Chromosomal and Plasmid-Based CRISPRi Platforms for Conditional Gene Silencing in Lactococcus lactis
Abstract
1. Introduction
2. Results
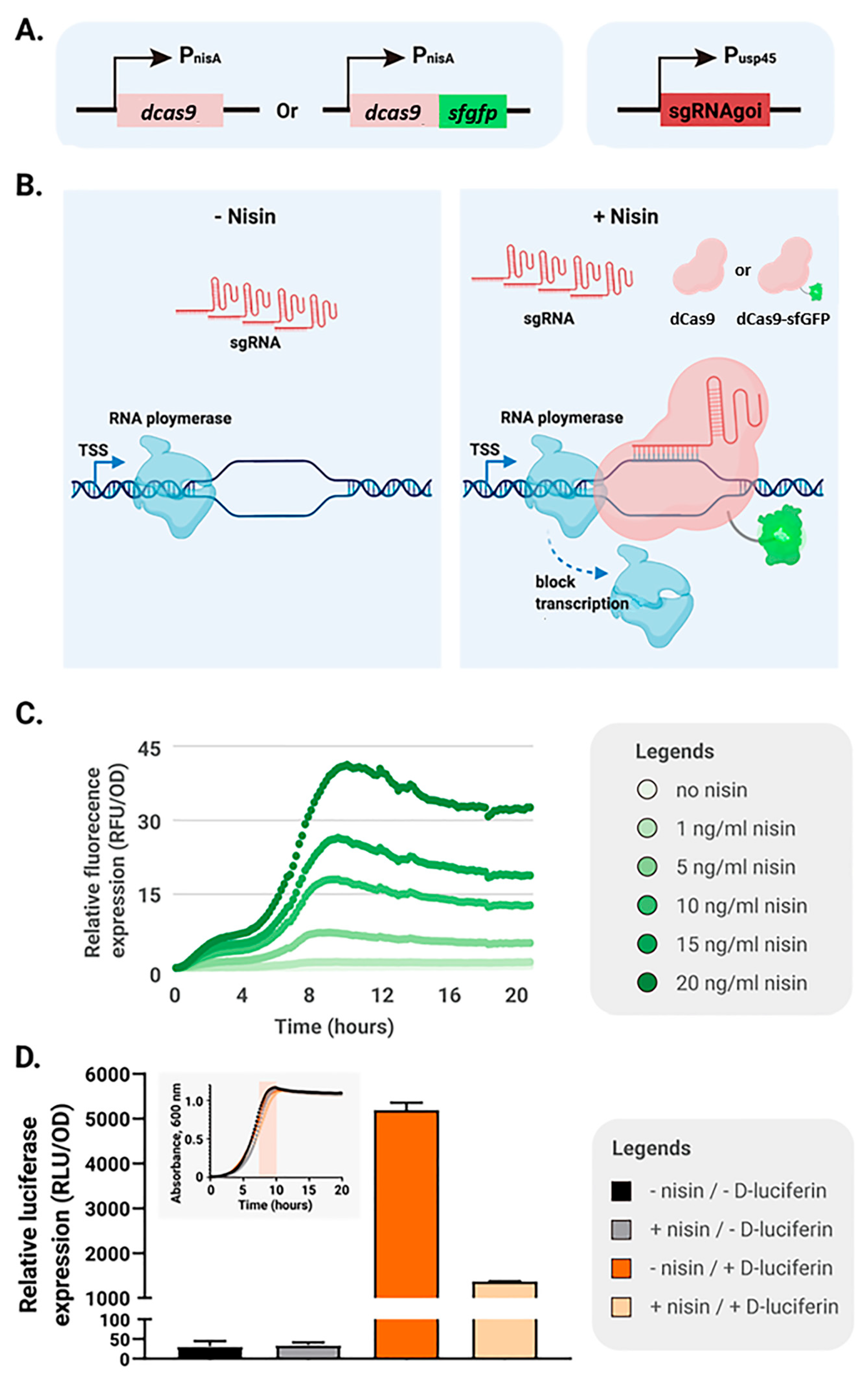
2.1. Establishment of an Inducible CRISPRi System with Fluorescently Tagged dCas9 in L. lactis
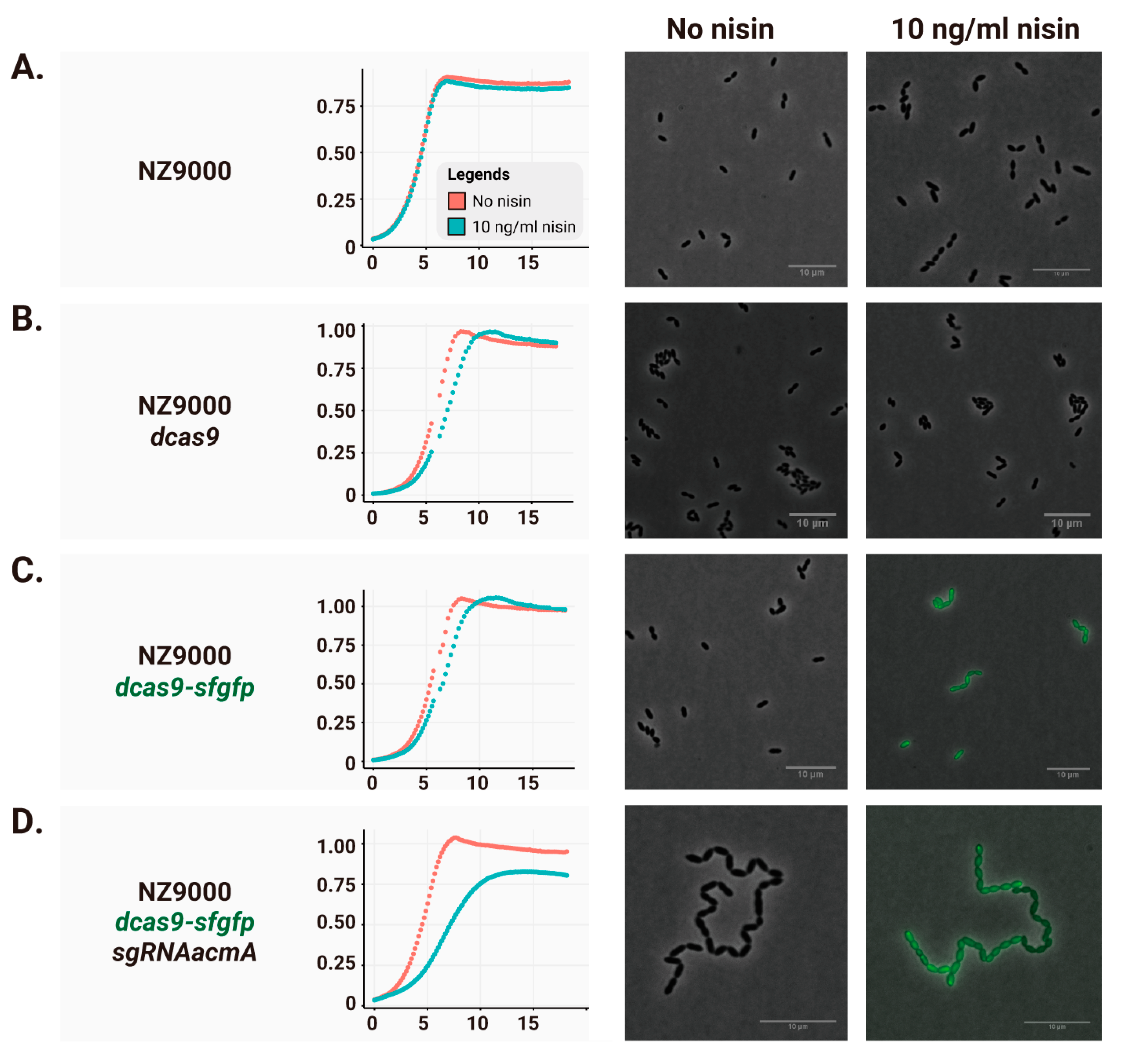
2.2. Leaky dCas9 Expression Causes Basal Gene Repression
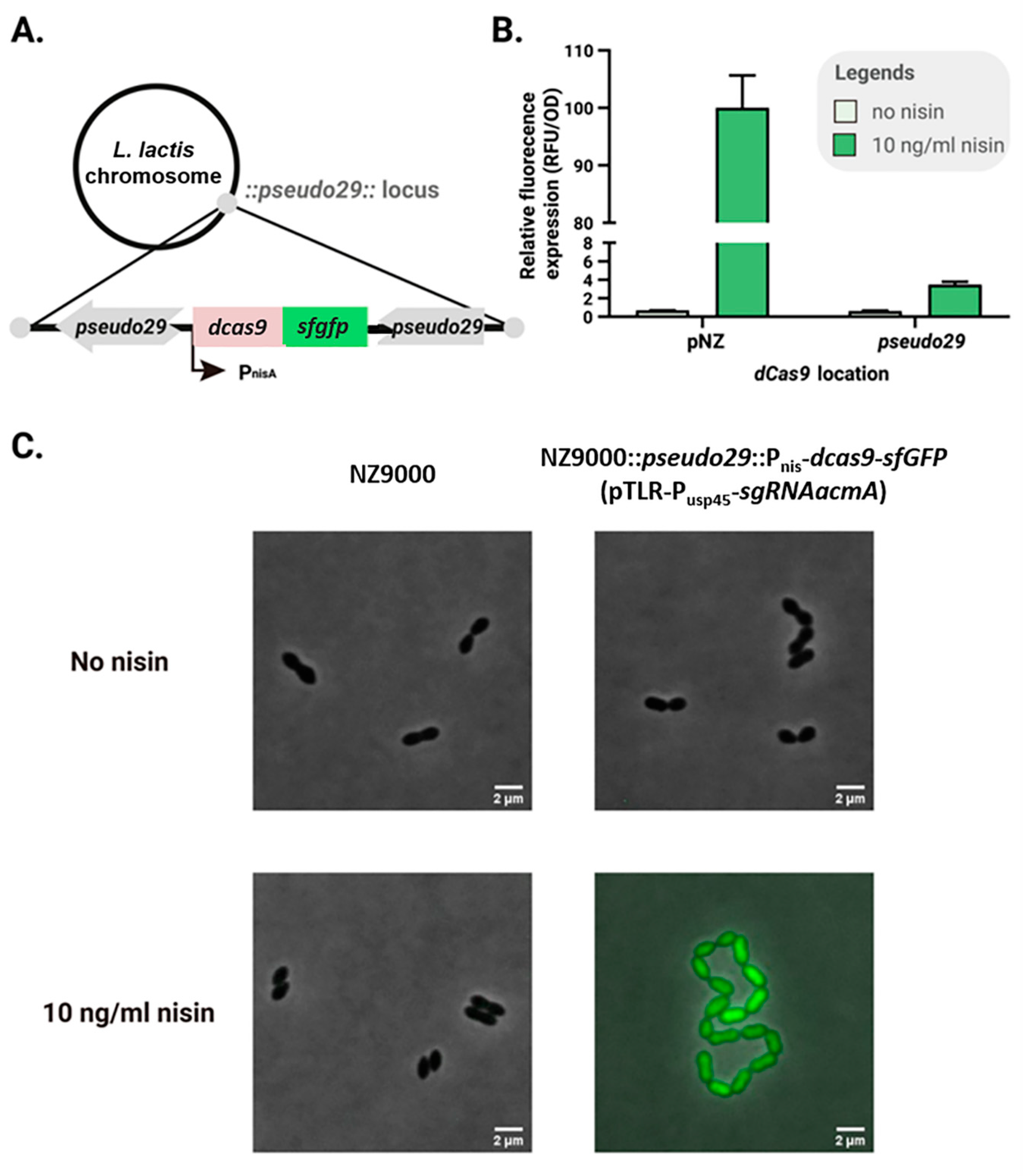
2.3. Reducing Leaky Expression Through Chromosomal dCas9-sfGFP Insertion
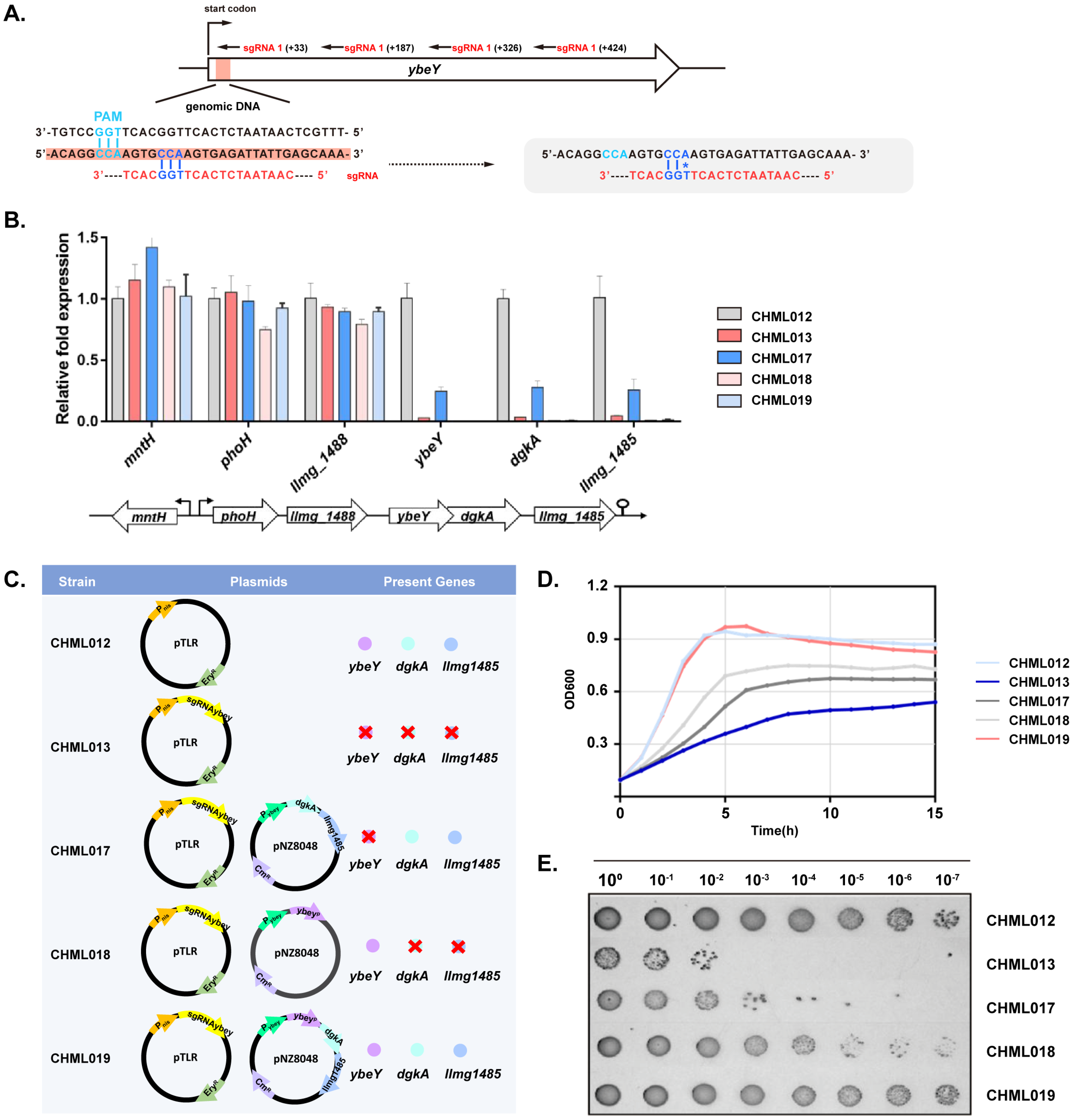
2.4. Application of Chromosomal-Based CRISPRi to Study ybeY Function
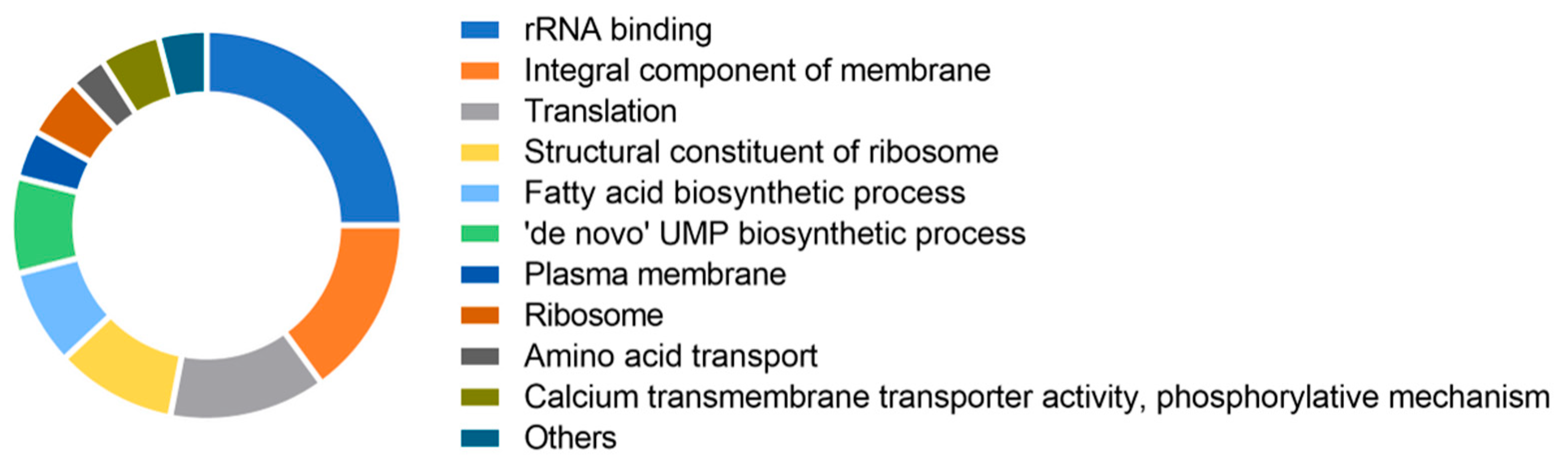
2.5. Silencing of ybeY Alters Ribosomal and Other Gene Expression
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media, and Culture Conditions
4.2. Recombinant DNA Techniques and Oligonucleotides
4.3. Construction of Plasmid-Based CRISPRi Systems
4.4. Quick-Fusion Cloning of sgRNA Genes
4.5. Construction of the Luc Reporter Strain
4.6. Construction of a Genome-Based CRISPRi System
4.7. Optical Density, Fluorescence, and Luminescence Measurements
4.8. Microscopy
4.9. Cell Viability Test
4.10. RNA Isolation and Quality Control
4.11. Real-Time PCR (RT-qPCR)
4.12. RNA Sequencing, Data Analysis and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LAB | lactic acid bacteria |
| NICE | nisin-controlled expression |
| CRISPR-Cas | clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins |
| DSB | double-strand break |
| crRNA | CRISPR RNA |
| tracrRNA | trans-activating RNA |
| dCas9 | catalytically inactive variant of Cas9 |
| CRISPRi | CRISPR interference |
| sfGFP | superfolder GFP |
| DEGs | differentially expressed genes |
| GO | Gene Ontology |
References
- Wessels, S.; Axelsson, L.; Bech Hansen, E.; De Vuyst, L.; Laulund, S.; Lähteenmäki, L.; Lindgren, S.; Mollet, B.; Salminen, S.; von Wright, A. The Lactic Acid Bacteria, the Food Chain, and Their Regulation. Trends Food Sci. Technol. 2004, 15, 498–505. [Google Scholar] [CrossRef]
- Sauer, M.; Russmayer, H.; Grabherr, R.; Peterbauer, C.K.; Marx, H. The Efficient Clade: Lactic Acid Bacteria for Industrial Chemical Production. Trends Biotechnol. 2017, 35, 756–769. [Google Scholar] [CrossRef]
