Omicron Subvariants Infection Kinetics and Nirmatrelvir Efficacy in Transgenic K18-hACE2 Mice
Abstract
1. Introduction
2. Results
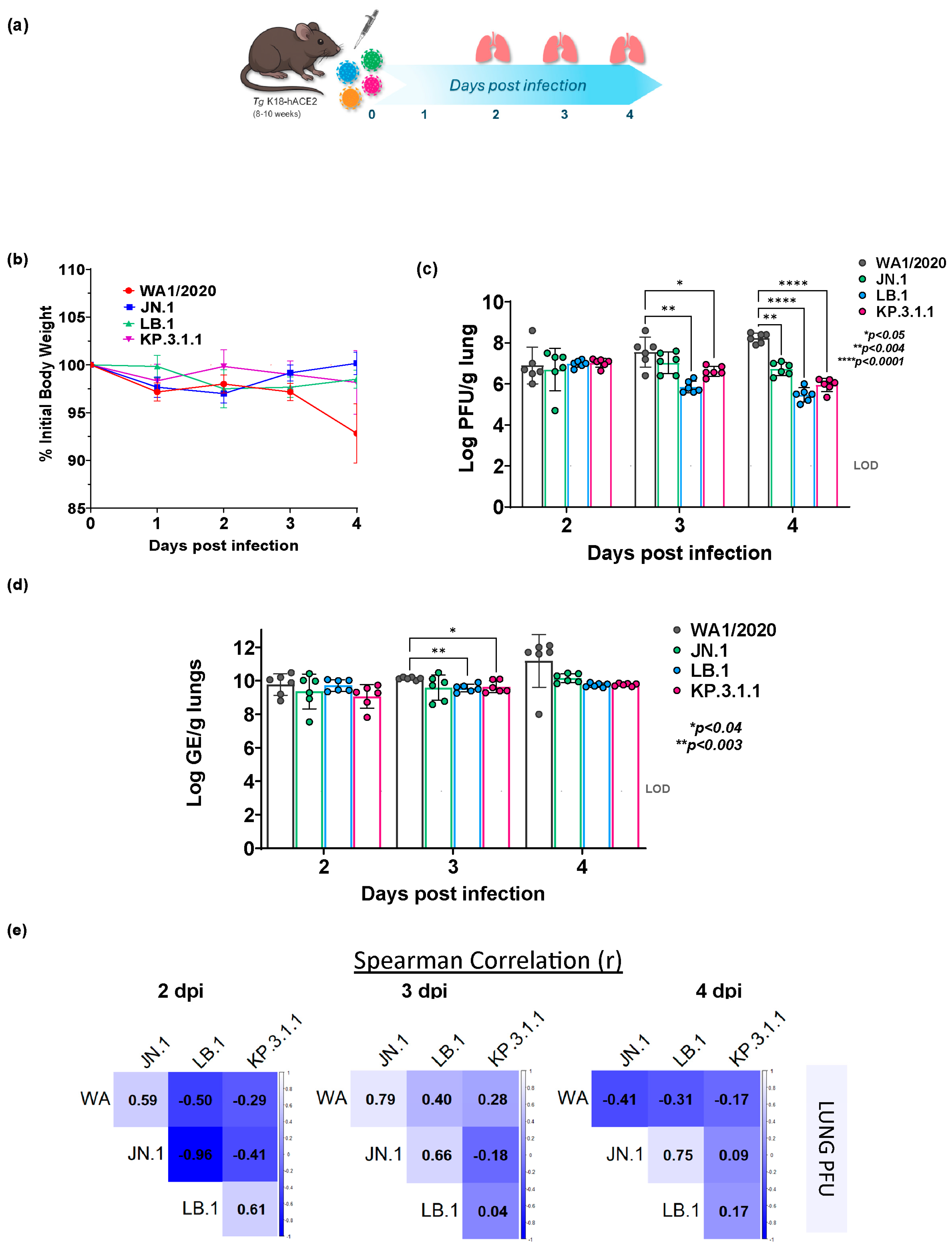
2.1. Altered Lung Infection Kinetics of Omicron Subvariants in K18-hACE2 Mice Compared to Parent WA1/2020
2.2. Divergent Temporal Patterns of Lung Viral Replication Among Omicron Subvariants and Parent WA1/2020
