Epigenetic Biomarkers for Cervical Cancer Progression: A Scoping Review
Abstract
1. Introduction
2. Materials and Methods
3. Results
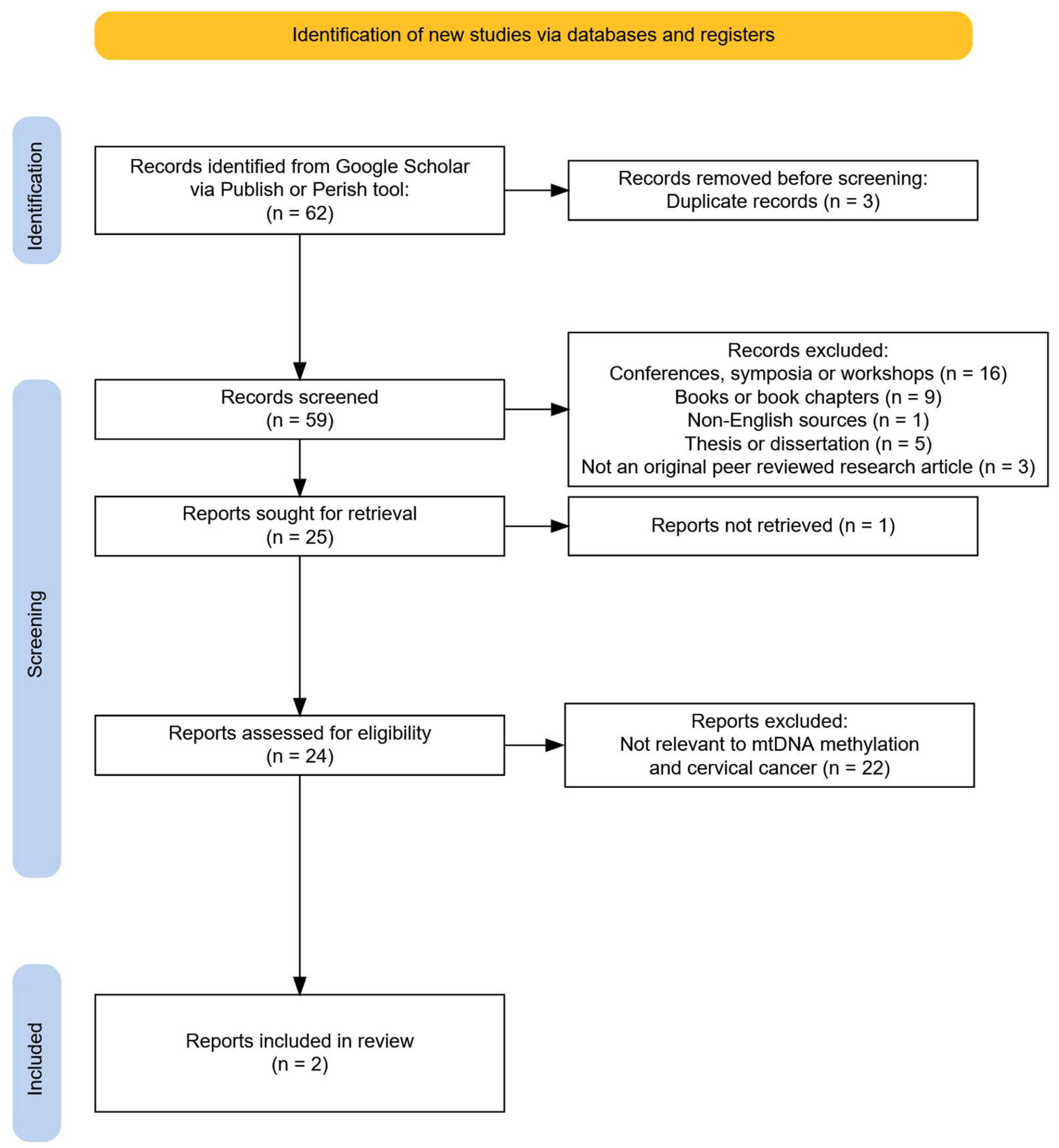
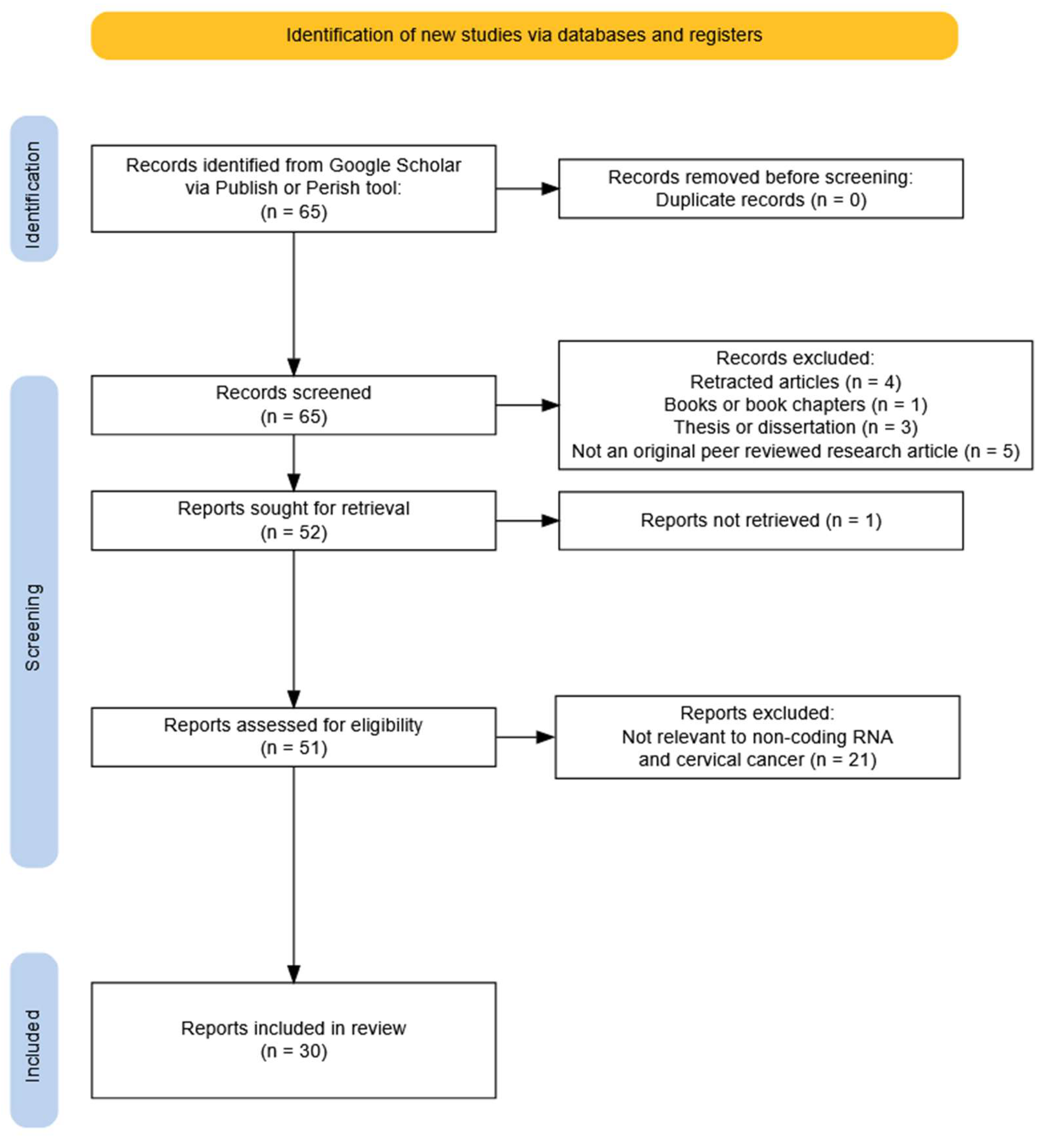
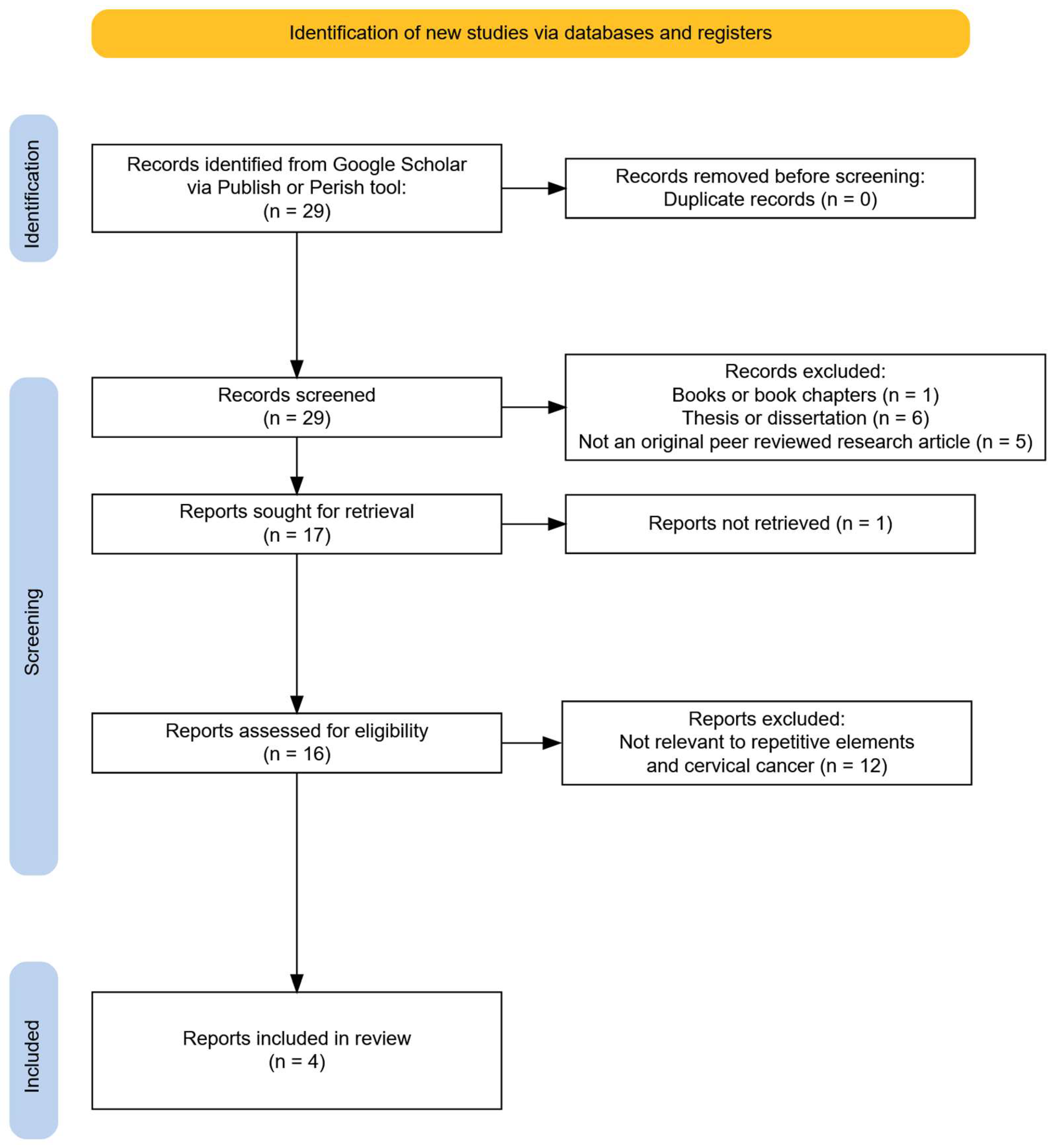
3.1. Study Selection Process
3.2. Methylation in Mitochondrial DNA
3.3. Histone Modifications
3.4. Non-Coding RNAs
3.4.1. lncRNA
3.4.2. circRNA
3.4.3. piRNA
3.4.4. miRNA
3.5. Methylation in Repetitive Elements
4. Discussion
4.1. Mitochondrial DNA Methylation: A Potential but Untapped Biomarker
4.2. Histone Modifications: A Reversible Contributor to Cervical Oncogenesis
4.3. Non-Coding RNAs: Expression as a Diagnostic and Prognostic Biomarker
4.4. Hypomethylation of Repetitive Elements: Biomarker of Genomic Instability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AcH3 | Histone H3 Acetylation |
| AMPK | AMP-Activated Protein Kinase |
| ARMD | Alu Recombination-Mediated Deletion |
| ATAC-seq | Assay For Transposase-Accessible Chromatin using Sequencing |
| BDNF-AS | BDNF antisense RNA |
| CCHE1 | Cervical Cancer High-Expressed lncRNA 1 |
| CHIP | Chromatin Immunoprecipitation |
| CIN | Cervical Intraepithelial Neoplasia |
| Circ-ITCH | Circular RNA itchy E3 ubiquitin protein ligase |
| circRNA | Circular RNA |
| CRNDE | Colorectal Neoplasia Differentially Expressed |
| CRUK | Cancer Research UK |
| CSRP2BP | Cysteine-Rich Protein 2-Binding Protein |
| CUT&Tag | Cleavage Under Targets and Tagmentation |
| DLEU1 | Deleted in Lymphocytic Leukemia 1 |
| DNMT1 | DNA-Methyltransferase 1 |
| DNMT3A | Methyltransferase 3A |
| EP300 | E1A Binding Protein P300 |
| ETS1 | E26 Transformation-Specific 1 |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FAM19A4 | Family with sequence similarity 19 (chemokine (C–C motif)-like) member A4 |
| H3K27ac | Histone H3 Lysine 27 Acetylation |
| H3K27me3 | Histone H3 Lysine 27 Trimethylation |
| H3K36 | Histone H3 Lysine 36 |
| H3K4me3 | Histone H3 Lysine 4 Trimethylation |
| H3K9 | Histone H3 Lysine 9 |
| H3K9ac | Histone H3 Lysine 9 Acetylation |
| H3K9me3 | Histone H3 Lysine 9 Trimethylation |
| HDAC | Histone Deacetylase |
| HERV | Human Endogenous Retrovirus |
| HPV | Human Papillomavirus |
| JMJD3 | Jumonji Domain-containing protein 3 |
| KCNMB 2-AS1 | Calcium-activated potassium channel subunit beta-2 Antisense RNA 1 |
| KDM2A | Lysine-Specific Demethylase 2A |
| KDM5A | Lysine-Specific Demethylase 5A |
| LINC00899 | Long Intergenic Non-protein Coding RNA 899 |
| LINE-1 | Long Interspersed Element 1 |
| LKB1 | Liver Kinase B1 |