- Solopova, A.; van Tilburg, A.Y.; Foito, A.; Allwood, J.W.; Stewart, D.; Kulakauskas, S.; Kuipers, O.P. Engineering Lactococcus lactis for the Production of Unusual Anthocyanins Using Tea as Substrate. Metab. Eng. 2019, 54, 160–169. [Google Scholar] [CrossRef]
- Wang, M.; Gao, Z.; Zhang, Y.; Pan, L. Lactic Acid Bacteria as Mucosal Delivery Vehicles: A Realistic Therapeutic Option. Appl. Microbiol. Biotechnol. 2016, 100, 5691–5701. [Google Scholar] [CrossRef]
- Kok, J.; van Gijtenbeek, L.A.; de Jong, A.; van der Meulen, S.B.; Solopova, A.; Kuipers, O.P. The Evolution of Gene Regulation Research in Lactococcus lactis. FEMS Microbiol. Rev. 2017, 41, S220–S243. [Google Scholar] [CrossRef]
- Mierau, I.; Kleerebezem, M. 10 Years of the Nisin-Controlled Gene Expression System (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef] [PubMed]
- de Ruyter, P.G.; Kuipers, O.P.; Beerthuyzen, M.M.; van Alen-Boerrigter, I.; de Vos, W.M. Functional Analysis of Promoters in the Nisin Gene Cluster of Lactococcus lactis. J. Bacteriol. 1996, 178, 3434–3439. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.; Buist, G.; Bolhuis, A.; ten Berge, A.; Kiel, J.; Mierau, I.; Dabrowska, M.; Venema, G.; Kok, J. A General System for Generating Unlabelled Gene Replacements in Bacterial Chromosomes. Mol. Gen. Genet. 1996, 253, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Solem, C.; Defoor, E.; Jensen, P.R.; Martinussen, J. Plasmid pCS1966, a New Selection/Counterselection Tool for Lactic Acid Bacterium Strain Construction Based on the oroP Gene, Encoding an Orotate Transporter from Lactococcus lactis. Appl. Environ. Microbiol. 2008, 74, 4772–4775. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR Interference: RNA-Directed Adaptive Immunity in Bacteria and Archaea. Nat. Rev. Genet. 2010, 11, 181–190. [Google Scholar] [CrossRef]
- Le Rhun, A.; Escalera-Maurer, A.; Bratovič, M.; Charpentier, E. CRISPR-Cas in Streptococcus Pyogenes. RNA Biol. 2019, 16, 380–389. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183, Correction in Cell 2021, 184, 844. [Google Scholar] [CrossRef]