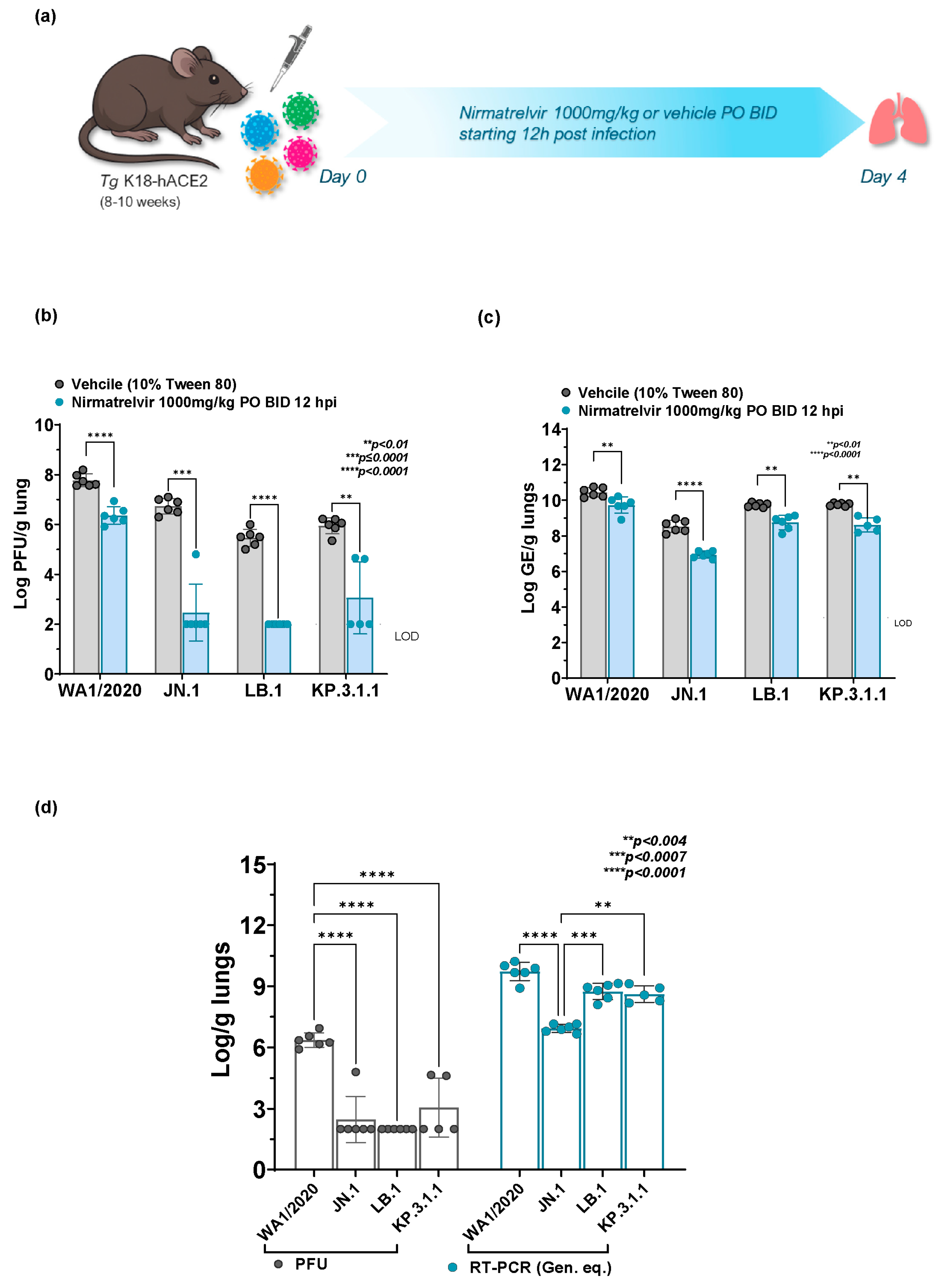
2.3. Broad In Vivo Efficacy of Nirmatrelvir Across WA1/2020 and Omicron Subvariants
2.4. Comparative Nirmatrelvir Susceptibility Reveals Enhanced Antiviral Response in Omicron Subvariants Infected Mice in Comparison to WA1/2020