| LMIC | Low And Middle-Income Countries |
| lncRNA | Long Non-Coding RNA |
| LSD1 | Lysine-Specific Demethylase 1 |
| m6A | N6-Methyladenosine |
| MAGI2-AS3 | Membrane associated guanylate kinase, WW and PDZ domain containing 2 Antisense RNA 3 |
| MEG3 | Maternally Expressed Gene 3 |
| METTL3 | Methyltransferase-like 3 |
| MIEN1 | Migration and Invasion Enhancer 1 |
| miR124-2 | MicroRNA-124 locus 2 |
| miR-16 | MicroRNA-16 |
| MIR210HG | MicroRNA-210 Host Gene |
| miRNA | MicroRNA |
| ML | Machine Learning |
| mtDNA | Mitochondrial DNA |
| ncRNA | Non-Coding RNA |
| nDNA | Nuclear DNA |
| NDUFA8 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 |
| NKILA | NF-KappaB Interacting LncRNA |
| PCAF | P300/CBP-associated factor |
| piRNA | Piwi-Interacting RNA |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PVT1 | Plasmacytoma variant translocation 1 |
| RARb2 | Retinoic Acid Receptor B2 |
| SAM | S-Adenosylmethionine |
| SINE | Short-Interspersed Element |
| SLC25A26 | Solute Carrier Family 25 Member 26 |
| STK11 | Serine/threonine kinase 11 |
| SUV39H1 | Suppressor of variegation 3–9 homolog 1 |
| Th1 | Type 1 T Helper |
| TP73-AS1 | Tumor Protein P73 Antisense RNA 1 |
| TSG | Tumour Suppressor Gene |
| TXNDC5 | Thioredoxin Domain Containing Protein 5 |
| UTX | Ubiquitously Transcribed tetratricopeptide repeat, X |
| WDR5 | WD Repeat Domain 5 |
| xAI | Explainable Artificial Intelligence |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical Cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Pecorino, L. Molecular Biology of Cancer: Mechanisms, Targets, and Therapeutics; Oxford University Press: Oxford, UK, 2021; pp. 245–246. ISBN 9780198833024. [Google Scholar]
- Biswas, S.; Rao, C.M. Epigenetics in Cancer: Fundamentals and Beyond. Pharmacol. Ther. 2017, 173, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Burdier, F.R.; Waheed, D.N.; Nedjai, B.; Steenbergen, R.D.M.; Poljak, M.; Baay, M.; Vorsters, A.; Van Keer, S. DNA Methylation as a Triage Tool for Cervical Cancer Screening—A Meeting Report. Prev. Med. Rep. 2024, 41, 102678. [Google Scholar] [CrossRef] [PubMed]
- Gilham, C.; Nedjai, B.; Scibior-Bentkowska, D.; Reuter, C.; Banwait, R.; Brentnall, A.R.; Cuzick, J.; Peto, J.; Lorincz, A.T. Long-Term Prediction by DNA Methylation of High-Grade Cervical Intraepithelial Neoplasia: Results of the ARTISTIC Cohort. Int. J. Cancer 2024, 155, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, S.; Pei, M.; Zhao, M.; Wang, L.; Jiang, Y.; Yang, T.; Zhao, J.; Song, L.; Yang, X. Crosstalk between Histone Modification and DNA Methylation Orchestrates the Epigenetic Regulation of the Costimulatory Factors, Tim-3 and Galectin-9, in Cervical Cancer. Oncol. Rep. 2019, 42, 2655–2669. