription factor |
| 25 | Glyma08g16630 | NAC transcription factor |
| 7,14,21 | Glyma01g36410 | ubiquitin–proteasome system protein |
| 20 | Glyma15g39830 | ubiquitin–proteasome system protein |
| 22 | Glyma08g08430 | ubiquitin–proteasome system protein |
| 27 | Glyma05g36490 | ubiquitin–proteasome system protein |
| 8,17,24 | Glyma17g04190 | ribosomal protein |
| 5 | Glyma02g01160 | SPL1 transcription factor |
| 30 | Glyma14g24710 | zinc finger-containing protein |
| 29 | Glyma04g01380 | NmrA domain-containing protein |
| 12 | Glyma11g20030 | heavy metal transporter protein |
| 16 | Glyma01g44680 | protein NETWORKED 1A-like |
| 18 | Glyma11g00910 | protein NETWORKED 1A-like |
| 15 | Glyma18g52221 | hypothetical protein |
| 2 | Glyma07g33050 | hypothetical protein |
| 4 | Glyma16g11160 | hypothetical protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Bu, Y.; Liu, Y.; Liu, G. Screening for GmRCD1-Interacting Proteins in Glycine Max and Characterization of the GmRCD1-GmNAC058 Interaction. Int. J. Mol. Sci. 2025, 26, 7760. https://doi.org/10.3390/ijms26167760
Li Y, Bu Y, Liu Y, Liu G. Screening for GmRCD1-Interacting Proteins in Glycine Max and Characterization of the GmRCD1-GmNAC058 Interaction. International Journal of Molecular Sciences. 2025; 26(16):7760. https://doi.org/10.3390/ijms26167760
Chicago/Turabian StyleLi, Yupeng, Youda Bu, Yun Liu, and Guobao Liu. 2025. "Screening for GmRCD1-Interacting Proteins in Glycine Max and Characterization of the GmRCD1-GmNAC058 Interaction" International Journal of Molecular Sciences 26, no. 16: 7760. https://doi.org/10.3390/ijms26167760
APA StyleLi, Y., Bu, Y., Liu, Y., & Liu, G. (2025). Screening for GmRCD1-Interacting Proteins in Glycine Max and Characterization of the GmRCD1-GmNAC058 Interaction. International Journal of Molecular Sciences, 26(16), 7760. https://doi.org/10.3390/ijms26167760






