Genome-Wide Identification and Functional Characterization of the LbaLHCB Gene Family Reveals Tissue-Specific Expression and Salt Stress Response in Lycium barbarum
Abstract
1. Introduction
2. Results
2.1. Systematic Evolution Analysis of LbaLHCB Proteins in Wolfberry
2.2. Analysis of Physicochemical Properties of LbaLHCB Proteins in Wolfberry
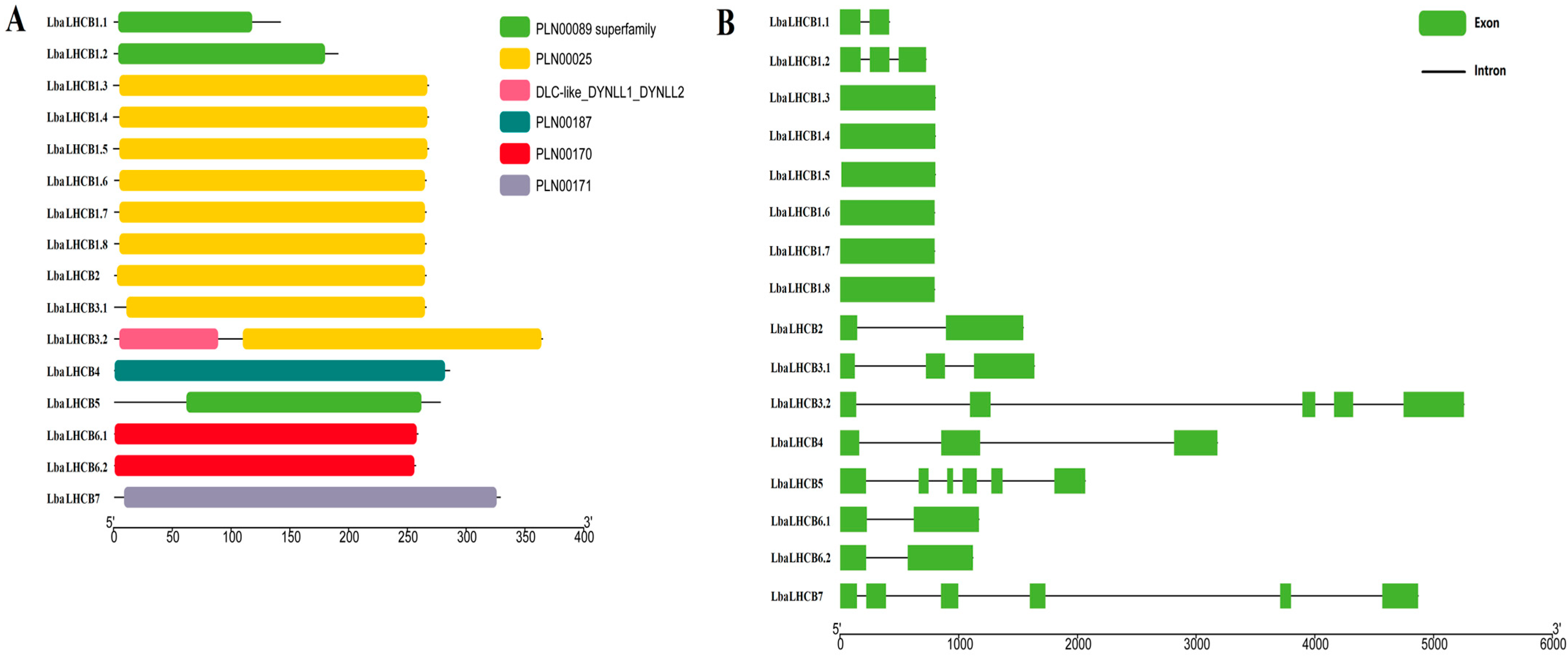
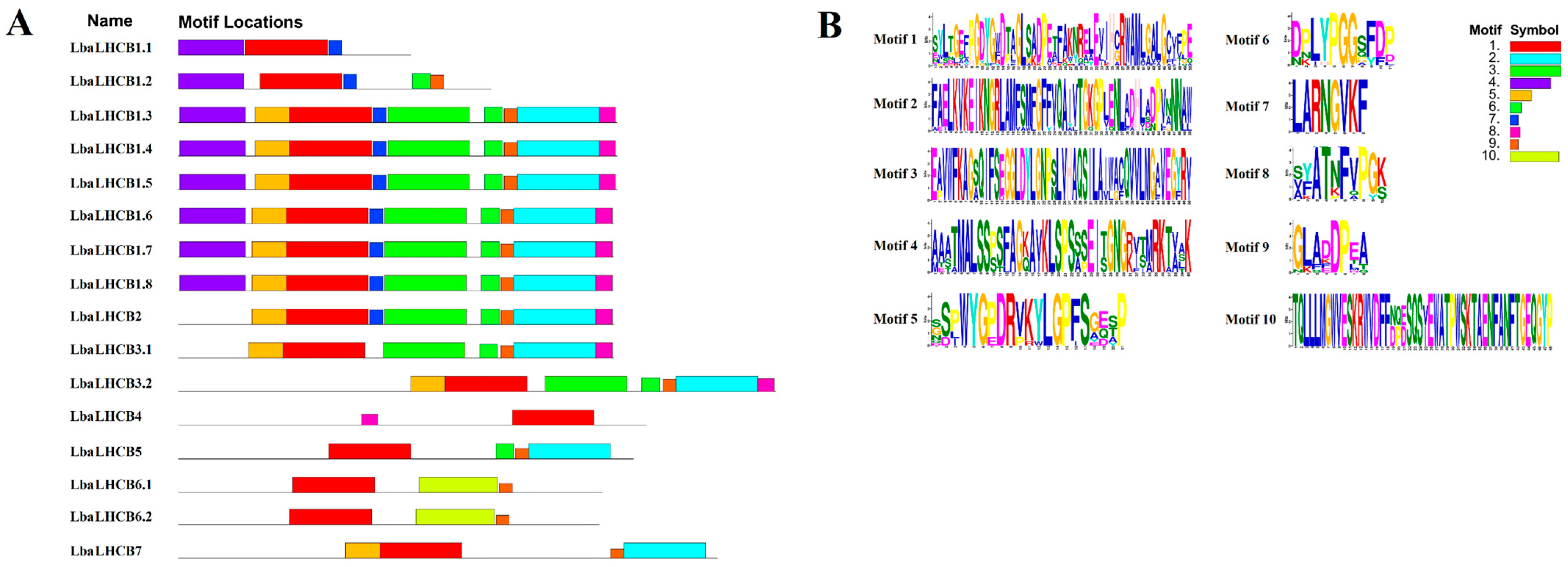
2.3. Analysis of LbaLHCB Gene Structure and Conserved Motifs in Wolfberry
2.4. Chromosomal Localization and Synteny Analysis of LbaLHCB Genes in Wolfberry
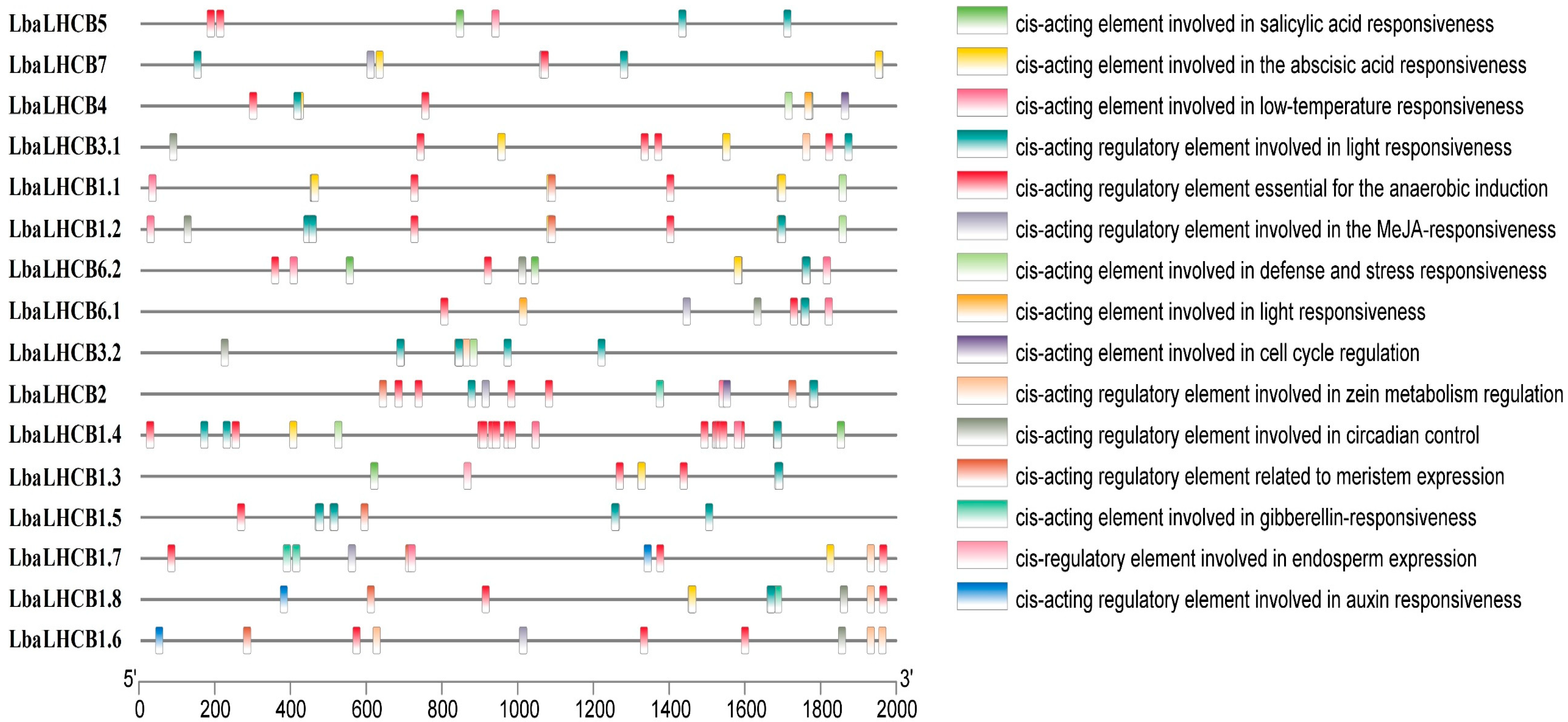
2.5. Analysis of Cis-Acting Elements in the Promoter Regions of LbaLHCB Genes in Wolfberry
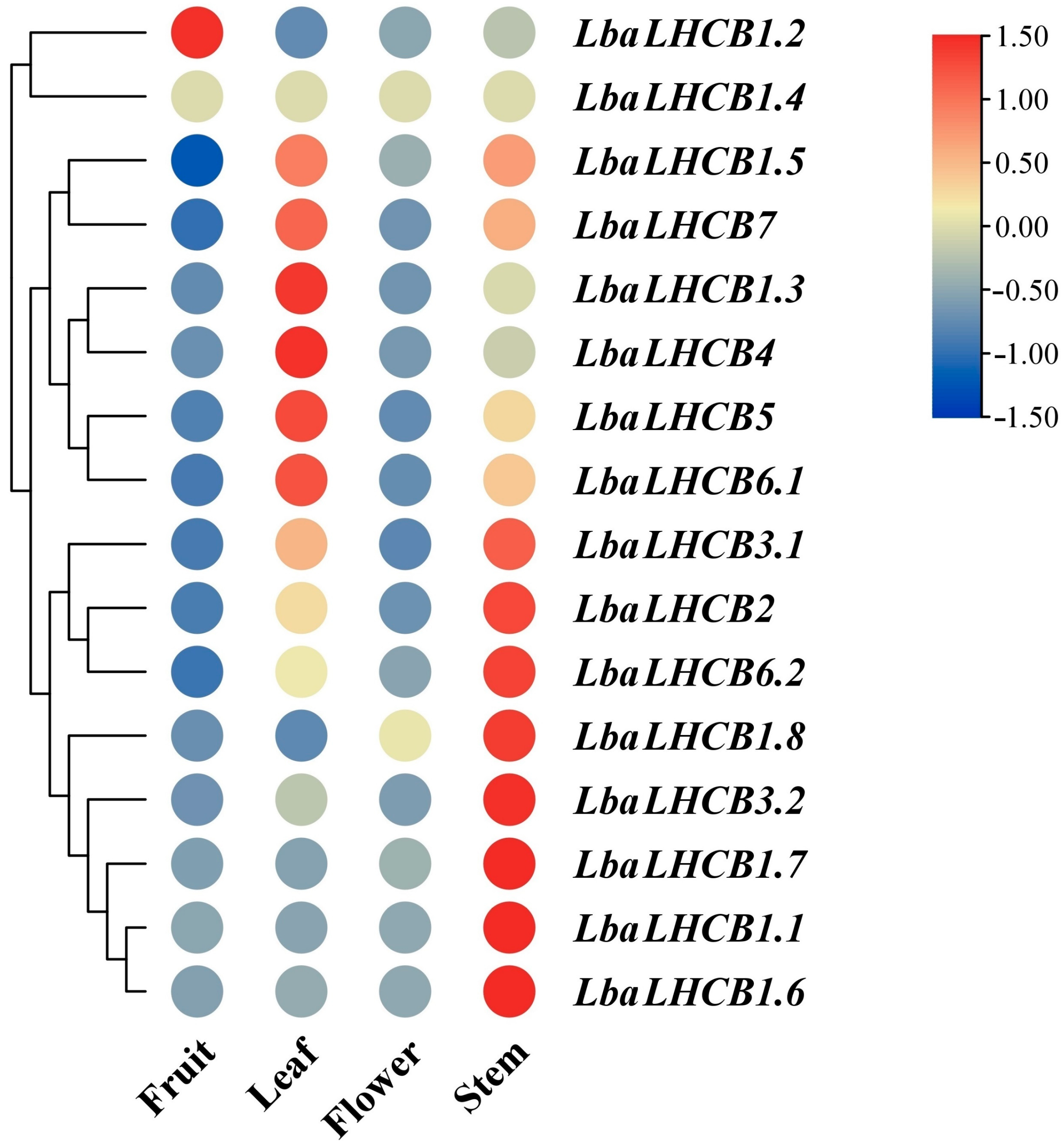
2.6. Heatmap Analysis of the Expression Patterns of LbaLHCBs in Different Tissues of Wolfberry
2.7. Heatmap Analysis of the Expression Patterns of LbaLHCBs Under Salt Stress of Wolfberry
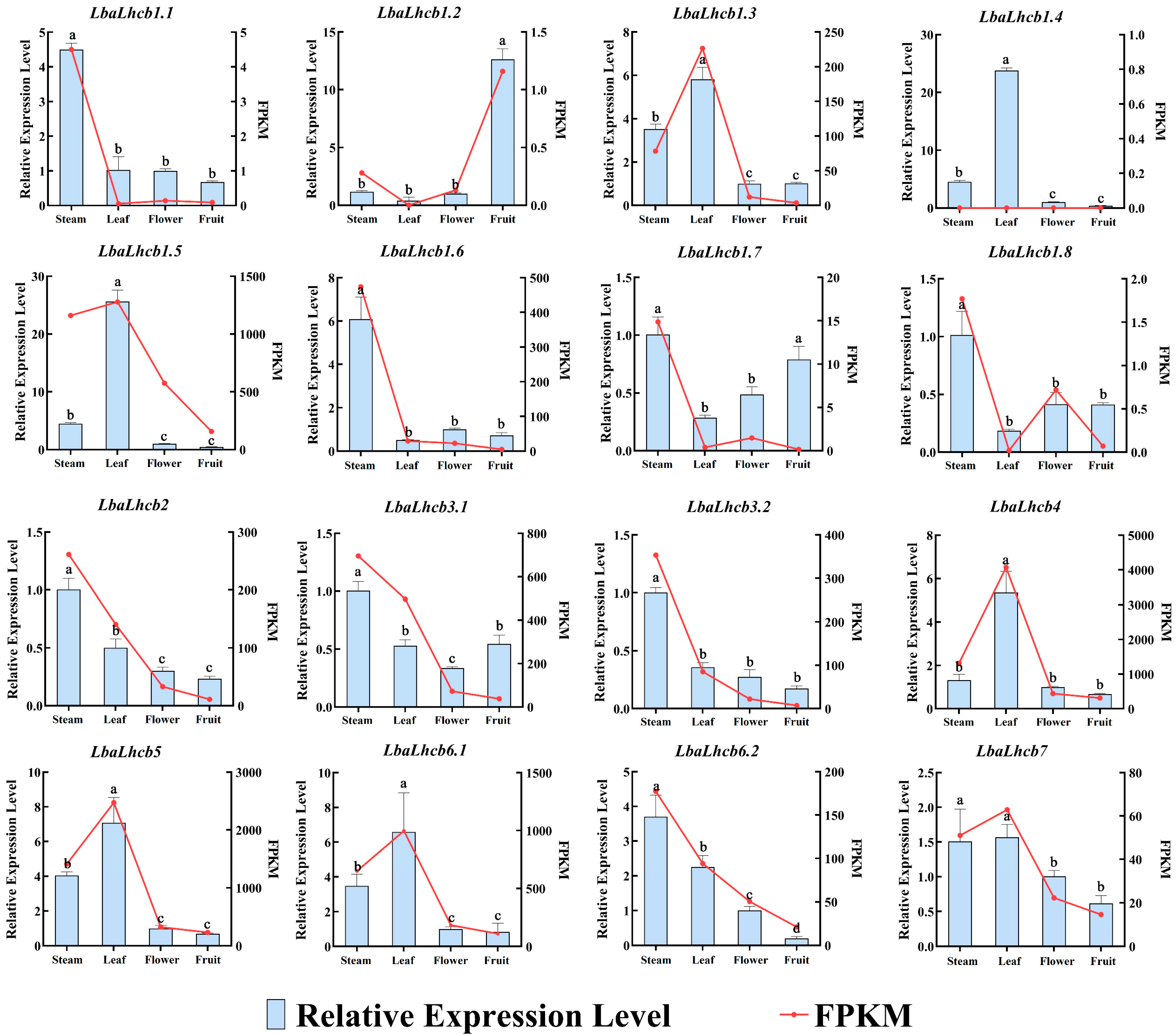
2.8. Expression Characteristics of LbaLHCB Genes in Different Tissues
2.9. qRT-PCR Validation of LbaLHCB Genes in Leaves Under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment Methods
4.2. Identification of LHCB Genes in the L. barbarum Genome
4.3. Phylogenetic Analysis of LbaLHCB Proteins
4.4. Gene Structure and Conserved Motif Analysis of the LbaLHCB Gene Family
4.5. Chromosomal Localization and Synteny Analysis of LbaLHCB Gene Family Members
4.6. Cis-Acting Element Analysis of LbaLHCB Genes
4.7. Expression Profiling of LbaLHCB Genes Based on RNA-Seq Data
4.8. RNA Extraction and RT-qPCR Analysis
4.9. Data Processing and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey-Serres, J.; Pierik, R.; Ruban, A.; Wingler, A. The Dynamic Plant: Capture, Transformation, and Management of Energy. Plant Physiol. 2018, 176, 961–966. [Google Scholar] [CrossRef]
- Pinnola, A.; Bassi, R. Molecular mechanisms involved in plant photoprotection. Biochem. Soc. Trans. 2018, 46, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, D.; van Amerongen, H.; Wientjes, E. Single chloroplast in folio imaging sheds light on photosystem energy redistribution during state transitions. Plant Physiol. 2023, 191, 1186–1198. [Google Scholar] [CrossRef]