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, H.; Tian, M.; Wang, D.; He, J.; Xu, T. DNA Methylation and Hydroxymethylation in Cervical Cancer: Diagnosis, Prognosis and Treatment. Front. Genet. 2020, 11, 347. [Google Scholar] [CrossRef]
- Bee, K.J.; Gradissimo, A.; Chen, Z.; Harari, A.; Schiffman, M.; Raine-Bennett, T.; Castle, P.E.; Clarke, M.; Wentzensen, N.; Burk, R.D. Genetic and Epigenetic Variations of Hpv52 in Cervical Precancer. Int. J. Mol. Sci. 2021, 22, 6463. [Google Scholar] [CrossRef]
- Sumiec, E.G.; Yim, Z.Y.; Mohy-Eldin, H.; Nedjai, B. The Current State of DNA Methylation Biomarkers in Self-Collected Liquid Biopsies for the Early Detection of Cervical Cancer: A Literature Review. Infect. Agents Cancer 2024, 19, 62. [Google Scholar] [CrossRef]
- Molina, M.A.; Carosi Diatricch, L.; Castany Quintana, M.; Melchers, W.J.G.; Andralojc, K.M. Cervical Cancer Risk Profiling: Molecular Biomarkers Predicting the Outcome of HrHPV Infection. Expert Rev. Mol. Diagn. 2020, 20, 1099–1120. [Google Scholar] [CrossRef]
- Bukowski, A.; Hoyo, C.; Graff, M.; Vielot, N.A.; Kosorok, M.R.; Brewster, W.R.; Maguire, R.L.; Murphy, S.K.; Nedjai, B.; Ladoukakis, E.; et al. Epigenome-Wide Differential Methylation and Differential Variability as Predictors of High-Grade Cervical Intraepithelial Neoplasia (CIN2+). Am. J. Epidemiol. 2025, 194, 1012–1022. [Google Scholar] [CrossRef]
- Ramírez, A.T.; Sánchez, G.I.; Nedjai, B.; Agudelo, M.C.; Brentnall, A.R.; Cuschieri, K.; Castañeda, K.M.; Cuzick, J.; Lorincz, A.T.; Sanchez, G.I.; et al. Effective Methylation Triage of HPV Positive Women with Abnormal Cytology in a Middle-Income Country. Int. J. Cancer 2021, 148, 1383–1393. [Google Scholar] [CrossRef]
- Kelly, H.; Benavente, Y.; Pavon, M.A.; De Sanjose, S.; Mayaud, P.; Lorincz, A.T. Performance of DNA Methylation Assays for Detection of High-Grade Cervical Intraepithelial Neoplasia (CIN2+): A Systematic Review and Meta-Analysis. Br. J. Cancer 2019, 121, 954–965. [Google Scholar] [CrossRef]
- Zhong, X.; Chen, D. Global Research Trends on Epigenetics and Cancer: A Bibliometric Analysis. Medicine 2025, 104, e43197. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, H.; Stolte, C.; Böhmer, G.; Hampl, M.; Hagemann, I.; Maier, E.; Denecke, A.; Hirchenhain, C.; Patzke, J.; Jentschke, M.; et al. Evaluation of CIN2/3 Lesion Regression in GynTect® DNA Methylation-Marker-Negative Patients in a Longitudinal Study. Cancers 2023, 15, 3951. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Harzing, A.W. Publish or Perish. Available online: https://harzing.com/resources/publish-or-perish (accessed on 1 February 2025).