- Westbrook, A.W.; Moo-Young, M.; Chou, C.P. Development of a CRISPR-Cas9 Tool Kit for Comprehensive Engineering of Bacillus Subtilis. Appl. Environ. Microbiol. 2016, 82, 4876–4895. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.-T.; Seo, S.-O.; Lynn, P.; Lu, T.; Jin, Y.-S.; Blaschek, H.P. Bacterial Genome Editing with CRISPR-Cas9: Deletion, Integration, Single Nucleotide Modification, and Desirable “Clean” Mutant Selection in Clostridium Beijerinckii as an Example. ACS Synth. Biol. 2016, 5, 721–732. [Google Scholar] [CrossRef]
- Schmitt, S.; Montalbán-López, M.; Peterhoff, D.; Deng, J.; Wagner, R.; Held, M.; Kuipers, O.P.; Panke, S. Analysis of Modular Bioengineered Antimicrobial Lanthipeptides at Nanoliter Scale. Nat. Chem. Biol. 2019, 15, 437–443. [Google Scholar] [CrossRef]
- Song, X.; Liu, L.; Liu, X.-X.; Xiong, Z.-Q.; Xie, C.-L.; Wang, S.-J.; Ai, L.-Z. Single-Plasmid Systems Based on CRISPR-Cas9 for Gene Editing in Lactococcus lactis. J. Dairy. Sci. 2021, 104, 10576–10585. [Google Scholar] [CrossRef]
- Oh, J.-H.; van Pijkeren, J.-P. CRISPR–Cas9-Assisted Recombineering in Lactobacillus Reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Škrlec, K.; Kocjan, J.; Olenic, M.; Štrukelj, B. Single Plasmid Systems for Inducible Dual Protein Expression and for CRISPR-Cas9/CRISPRi Gene Regulation in Lactic Acid Bacterium Lactococcus lactis. Sci. Rep. 2018, 8, 1009. [Google Scholar] [CrossRef] [PubMed]
- Overkamp, W.; Beilharz, K.; Detert Oude Weme, R.; Solopova, A.; Karsens, H.; Kovács, Á.T.; Kok, J.; Kuipers, O.P.; Veening, J.-W. Benchmarking Various Green Fluorescent Protein Variants in Bacillus Subtilis, Streptococcus Pneumoniae, and Lactococcus lactis for Live Cell Imaging. Appl. Environ. Microbiol. 2013, 79, 6481–6490. [Google Scholar] [CrossRef] [PubMed]
- van Gijtenbeek, L.A.; Robinson, A.; van Oijen, A.M.; Poolman, B.; Kok, J. On the Spatial Organization of mRNA, Plasmids, and Ribosomes in a Bacterial Host Overexpressing Membrane Proteins. PLOS Genet. 2016, 12, e1006523. [Google Scholar] [CrossRef] [PubMed]