- Kılıç, M.; Käpylä, V.; Gollan, P.J.; Aro, E.M.; Rintamäki, E. PSI Photoinhibition and Changing CO2 Levels Initiate Retrograde Signals to Modify Nuclear Gene Expression. Antioxidants 2023, 12, 1902. [Google Scholar] [CrossRef]
- Barber, J. Photosystem II: The water splitting enzyme of photosynthesis and the origin of oxygen in our atmosphere. Q. Rev. Biophys. 2016, 49, e14, Correction in Q. Rev. Biophys. 2016, 49, e16. [Google Scholar] [CrossRef]
- Shen, J.R. The Structure of Photosystem II and the Mechanism of Water Oxidation in Photosynthesis. Annu. Rev. Plant Biol. 2015, 66, 23–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Melø, T.B.; Li, H.; Naqvi, K.R.; Yang, C.H. The inter-monomer interface of the major light-harvesting chlorophyll a/b complexes of photosystem II (LHCII) influences the chlorophyll triplet distribution. J. Plant Physiol. 2014, 171, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, X.; Sang, M.; Chen, D.; Wang, K.; Lin, R.; Lu, C.; Shen, J.R.; Kuang, T. Spectral and functional studies on siphonoxanthin-type light-harvesting complex of photosystem II from Bryopsis corticulans. Photosynth. Res. 2013, 117, 267–279. [Google Scholar] [CrossRef]
- Lan, Y.H.; Song, Y.; Zhao, F.; Cao, Y.; Luo, D.L.; Qiao, D.R.; Cao, Y.; Xu, H. Phylogenetic, Structural and Functional Evolution of the LHC Gene Family in Plant Species. Int. J. Mol. Sci. 2022, 24, 488. [Google Scholar] [CrossRef]
- Neilson, J.A.D.; Rangsrikitphoti, P.; Durnford, D.G. Evolution and regulation of Bigelowiella natans light-harvesting antenna system. J. Plant Physiol. 2017, 217, 68–76. [Google Scholar] [CrossRef]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef] [PubMed]
- Ilíková, I.; Ilík, P.; Opatíková, M.; Arshad, R.; Nosek, L.; Karlický, V.; Kučerová, Z.; Roudnický, P.; Pospíšil, P.; Lazár, D.; et al. Towards spruce-type photosystem II: Consequences of the loss of light-harvesting proteins LHCB3 and LHCB6 in Arabidopsis. Plant Physiol. 2021, 187, 2691–2715. [Google Scholar] [CrossRef] [PubMed]