- Sun, C.; Reimers, L.L.; Burk, R.D. Methylation of HPV16 Genome CpG Sites Is Associated with Cervix Precancer and Cancer. Gynecol. Oncol. 2011, 121, 59–63. [Google Scholar] [CrossRef]
- Menga, A.; Palmieri, E.M.; Cianciulli, A.; Infantino, V.; Mazzone, M.; Scilimati, A.; Palmieri, F.; Castegna, A.; Iacobazzi, V. SLC25A26 Overexpression Impairs Cell Function via MtDNA Hypermethylation and Rewiring of Methyl Metabolism. FEBS J. 2017, 284, 967–984. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Castegna, A.; Infantino, V.; Andria, G. Mitochondrial DNA Methylation as a Next-Generation Biomarker and Diagnostic Tool. Mol. Genet. Metab. 2013, 110, 25–34. [Google Scholar] [CrossRef]
- Dong, Z.; Pu, L.; Cui, H. Mitoepigenetics and Its Emerging Roles in Cancer. Front. Cell Dev. Biol. 2020, 8, 4. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New Insights into Nucleosome and Chromatin Structure: An Ordered State or a Disordered Affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef]
- Woodcock, C.L.; Skoultchi, A.I.; Fan, Y. Role of Linker Histone in Chromatin Structure and Function: H1 Stoichiometry and Nucleosome Repeat Length. Chromosome Res. 2006, 14, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.B.; Horvath, S.; Shi, T.; Yu, H.; Tze, S.; Grunstein, M.; Kurdistani, S.K. Global Histone Modification Patterns Predict Risk of Prostate Cancer Recurrence. Nature 2005, 435, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, S.; Zhao, M.; Yang, T.; Quan, S.; Yang, Q.; Song, L.; Yang, X. SUV39H1-DNMT3A-Mediated Epigenetic Regulation of Tim-3 and Galectin-9 in the Cervical Cancer. Cancer Cell Int. 2020, 20, 325. [Google Scholar] [CrossRef]
- Chen, D.; Cai, B.; Zhu, Y.; Ma, Y.; Yu, X.; Xiong, J.; Shen, J.; Tie, W.; Zhang, Y.; Guo, F. Targeting Histone Demethylases JMJD3 and UTX: Selenium as a Potential Therapeutic Agent for Cervical Cancer. Clin. Epigenet. 2024, 16, 51. [Google Scholar] [CrossRef]