- García-Cayuela, T.; de Cadiñanos, L.P.G.; Mohedano, M.L.; de Palencia, P.F.; Boden, D.; Wells, J.; Peláez, C.; López, P.; Requena, T. Fluorescent Protein Vectors for Promoter Analysis in Lactic Acid Bacteria and Escherichia Coli. Appl. Microbiol. Biotechnol. 2012, 96, 171–181. [Google Scholar] [CrossRef]
- de Ruyter, P.G.; Kuipers, O.P.; de Vos, W.M. Controlled Gene Expression Systems for Lactococcus lactis with the Food-Grade Inducer Nisin. Appl. Environ. Microbiol. 1996, 62, 3662–3667. [Google Scholar] [CrossRef]
- Buist, G.; Venema, G.; Kok, J. Autolysis of Lactococcus lactis Is Influenced by Proteolysis. J. Bacteriol. 1998, 180, 22. [Google Scholar] [CrossRef]
- Wegmann, U.; O’Connell-Motherway, M.; Zomer, A.; Buist, G.; Shearman, C.; Canchaya, C.; Ventura, M.; Goesmann, A.; Gasson, M.J.; Kuipers, O.P.; et al. Complete Genome Sequence of the Prototype Lactic Acid Bacterium Lactococcus lactis Subsp. Cremoris MG1363. J. Bacteriol. 2007, 189, 3256–3270. [Google Scholar] [CrossRef]
- Xia, Y.; Xu, C.; Wang, D.; Weng, Y.; Jin, Y.; Bai, F.; Cheng, Z.; Kuipers, O.P.; Wu, W. YbeY Controls the Type III and Type VI Secretion Systems and Biofilm Formation through RetS in Pseudomonas Aeruginosa. Appl. Environ. Microbiol. 2020, 87, 5. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Weng, Y.; Xu, C.; Wang, D.; Pan, X.; Tian, Z.; Xia, B.; Li, H.; Chen, R.; Liu, C.; et al. Endoribonuclease YbeY Is Essential for RNA Processing and Virulence in Pseudomonas Aeruginosa. mBio 2020, 11, 3. [Google Scholar] [CrossRef]
- Saramago, M.; Peregrina, A.; Robledo, M.; Matos, R.G.; Hilker, R.; Serrania, J.; Becker, A.; Arraiano, C.M.; Jiménez-Zurdo, J.I. Sinorhizobium Meliloti YbeY Is an Endoribonuclease with Unprecedented Catalytic Features, Acting as Silencing Enzyme in Riboregulation. Nucleic Acids Res. 2017, 45, 1371–1391. [Google Scholar] [CrossRef]
- Jacob, A.I.; Köhrer, C.; Davies, B.W.; RajBhandary, U.L.; Walker, G.C. Conserved Bacterial RNase YbeY Plays Key Roles in 70S Ribosome Quality Control and 16S rRNA Maturation. Mol. Cell 2013, 49, 427–438. [Google Scholar] [CrossRef]