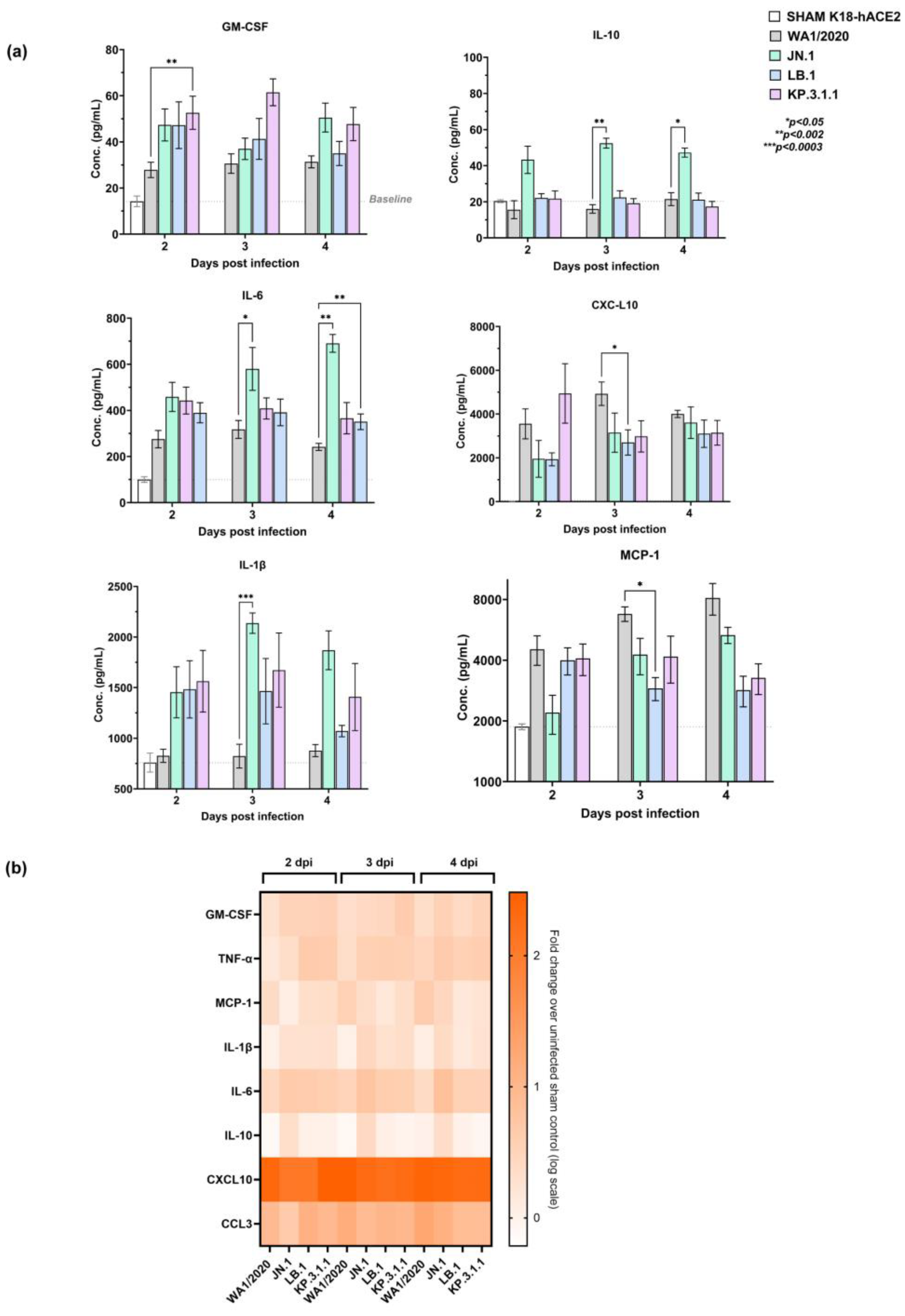
2.5. Early Pulmonary Immune Response Analysis Reveals Distinct Pattern in Omicron Subvariants Infected Mice
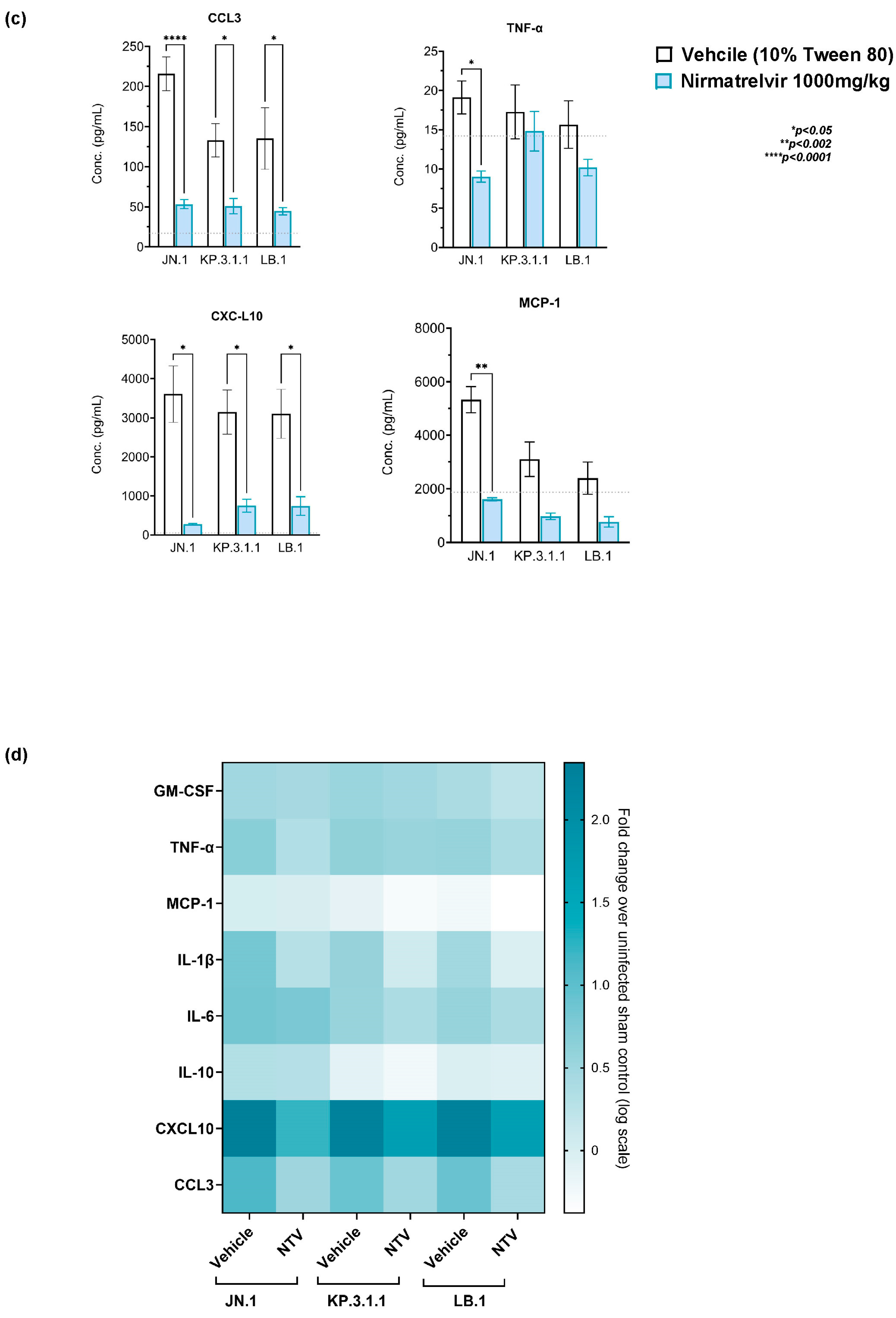
2.6. Nirmatrelvir Treatment Attenuates Inflammatory Cytokine and Chemokine Responses in Omicron-Infected Mice
3. Discussion
4. Materials and Methods
4.1. Cell and Virus Culture
4.2. In Vitro Antiviral Assay
4.3. In Vivo Studies
4.4. Viral Load Assessment
4.4.1. Plaque Assay
4.4.2. RNA Extraction and RT-qPCR
4.5. Chemokine and Cytokine Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Report. 2024. Available online: https://data.who.int/dashboards/covid19/cases (accessed on 25 May 2020).