- Ou, R.; Zhu, L.; Zhao, L.; Li, W.; Tao, F.; Lu, Y.; He, Q.; Li, J.; Ren, Y.; Xu, Y. HPV16 E7-Induced Upregulation of KDM2A Promotes Cervical Cancer Progression by Regulating MiR-132–Radixin Pathway. J. Cell Physiol. 2019, 234, 2659–2671. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, H.; Zhang, Q.; Shi, Z.; Zhang, Y.; Zhao, L.; Ren, Y.; Ou, R.; Xu, Y. Human Papillomavirus Type 16 E7 Oncoprotein-Induced Upregulation of Lysine-Specific Demethylase 5A Promotes Cervical Cancer Progression by Regulating the MicroRNA-424–5p/Suppressor of Zeste 12 Pathway. Exp. Cell Res. 2020, 396, 112277. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Chen, C.; Zhang, J.; Qian, W.; Dong, Y.; Liu, Z.; Zhang, X.; Wang, X.; Zhang, Z.; et al. LSD1 Binds to HPV16 E7 and Promotes the Epithelial-Mesenchymal Transition in Cervical Cancer by Demethylating Histones at the Vimentin Promoter. Oncotarget 2017, 8, 11329. [Google Scholar] [CrossRef][Green Version]
- Beyer, S.; Zhu, J.; Mayr, D.; Kuhn, C.; Schulze, S.; Hofmann, S.; Dannecker, C.; Jeschke, U.; Kost, B.P. Histone H3 Acetyl K9 and Histone H3 Tri Methyl K4 as Prognostic Markers for Patients with Cervical Cancer. Int. J. Mol. Sci. 2017, 18, 477. [Google Scholar] [CrossRef]
- Pan, B.; Liu, C.; Su, J.; Xia, C. Activation of AMPK Inhibits Cervical Cancer Growth by Hyperacetylation of H3K9 through PCAF. Cell Commun. Signal. 2024, 22, 306. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Wu, J.; Tian, Y.; Zhou, H.; Zhou, Y.; Hu, W.; Zhao, W.; Wei, H.; Ling, B.; Ma, C. Targeting of Histone Deacetylases to Reactivate Tumour Suppressor Genes and Its Therapeutic Potential in a Human Cervical Cancer Xenograft Model. PLoS ONE 2013, 8, e80657. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.Y.; Huo, F.C.; Du, W.Q.; Sun, X.L.; Jiang, X.; Zhang, L.S.; Pei, D.S. METTL3 Potentiates Progression of Cervical Cancer by Suppressing ER Stress via Regulating M6A Modification of TXNDC5 MRNA. Oncogene 2022, 41, 4420–4432. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Tang, H.; He, Q.; Sun, J.; Yang, Y.; Kong, L.; Wang, Y. NDUFA8 Is Transcriptionally Regulated by EP300/H3K27ac and Promotes Mitochondrial Respiration to Support Proliferation and Inhibit Apoptosis in Cervical Cancer. Biochem. Biophys. Res. Commun. 2024, 693, 149374. [Google Scholar] [CrossRef]
- Yang, X.; Sun, F.; Gao, Y.; Li, M.Y.; Liu, M.; Wei, Y.; Jie, Q.; Wang, Y.; Mei, J.; Mei, J.; et al. Histone Acetyltransferase CSRP2BP Promotes the Epithelial–Mesenchymal Transition and Metastasis of Cervical Cancer Cells by Activating N-Cadherin. J. Exp. Clin. Cancer Res. 2023, 42, 268. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-Coding RNAs as Regulators in Epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and Their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Yang, J.P.; Yang, X.J.; Xiao, L.; Wang, Y. Long Noncoding RNA PVT1 as a Novel Serum Biomarker for Detection of Cervical Cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3980–3986. [Google Scholar]