- Pandey, S.P.; Minesinger, B.K.; Kumar, J.; Walker, G.C. A Highly Conserved Protein of Unknown Function in Sinorhizobium Meliloti Affects sRNA Regulation Similar to Hfq. Nucleic Acids Res. 2011, 39, 4691–4708. [Google Scholar] [CrossRef] [PubMed]
- Hoekzema, M.; Romilly, C.; Holmqvist, E.; Wagner, E.G.H. Hfq-dependent mRNA Unfolding Promotes sRNA-based Inhibition of Translation. EMBO J. 2019, 38, e101199. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Gimpel, M.; Wildenhain, T.; Brantl, S. A New Role for CsrA: Promotion of Complex Formation between an sRNA and Its mRNA Target in Bacillus Subtilis. RNA Biol. 2019, 16, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Wang, C.; Drewry, L.L.; Vogel, J. Molecular Mechanism of mRNA Repression in Trans by a ProQ-dependent Small RNA. EMBO J. 2017, 36, 1029–1045. [Google Scholar] [CrossRef]
- Hämmerle, H.; Amman, F.; Večerek, B.; Stülke, J.; Hofacker, I.; Bläsi, U. Impact of Hfq on the Bacillus Subtilis Transcriptome. PLoS ONE 2014, 9, e98661. [Google Scholar] [CrossRef]
- Gutiérrez, P.; Li, Y.; Osborne, M.J.; Pomerantseva, E.; Liu, Q.; Gehring, K. Solution Structure of the Carbon Storage Regulator Protein CsrA from Escherichia Coli. J. Bacteriol. 2005, 187, 3496–3501. [Google Scholar] [CrossRef]
- Vercruysse, M.; Köhrer, C.; Davies, B.W.; Arnold, M.F.F.; Mekalanos, J.J.; RajBhandary, U.L.; Walker, G.C. The Highly Conserved Bacterial RNase YbeY Is Essential in Vibrio Cholerae, Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing. PLOS Pathog. 2014, 10, e1004175. [Google Scholar] [CrossRef]
- Kobayashi, K.; Ehrlich, S.D.; Albertini, A.; Amati, G.; Andersen, K.K.; Arnaud, M.; Asai, K.; Ashikaga, S.; Aymerich, S.; Bessieres, P.; et al. Essential Bacillus Subtilis Genes. Proc. Natl. Acad. Sci. USA 2003, 100, 4678–4683. [Google Scholar] [CrossRef]
- Akerley, B.J.; Rubin, E.J.; Novick, V.L.; Amaya, K.; Judson, N.; Mekalanos, J.J. A Genome-Scale Analysis for Identification of Genes Required for Growth or Survival of Haemophilus Influenzae. Proc. Natl. Acad. Sci. USA 2002, 99, 966–971. [Google Scholar] [CrossRef]