- Longoni, P.; Douchi, D.; Cariti, F.; Fucile, G.; Goldschmidt-Clermont, M. Phosphorylation of the Light-Harvesting Complex II Isoform Lhcb2 Is Central to State Transitions. Plant Physiol. 2015, 169, 2874–2883. [Google Scholar] [CrossRef] [PubMed]
- Sattari Vayghan, H.; Nawrocki, W.J.; Schiphorst, C.; Tolleter, D.; Hu, C.; Douet, V.; Glauser, G.; Finazzi, G.; Croce, R.; Wientjes, E.; et al. Photosynthetic Light Harvesting and Thylakoid Organization in a CRISPR/Cas9 Arabidopsis thaliana LHCB1 Knockout Mutant. Front. Plant Sci. 2022, 13, 833032. [Google Scholar] [CrossRef]
- Peterson, R.B.; Schultes, N.P. Light-harvesting complex B7 shifts the irradiance response of photosynthetic light-harvesting regulation in leaves of Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 311–318. [Google Scholar] [CrossRef]
- Chen, Y.E.; Ma, J.; Wu, N.; Su, Y.Q.; Zhang, Z.W.; Yuan, M.; Zhang, H.Y.; Zeng, X.Y.; Yuan, S. The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 2018, 130, 267–276. [Google Scholar] [CrossRef]
- Guardini, Z.; Gomez, R.L.; Caferri, R.; Dall’Osto, L.; Bassi, R. Loss of a single chlorophyll in CP29 triggers re-organization of the Photosystem II supramolecular assembly. Biochim. Biophys. Acta (BBA)-Bioenerg. 2022, 1863, 148555. [Google Scholar] [CrossRef]
- Bianchetti, R.; Bellora, N.; de Haro, L.A.; Zuccarelli, R.; Rosado, D.; Freschi, L.; Rossi, M.; Bermudez, L. Phytochrome-Mediated Light Perception Affects Fruit Development and Ripening Through Epigenetic Mechanisms. Front. Plant Sci. 2022, 13, 870974. [Google Scholar] [CrossRef]
- Huang, T.; Liu, H.; Tao, J.P.; Zhang, J.Q.; Zhao, T.M.; Hou, X.L.; Xiong, A.S.; You, X. Low light intensity elongates period and defers peak time of photosynthesis: A computational approach to circadian-clock-controlled photosynthesis in tomato. Hortic. Res. 2023, 10, uhad077. [Google Scholar] [CrossRef]
- Hu, Z.H.; Zhang, N.; Qin, Z.Y.; Li, J.W.; Tao, J.P.; Yang, N.; Chen, Y.; Kong, J.Y.; Luo, W.; Chen, X.; et al. Circadian rhythm response and its effect on photosynthetic characteristics of the Lhcb family genes in tea plant. BMC Plant Biol. 2024, 24, 333. [Google Scholar] [CrossRef]