- Wang, C.; Zou, H.; Yang, H.; Wang, L.; Chu, H.; Jiao, J.; Wang, Y.; Chen, A. Long Non-Coding RNA Plasmacytoma Variant Translocation 1 Gene Promotes the Development of Cervical Cancer via the NF-ΚB Pathway. Mol. Med. Rep. 2019, 20, 2433–2440. [Google Scholar] [CrossRef]
- Barr, J.A.; Hayes, K.E.; Brownmiller, T.; Harold, A.D.; Jagannathan, R.; Lockman, P.R.; Khan, S.; Martinez, I. Long Non-Coding RNA FAM83H-AS1 Is Regulated by Human Papillomavirus 16 E6 Independently of P53 in Cervical Cancer Cells. Sci. Rep. 2019, 9, 3662. [Google Scholar] [CrossRef]
- Hu, X.-L.; Huang, X.-T.; Zhang, J.-N.; Liu, J.; Wen, L.-J.; Xu, X.; Zhou, J.-Y. Long Noncoding RNA MIR210HG Is Induced by Hypoxia-Inducible Factor 1α and Promotes Cervical Cancer Progression. Am. J. Cancer Res. 2022, 12, 2783–2797. [Google Scholar] [PubMed]
- Yang, M.; Zhai, X.; Xia, B.; Wang, Y.; Lou, G. Long Noncoding RNA CCHE1 Promotes Cervical Cancer Cell Proliferation via Upregulating PCNA. Tumor Biol. 2015, 36, 7615–7622. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Fang, X.; Xie, Q.; Wang, Y.; Lin, Z.; Lin, R.; Yao, T. Long Non-Coding RNA AK001903 Regulates Tumor Progression in Cervical Cancer. Oncol. Lett. 2021, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Fan, L.P. Long Non-Coding RNA CRNDE Enhances Cervical Cancer Progression by Suppressing PUMA Expression. Biomed. Pharmacother. 2019, 117, 108726. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.; Wu, D.; Zhang, D.; Sun, M. Long Noncoding RNA KCNMB2-AS1 Stabilized by N6-Methyladenosine Modification Promotes Cervical Cancer Growth Through Acting as a Competing Endogenous RNA. Cell Transpl. 2020, 29, 963689720964382. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, J. Elevated Expression Levels of Long Non-Coding RNA, Loc554202, Are Predictive of Poor Prognosis in Cervical Cancer. Tohoku J. Exp. Med. 2017, 243, 165–172. [Google Scholar] [CrossRef][Green Version]
- Guo, X.; Li, L.; Gao, Z.; Zhou, H.; Li, J.; Wang, Q. The Long Non-Coding RNA PTTG3P Promotes Growth and Metastasis of Cervical Cancer through PTTG1. Aging 2019, 11, 1333–1341. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, W.; Zhang, C.; Liu, X.; Lv, J.; Li, X.; Zhao, L.; Li, W.; Li, J.; Ren, Y.; et al. Long Non-Coding RNA RP11-552M11.4 Favors Tumorigenesis and Development of Cervical Cancer via Modulating MiR-3941/ATF1 Signaling. Int. J. Biol. Macromol. 2019, 130, 24–33. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, B.; Wang, S.; Li, X.; Fan, T. Long Non-Coding RNA TP73 Antisense RNA 1 Facilitates the Proliferation and Migration of Cervical Cancer Cells via Regulating MicroRNA-607/Cyclin D2. Mol. Med. Rep. 2019, 20, 3371–3378. [Google Scholar] [CrossRef]