- Davies, B.W.; Köhrer, C.; Jacob, A.I.; Simmons, L.A.; Zhu, J.; Aleman, L.M.; RajBhandary, U.L.; Walker, G.C. Role of Escherichia Coli YbeY, a Highly Conserved Protein, in rRNA Processing. Mol. Microbiol. 2010, 78, 506–518. [Google Scholar] [CrossRef]
- Pandey, S.P.; Winkler, J.A.; Li, H.; Camacho, D.M.; Collins, J.J.; Walker, G.C. Central Role for RNase YbeY in Hfq-Dependent and Hfq-Independent Small-RNA Regulation in Bacteria. BMC Genom. 2014, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Biswas, I.; Gruss, A.; Ehrlich, S.D.; Maguin, E. High-Efficiency Gene Inactivation and Replacement System for Gram-Positive Bacteria. J. Bacteriol. 1993, 175, 3628–3635. [Google Scholar] [CrossRef]
- de Jong, A.; Kuipers, O.P.; Kok, J. FUNAGE-Pro: Comprehensive Web Server for Gene Set Enrichment Analysis of Prokaryotes. Nucleic Acids Res. 2022, 50, W330–W336. [Google Scholar] [CrossRef]
- van der Meulen, S.B.; Hesseling-Meinders, A.; de Jong, A.; Kok, J. The Protein Regulator ArgR and the sRNA Derived from the 3’-UTR Region of Its Gene, ArgX, Both Regulate the Arginine Deiminase Pathway in Lactococcus lactis. PLoS ONE 2019, 14, e0218508. [Google Scholar] [CrossRef] [PubMed]
- van der Meulen, S.B.; de Jong, A.; Kok, J. Transcriptome Landscape of Lactococcus lactis Reveals Many Novel RNAs Including a Small Regulatory RNA Involved in Carbon Uptake and Metabolism. Taylor Fr. 2016, 13, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2018, 13, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gallay, C.; Kjos, M.; Domenech, A.; Slager, J.; van Kessel, S.P.; Knoops, K.; Sorg, R.A.; Zhang, J.-R.; Veening, J.-W. High-throughput CRISPRi Phenotyping Identifies New Essential Genes in Streptococcus Pneumoniae. Mol. Syst. Biol. 2017, 13, 931. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR Interference (CRISPRi) for Sequence-Specific Control of Gene Expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Zuckermann, M.; Hlevnjak, M.; Yazdanparast, H.; Zapatka, M.; Jones, D.T.W.; Lichter, P.; Gronych, J. A Novel Cloning Strategy for One-Step Assembly of Multiplex CRISPR Vectors. Sci. Rep. 2018, 8, 17499. [Google Scholar] [CrossRef]