- Sun, W.L.; Shahrajabian, M.H.; Cheng, Q. Health benefits of wolfberry (Gou Qi Zi, Fructus barbarum L.) on the basis of ancient Chineseherbalism and Western modern medicine. Avicenna J. Phytomed. 2021, 11, 109–119. [Google Scholar]
- Ma, Y.P.; Xie, Y.; Ha, R.; Cao, B.; Song, L.H. Effects of Elevated CO2 on Photosynthetic Accumulation, Sucrose Metabolism-Related Enzymes, and Genes Identification in Goji Berry (Lycium barbarum L.). Front. Plant Sci. 2021, 12, 643555. [Google Scholar] [CrossRef]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010, 76, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wan, R.; Shi, Z.G.; Ma, W.L.; Wang, H.; Chen, Y.W.; Bo, J.H.; Li, Y.X.; An, W.; Qin, K.; et al. Transcriptomics and Metabolomics Reveal the Critical Genes of Carotenoid Biosynthesis and Color Formation of Goji (Lycium barbarum L.) Fruit Ripening. Plants 2023, 12, 2791. [Google Scholar] [CrossRef]
- Liu, X.X.; Fan, W.Q.; Jiao, H.H.; Gao, H.; Tang, J.N.; Zhu, J.Z.; Yue, S.J.; Zheng, R. Comparative analysis of differentially expressed genes for biosynthesis of active ingredients in fruits of different cultivars of Lycium barbarum L. based on transcriptome sequencing. Sheng Wu Gong Cheng Xue Bao 2023, 39, 3015–3036. [Google Scholar] [CrossRef]
- Fatchurrahman, D.; Amodio, M.; De Chiara, M.; Mastrandrea, L.; Colelli, G. Characterization and postharvest behavior of goji berry (Lycium barbarum L.) during ripening. Postharvest Biol. Technol. 2022, 191, 111975. [Google Scholar] [CrossRef]
- Ma, R.X.; Sun, X.Z.; Yang, C.; Fan, Y.L. Integrated transcriptome and metabolome provide insight into flavonoid variation in goji berries (Lycium barbarum L.) from different areas in China. Plant Physiol. Biochem. 2023, 199, 107722. [Google Scholar] [CrossRef]
- Islam, T.; Yu, X.; Badwal, T.S.; Xu, B. Comparative studies on phenolic profiles, antioxidant capacities and carotenoid contents of red goji berry (Lycium barbarum) and black goji berry (Lycium ruthenicum). Chem. Cent. J. 2017, 11, 59. [Google Scholar] [CrossRef]