- Song, Z.; Xing, F.; Jiang, H.; He, Y.; Lv, J. Long Non-Coding RNA TP73-AS1 Predicts Poor Prognosis and Regulates Cell Proliferation and Migration in Cervical Cancer. Arch. Med. Sci. 2022, 18, 523–534. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, F.; Wu, X.; Zhao, W.; Xia, Q. The Expression and Clinical Significance of Serum Exosomal-Long Non-Coding RNA DLEU1 in Patients with Cervical Cancer. Ann. Med. 2025, 57, 2442537. [Google Scholar] [CrossRef]
- Hou, S.; Zhang, X.; Yang, J. Long Non-Coding RNA ABHD11-AS1 Facilitates the Progression of Cervical Cancer by Competitively Binding to MiR-330–5p and Upregulating MARK2. Exp. Cell Res. 2022, 410, 112929. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, Z.; Gao, Y.; Yao, T. Downregulation of Long Noncoding RNA MEG3 Is Associated with Poor Prognosis and Promoter Hypermethylation in Cervical Cancer. J. Exp. Clin. Cancer Res. 2017, 36, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qu, J. Long Non-Coding RNA MEG3 Suppresses Survival, Migration, and Invasion of Cervical Cancer. OncoTargets Ther. 2018, 11, 4999–5007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, C.; Yan, T.; Wang, J.; Liang, W. Long Noncoding RNA BDNF-AS Is Downregulated in Cervical Cancer and Has Anti-Cancer Functions by Negatively Associating with BDNF. Arch. Biochem. Biophys. 2018, 646, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gu, Y.; Gu, Y.; Wu, J. Long Noncoding RNA LINC00899/MiR-944/ESR1 Axis Regulates Cervical Cancer Cell Proliferation, Migration, and Invasion. J. Interferon Cytokine Res. 2021, 41, 220–233. [Google Scholar] [CrossRef]
- Wang, F.; Jiang, X.; Wang, P. NF-κB Interaction Long Non-coding RNA Inhibits Migration, Invasion and Epithelial-mesenchymal Transition of Cervical Cancer Cells through Inhibiting NF-κB Signaling Pathways. Exp. Ther. Med. 2020, 20, 1039–1047. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Q.; Zhang, Q. The Mechanism of HPV-Mediated DNA Methylation in the Promoter Region of Long-Chain Non-Coding RNA MAGI2-AS3 in the Occurrence and Development of Cervical Cancer. Clin. Exp. Obs. Gynecol. 2024, 51, 140. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, S.; Chen, W.; Dong, X.; Zhang, R.; Ye, H.; Mei, X.; Liu, H.; Fang, Y.; He, C. Circular Rna Circypel2: A Novel Biomarker in Cervical Cancer. Genes 2022, 13, 38. [Google Scholar] [CrossRef]