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef]
- Zhang, W.; Davis, B.D.; Chen, S.S.; Martinez, J.M.; Plummer, J.T.; Vail, E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021, 325, 1324–1326. [Google Scholar] [CrossRef]
- Kaku, Y.; Yo, M.S.; Tolentino, J.E.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants. Lancet Infect. Dis. 2024, 24, e482–e483. [Google Scholar] [CrossRef]
- Kaku, Y.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP.3.1.1 variant. Nature 2025, 637, 921–929. [Google Scholar] [CrossRef]
- Jian, F.; Wang, J.; Yisimayi, A.; Song, W.; Xu, Y.; Chen, X.; Niu, X.; Yang, S.; Yu, Y.; Wang, P.; et al. Evolving antibody response to SARS-CoV-2 antigenic shift from XBB to JN. 1. Nature 2024, 637, 921–929. [Google Scholar] [CrossRef]
- Yang, H.; Guo, H.; Wang, A.; Cao, L.; Fan, Q.; Jiang, J.; Wang, M.; Lin, L.; Ge, X.; Wang, H.; et al. Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants. Nat. Commun. 2024, 15, 7715. [Google Scholar] [CrossRef]
- Wang, Q.; Mellis, I.A.; Ho, J.; Bowen, A.; Kowalski-Dobson, T.; Valdez, R.; Katsamba, P.S.; Wu, M.; Lee, C.; Shapiro, L.; et al. Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages. Emerg. Microbes Infect. 2024, 13, 2402880. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. CDC JN.1 Update. Available online: https://www.cdc.gov/ncird/whats-new/JN.1-update-2024-01-05.html (accessed on 5 January 2024).
- Taylor, A.L.; Starr, T.N. Deep mutational scanning of SARS-CoV-2 Omicron BA.2.86 and epistatic emergence of the KP.3 variant. Virus Evol. 2024, 10, veae067. [Google Scholar] [CrossRef]
- Sacco, M.D.; Hu, Y.; Gongora, M.V.; Meilleur, F.; Kemp, M.T.; Zhang, X.; Wang, J.; Chen, Y. The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. Cell Res. 2022, 32, 498–500. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Mahale, P.; Malden, D.; Hong, V.; Ackerson, B.K.; Lewin, B.J.; Link-Gelles, R.; Feldstein, L.R.; Lipsitch, M.; Tartof, S.Y. Immune escape and attenuated severity associated with the SARS-CoV-2 BA.2.86/JN.1 lineage. Nat. Commun. 2024, 15, 8550. [Google Scholar] [CrossRef]
- He, P.; Liu, B.; Gao, X.; Yan, Q.; Pei, R.; Sun, J.; Chen, Q.; Hou, R.; Li, Z.; Zhang, Y.; et al. SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope. Nat. Microbiol. 2022, 7, 1635–1649. [Google Scholar] [CrossRef]
- Reuschl, A.K.; Thorne, L.G.; Whelan, M.V.X.; Ragazzini, R.; Furnon, W.; Cowton, V.M.; De Lorenzo, G.; Mesner, D.; Turner, J.L.; Dowgier, G.; et al. Evolution of enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants. Nat. Microbiol. 2024, 9, 451–463. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Ullrich, S.; Ekanayake, K.B.; Otting, G.; Nitsche, C. Main protease mutants of SARS-CoV-2 remain susceptible to Nirmatrelvir. Nat. Commun. 2022, 13, 3205. [Google Scholar] [CrossRef]
- Meyer, C.; Garzia, A.; Miller, M.W.; Huggins, D.J.; Myers, R.W.; Hoffmann, H.H.; Ashbrook, A.W.; Jannath, S.Y.; Liverton, N.; Kargman, S.; et al. Small-molecule inhibition of SARS-CoV-2 NSP14 RNA cap methyltransferase. Nature 2025, 637, 1178–1185. [Google Scholar] [CrossRef]
- Jiang, R.D.; Liu, M.Q.; Chen, Y.; Shan, C.; Zhou, Y.W.; Shen, X.R.; Li, Q.; Zhang, L.; Zhu, Y.; Si, H.R.; et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 2020, 182, 50–58.e8. [Google Scholar] [CrossRef]
- McCray, P.B., Jr.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef]
- Rosales, R.; McGovern, B.L.; Rodriguez, M.L.; Leiva-Rebollo, R.; Diaz-Tapia, R.; Benjamin, J.; Rai, D.K.; Cardin, R.D.; Anderson, A.S.; Alshammary, H.; et al. Nirmatrelvir and molnupiravir maintain potent In vitro and in vivo antiviral activity against circulating SARS-CoV-2 omicron subvariants. Virus Res. 2024, 330, 199053. [Google Scholar] [CrossRef]