- Liang, Y.F.; Feng, D.P.; Sun, Z.J.; Ye, P.; Liang, S.F.; Shi, T.Y. Effects of water-fertiliser coupling on the photosynthesis and quality of Lycium barbarum based on predicted crop evapotranspiration (ETc). Sci. Rep. 2024, 14, 31405. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Qi, G.P.; Ma, Y.L.; Yin, M.H.; Wang, J.H.; Kang, Y.X.; Jia, Q.; Gao, Y.L.; Tian, R.R.; Zhang, R.; et al. Effects of Water and Nitrogen Control on the Growth Physiology, Yields, and Economic Benefits of Lycium barbarum Plants in a Lycium barbarum + Alfalfa System. Plants 2024, 13, 1095. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, H.; Ono, T.; Minagawa, J. Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 2001, 42, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, C.; Wang, X.; Ma, Q.; Fan, S.; Zhang, C. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2.3 on chlorophyll a synthesis. Plant Biol. 2021, 23, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.Y.; Kim, D.J.; Baek, J.M.; Choi, H.K.; Ellis, L.C.; Küester, H.; McCombie, W.R.; Peng, H.M.; Cook, D.R. Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol. 2003, 131, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X.W. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef]
- Rodgers-Melnick, E.; Mane, S.P.; Dharmawardhana, P.; Slavov, G.T.; Crasta, O.R.; Strauss, S.H.; Brunner, A.M.; Difazio, S.P. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012, 22, 95–105. [Google Scholar] [CrossRef]
- Klimmek, F.; Sjödin, A.; Noutsos, C.; Leister, D.; Jansson, S. Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 2006, 140, 793–804. [Google Scholar] [CrossRef]
- Wei, Y.C.; Lu, X.; Bao, J.Y.; Zhang, C.C.; Yan, H.K.; Li, K.; Gong, M.S.; Li, S.; Ma, S.Y. Identification and expression analysis of chlorophyll a/b binding protein gene family in grape (Vitis vinifera). Physiol. Mol. Biol. Plants 2022, 28, 1147–1158. [Google Scholar] [CrossRef]
- Chory, J.; Wu, D. Weaving the complex web of signal transduction. Plant Physiol. 2001, 125, 77–80. [Google Scholar] [CrossRef]