- Taylor, R.G.; Walker, D.C.; Mclnnes, R.R. E. Coli Host Strains Significantly Affect the Quality of Small Scale Plasmid DNA Preparations Used for Sequencing. Nucleic Acids Res. 1993, 21, 1677–1678. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Chapter 8. In Vitro Amplification of DNA by the Polymerase Chain Reaction. In Molecular Cloning; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 2, ISBN 0-87969-576-5. [Google Scholar]
- Van Die, I.M.; Bergmans, H.E.N.; Hoekstra, W.P.M. Transformation in Escherichia coli: Studies On The Role Of The Heat Shock In Induction Of Competence. Microbiology 1983, 129, 663–670. [Google Scholar] [CrossRef]
- Holo, H.; Nes, I.F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. Cremoris Grown with Glycine in Osmotically Stabilized Media. Appl. Environ. Microbiol. 1989, 55, 3119–3123. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Yan, M.; Zhou, S.-R.; Xue, H.-W. CRISPR Primer Designer: Design Primers for Knockout and Chromosome Imaging CRISPR-Cas System. J. Integr. Plant Biol. 2015, 57, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Sorg, R.A.; Kuipers, O.P.; Veening, J.-W. Gene Expression Platform for Synthetic Biology in the Human Pathogen Streptococcus Pneumoniae. ACS Synth. Biol. 2015, 4, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.J.P.; Santos, C.S.; Pacheco, L.G.C. Bacterial Reference Genes for Gene Expression Studies by RT-qPCR: Survey and Analysis. Antonie Van Leeuwenhoek 2015, 108, 685–693. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; van der Meulen, S.; Kuipers, O.P.; Kok, J. T-REx: Transcriptome Analysis Webserver for RNA-Seq Expression Data. BMC Genom. 2015, 16, 663. [Google Scholar] [CrossRef]

| Strains | Relevant Genotype and Plasmids | Source |
|---|---|---|
| DH5α | DH5α | [49] |
| NZ9000 | MG1363, pepN::nisRK | [23] |
| CHML001 | NZ9000 (pNZ-PnisA-dcas9) | This study |
| CHML003 | NZ9000 (pNZ-PnisA-dcas9-sfgfp) | This study |
| CHML004 | NZ9000 (pNZ-PnisA-dcas9-sfgfp & pTLR-Pusp45-sgRNAluc) | This study |
| CHML005 | NZ9000 (pNZ-PnisA-dcas9-sfgfp & pTLR-Pusp45-sgRNAacmA) | This study |
| CHML007 | NZ9000 pseudo10::Pusp45-luc | This study |