- Alvarez, N.; Gonzalez-Jimenez, I.; Rasheed, R.; Goldgirsh, K.; Park, S.; Perlin, D.S. Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Variants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections. Virus 2025, 17, 918. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255, Erratum in Signal Transduct. Target. Ther. 2021, 6, 326. [Google Scholar] [CrossRef]
- Zaderer, V.; Abd El Halim, H.; Wyremblewsky, A.L.; Lupoli, G.; Dächert, C.; Muenchhoff, M.; Graf, A.; Blum, H.; Lass-Flörl, C.; Keppler, O.T.; et al. Omicron subvariants illustrate reduced respiratory tissue penetration, cell damage and inflammatory responses in human airway epithelia. Front. Immunol. 2023, 14, 1258268. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, V.W.; Layton, M.E.; Parish, C.A.; Perkins, J.J.; Schreier, J.D.; Wang, Y.; Adam, G.C.; Alvarez, N.; Bahmanjah, S.; Bahnck-Teets, C.M.; et al. Invention of mK-7845, a SARS-CoV-2 3CL protease inhibitor employing a novel difluorinated glutamine mimic. J. Med. Chem. 2024, 67, 3935–3958. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, N.; Adam, G.C.; Howe, J.A.; Sharma, V.; Zimmerman, M.D.; Dolgov, E.; Rasheed, R.; Nizar, F.; Sahay, K.; Nelson, A.M.; et al. Novel pan-coronavirus 3CL protease inhibitor MK-7845: Biological and pharmacological profiling. Viruses 2024, 16, 1158. [Google Scholar] [CrossRef]
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, e105. [Google Scholar]
- Corman, V.M.; Müller, M.; Costabel, U.; Timm, J.; Binger, T.; Meyer, B.; Kreher, P.; Lattwein, E.; Eschbach-Bludau, M.; Nitsche, A.; et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012, 17, 20334. [Google Scholar] [CrossRef]
| Strain | Infection Dose PFU/Mouse |
|---|---|
| WA1/2020 | 2.85 × 105 |
| JN.1 | 1.2 × 105 |
| LB.1 | 1.25 × 104 |
| KP.3.1.1 | 4.6 × 104 |
| WA1/2020 | 2.85 × 105 |
| JN.1 | 1.2 × 105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, V.; Dolgov, E.; Tillery, T.; Mendez Romero, C.; Rojas-Triana, A.; Villalba Guzman, D.M.; Goldgirsh, K.; Rasheed, R.; Gonzalez-Jimenez, I.; Alvarez, N.; et al. Omicron Subvariants Infection Kinetics and Nirmatrelvir Efficacy in Transgenic K18-hACE2 Mice. Int. J. Mol. Sci. 2025, 26, 9509. https://doi.org/10.3390/ijms26199509
Sharma V, Dolgov E, Tillery T, Mendez Romero C, Rojas-Triana A, Villalba Guzman DM, Goldgirsh K, Rasheed R, Gonzalez-Jimenez I, Alvarez N, et al. Omicron Subvariants Infection Kinetics and Nirmatrelvir Efficacy in Transgenic K18-hACE2 Mice. International Journal of Molecular Sciences. 2025; 26(19):9509. https://doi.org/10.3390/ijms26199509
Chicago/Turabian StyleSharma, Vijeta, Enriko Dolgov, Taylor Tillery, Camila Mendez Romero, Alberto Rojas-Triana, Diana M. Villalba Guzman, Kira Goldgirsh, Risha Rasheed, Irene Gonzalez-Jimenez, Nadine Alvarez, and et al. 2025. "Omicron Subvariants Infection Kinetics and Nirmatrelvir Efficacy in Transgenic K18-hACE2 Mice" International Journal of Molecular Sciences 26, no. 19: 9509. https://doi.org/10.3390/ijms26199509
APA StyleSharma, V., Dolgov, E., Tillery, T., Mendez Romero, C., Rojas-Triana, A., Villalba Guzman, D. M., Goldgirsh, K., Rasheed, R., Gonzalez-Jimenez, I., Alvarez, N., Park, S., Murugan, M., Nelson, A. M., & Perlin, D. S. (2025). Omicron Subvariants Infection Kinetics and Nirmatrelvir Efficacy in Transgenic K18-hACE2 Mice. International Journal of Molecular Sciences, 26(19), 9509. https://doi.org/10.3390/ijms26199509