- Jiao, Y.L.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Xu, Z.S.; Zhuang, J. De novo assembly and transcriptome characterization: Novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol. 2014, 14, 277. [Google Scholar] [CrossRef]
- Wang, Y.H.; Liu, P.Z.; Liu, H.; Zhang, R.R.; Liang, Y.; Xu, Z.S.; Li, X.J.; Luo, Q.; Tan, G.F.; Wang, G.L.; et al. Telomere-to-telomere carrot (Daucus carota) genome assembly reveals carotenoid characteristics. Hortic. Res. 2023, 10, uhad103. [Google Scholar] [CrossRef]
- Li, Y.P.; Su, L.Y.; Huang, T.; Liu, H.; Tan, S.S.; Deng, Y.J.; Wang, Y.H.; Xiong, A.S. The telomere-to-telomere genome of Pucai (Typha angustifolia L.): A distinctive semiaquatic vegetable with lignin and chlorophyll as quality characteristics. Hortic. Res. 2025, 12, uhaf079. [Google Scholar] [CrossRef]
- Zhao, J.H.; Xu, Y.H.; Li, H.X.; Zhu, X.L.; Yin, Y.; Zhang, X.Y.; Qin, X.Y.; Zhou, J.; Duan, L.Y.; Liang, X.J.; et al. ERF5.1 modulates carotenoid accumulation by interacting with CCD4.1 in Lycium. Hortic. Res. 2023, 10, uhad230. [Google Scholar] [CrossRef]
- Zhao, Q.C.; Jing, W.K.; Fu, X.J.; Yang, R.Y.; Zhu, C.Y.; Zhao, J.X.; Choisy, P.; Xu, T.; Ma, N.; Zhao, L.J.; et al. TSPO-induced degradation of the ethylene receptor RhETR3 promotes salt tolerance in rose (Rosa hybrida). Hortic. Res. 2024, 11, uhae040. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.R.; Yang, W.J.; Jing, W.K.; Shahid, M.O.; Liu, Y.M.; Qiu, X.H.; Choisy, P.; Xu, T.; Ma, N.; Gao, J.P.; et al. Multi-omics analysis reveals key regulatory defense pathways and genes involved in salt tolerance of rose plants. Hortic. Res. 2024, 11, uhae068. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, X.; Tao, H.; Wang, P. Growth performance and physiological response in the halophyte Lycium barbarum grown at salt-affected soil. Ann. Appl. Biol. 2006, 149, 263–269. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; He, K.N.; Zhang, T.; Tang, D.; Li, R.J.; Jia, S.F. Physiological responses of Goji berry (Lycium barbarum L.) to saline-alkaline soil from Qinghai region, China. Sci. Rep. 2019, 9, 12057. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Hu, Z.H.; Huang, T.; Zhang, N.; Chen, C.; Yang, K.X.; Sun, M.Z.; Yang, N.; Chen, Y.; Tao, J.P.; Liu, H.; et al. Interference of skeleton photoperiod in circadian clock and photosynthetic efficiency of tea plant: In-depth analysis of mathematical model. Hortic. Res. 2024, 11, uhae226. [Google Scholar] [CrossRef]
- Jiang, Q.; Xu, Z.S.; Wang, F.; Li, M.Y.; Ma, J.; Xiong, A.S. Effects of abiotic stresses on the expression of Lhcb1 gene and photosynthesis of Oenanthe javanica and Apium graveolens. Biol Plant. 2014, 58, 256–264. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Cao, X.Y.; Ma, Y.N.; Qin, X.Y.; Bai, X.R.; Zhang, X.Y.; Xiong, A.S.; Yin, Y.; Zheng, R. Genome-Wide Identification of bZIP Gene Family in Lycium barbarum and Expression During Fruit Development. Int. J. Mol. Sci. 2025, 26, 4665. [Google Scholar] [CrossRef]
- Huang, S.D.; Kang, Z.; Xu, Z.L. Robust deep k-means: An effective and simple method for data clustering. Pattern Recognit. 2021, 117, 107996. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2021, 29, e45. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Gene Accession | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| LbaLHCB1.1 | Lba04g02519 | 141 | 15,204.67 | 9.39 | 22.49 | 92.84 | 0.15 | Chloroplast |
| LbaLHCB1.2 | Lba04g02521 | 190 | 20,133.58 | 8.59 | 20.06 | 100.68 | 0.287 | Chloroplast |
| LbaLHCB1.3 | Lba11g02117 | 267 | 28,363.39 | 5.47 | 26.45 | 80.04 | 0.023 | Chloroplast |
| LbaLHCB1.4 | Lba11g02063 | 267 | 28,183.19 | 5.46 | 23.64 | 80.45 | 0.042 | Chloroplast |
| LbaLHCB1.5 | Lba11g02575 | 267 | 28,357.31 | 5.47 | 23.13 | 77.15 | 0.015 | Chloroplast |
| LbaLHCB1.6 | Lba12g01281 | 265 | 28,041.94 | 5.14 | 28.11 | 80.34 | 0.045 | Chloroplast |
| LbaLHCB1.7 | Lba12g01279 | 265 | 28,071.96 | 5.15 | 29.15 | 79.25 | 0.027 | Chloroplast |
| LbaLHCB1.8 | Lba12g01278 | 265 | 28,061.92 | 5.15 | 28.5 | 79.25 | 0.03 | Chloroplast |
| LbaLHCB2 | Lba11g01177 | 265 | 28,742.73 | 5.48 | 27.24 | 78.08 | −0.046 | Chloroplast |