| CHML008 | NZ9000 pseudo10::Pusp45-luc (pNZ-PnisA-dcas9-sfgfp & pTLR-Pusp45-sgRNAluc) | This study |
| CHML009 | NZ9000 pseudo29::PnisA-dcas9 | This study |
| CHML010 | NZ9000 pseudo29::PnisA-dcas9-sfgfp | This study |
| CHML011 | NZ9000 pseudo29::PnisA-dcas9-sfgfp (pTLR-Pusp45-sgRNAacmA) | This study |
| CHML012 | NZ9000 pseudo29::PnisA-dcas9 (pTLR) | This study |
| CHML013 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey) | This study |
| CHML014 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey2) | This study |
| CHML015 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey3) | This study |
| CHML016 | NZ9000 pseudo29::PnisA-dcas9(pTLR-Pusp45-sgRNAybey4) | This study |
| CHML017 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey & pNZ-Pybey-dgkA-llmg1485) | This study |
| CHML018 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey & pNZ-Pybey-ybeyp) | This study |
| CHML019 | NZ9000 pseudo29::PnisA-dcas9 (pTLR-Pusp45-sgRNAybey & pNZ-Pybey- ybeyp-dgkA-llmg1485) | This study |
| CHML020 | NZ9000 pseudo29::PnisA-dcas9, pseudo10::Pybey-dgkA-llmg1485 (pTLR & pNZ-Pusp45-sgRNAybey) | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Liu, M.; Kok, J. Chromosomal and Plasmid-Based CRISPRi Platforms for Conditional Gene Silencing in Lactococcus lactis. Int. J. Mol. Sci. 2025, 26, 9516. https://doi.org/10.3390/ijms26199516
Huang C, Liu M, Kok J. Chromosomal and Plasmid-Based CRISPRi Platforms for Conditional Gene Silencing in Lactococcus lactis. International Journal of Molecular Sciences. 2025; 26(19):9516. https://doi.org/10.3390/ijms26199516
Chicago/Turabian StyleHuang, Chenxi, Meishan Liu, and Jan Kok. 2025. "Chromosomal and Plasmid-Based CRISPRi Platforms for Conditional Gene Silencing in Lactococcus lactis" International Journal of Molecular Sciences 26, no. 19: 9516. https://doi.org/10.3390/ijms26199516
APA StyleHuang, C., Liu, M., & Kok, J. (2025). Chromosomal and Plasmid-Based CRISPRi Platforms for Conditional Gene Silencing in Lactococcus lactis. International Journal of Molecular Sciences, 26(19), 9516. https://doi.org/10.3390/ijms26199516