| LbaLHCB3.1 | Lba04g02330 | 265 | 28,630.76 | 5.1 | 16.62 | 85.4 | 0.049 | Chloroplast |
| LbaLHCB3.2 | Lba11g00344 | 364 | 39,390.05 | 4.82 | 16.48 | 85.25 | 0.092 | Chloroplast |
| LbaLHCB4 | Lba03g00842 | 285 | 31,131.64 | 5.61 | 33.44 | 88.77 | −0.057 | Chloroplast |
| LbaLHCB5 | Lba01g01890 | 277 | 29,726.22 | 5.71 | 39.34 | 88.52 | −0.011 | Chloroplast |
| LbaLHCB6.1 | Lba06g02886 | 258 | 27,378.43 | 6.15 | 24.63 | 86.78 | 0.097 | Chloroplast |
| LbaLHCB6.2 | Lba06g02885 | 256 | 27,265.4 | 6.16 | 23.1 | 88.2 | 0.145 | Chloroplast |
| LbaLHCB7 | Lba02g02883 | 328 | 36,347.12 | 7.74 | 39.31 | 103.2 | 0.078 | Chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.-H.; Yin, Y.; Wang, L.-X.; Zhang, N.; Wang, Y.-H.; Zhuang, J.; Xiong, A.-S. Genome-Wide Identification and Functional Characterization of the LbaLHCB Gene Family Reveals Tissue-Specific Expression and Salt Stress Response in Lycium barbarum. Int. J. Mol. Sci. 2025, 26, 9523. https://doi.org/10.3390/ijms26199523
Hu Z-H, Yin Y, Wang L-X, Zhang N, Wang Y-H, Zhuang J, Xiong A-S. Genome-Wide Identification and Functional Characterization of the LbaLHCB Gene Family Reveals Tissue-Specific Expression and Salt Stress Response in Lycium barbarum. International Journal of Molecular Sciences. 2025; 26(19):9523. https://doi.org/10.3390/ijms26199523
Chicago/Turabian StyleHu, Zhi-Hang, Yue Yin, Li-Xiang Wang, Nan Zhang, Ya-Hui Wang, Jing Zhuang, and Ai-Sheng Xiong. 2025. "Genome-Wide Identification and Functional Characterization of the LbaLHCB Gene Family Reveals Tissue-Specific Expression and Salt Stress Response in Lycium barbarum" International Journal of Molecular Sciences 26, no. 19: 9523. https://doi.org/10.3390/ijms26199523
APA StyleHu, Z.-H., Yin, Y., Wang, L.-X., Zhang, N., Wang, Y.-H., Zhuang, J., & Xiong, A.-S. (2025). Genome-Wide Identification and Functional Characterization of the LbaLHCB Gene Family Reveals Tissue-Specific Expression and Salt Stress Response in Lycium barbarum. International Journal of Molecular Sciences, 26(19), 9523. https://doi.org/10.3390/ijms26199523