- Meng, L.; Jia, X.; Yu, W.; Wang, C.; Chen, J.; Liu, F. Circular RNA UBAP2 Contributes to Tumor Growth and Metastasis of Cervical Cancer via Modulating MiR-361-3p/SOX4 Axis. Cancer Cell Int. 2020, 20, 357. [Google Scholar] [CrossRef]
- Song, T.F.; Xu, A.L.; Chen, X.H.; Gao, J.Y.; Gao, F.; Kong, X.C. Circular RNA CircRNA_101996 Promoted Cervical Cancer Development by Regulating MiR-1236-3p/TRIM37 Axis. Kaohsiung J. Med. Sci. 2021, 37, 547–561. [Google Scholar] [CrossRef]
- Xie, H.; Wang, J.; Wang, B. Circular RNA Circ_0003221 Promotes Cervical Cancer Progression by Regulating MiR-758-3p/CPEB4 Axis. Cancer Manag. Res. 2021, 13, 5337–5350. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guo, R.; Liu, Q.; Sun, J.; Wang, H. Circular RNA Circ-ITCH Inhibits the Malignant Behaviors of Cervical Cancer by MicroRNA-93-5p/FOXK2 Axis. Reprod. Sci. 2020, 27, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Li, Z.; Luo, X.; Wang, D.; Zhou, Y.; Zhao, J.; Gao, S.; Yang, Y.; Fu, W.; Kong, L.; et al. PiRNA-14633 Promotes Cervical Cancer Cell Malignancy in a METTL14-Dependent M6A RNA Methylation Manner. J. Transl. Med. 2022, 20, 51. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Han, Y.; Zhu, G.; Li, G.; Wu, X. Krüppel-like Factor 5-Induced Overexpression of Long Non-Coding RNA DANCR Promotes the Progression of Cervical Cancer via Repressing MicroRNA-145-3p to Target ZEB1. Cell Cycle 2021, 20, 1441–1454. [Google Scholar] [CrossRef]
- Cheng, X.; Shen, C.; Liao, Z. High Expression of Circular RNA-Mitochondrial TRNA Translation Optimization 1 Assists the Diagnosis of High-Risk Human Papillomavirus Infection in Cervical Cancer. J. Low. Genit. Tract Dis. 2022, 26, 212–218. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, J.; He, Z.; Wu, S. Long Noncoding RNA WT1-AS Inhibits the Progression of Cervical Cancer by Sponging MiR-205. Cancer Biother. Radiopharm. 2021, 36, 491–500. [Google Scholar] [CrossRef]
- Criscione, S.W.; Zhang, Y.; Thompson, W.; Sedivy, J.M.; Neretti, N. Transcriptional Landscape of Repetitive Elements in Normal and Cancer Human Cells. BMC Genom. 2014, 15, 583. [Google Scholar] [CrossRef]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Rajendiran, S.; Gibbs, L.D.; Van Treuren, T.; Klinkebiel, D.L.; Vishwanatha, J.K. MIEN1 Is Tightly Regulated by SINE Alu Methylation in Its Promoter. Oncotarget 2016, 7, 65307–65319. [Google Scholar] [CrossRef]
- Curty, G.; Menezes, A.N.; Brant, A.C.; de Mulder Rougvie, M.; Moreira, M.Â.M.; Soares, M.A. Expression of Retroelements in Cervical Cancer and Their Interplay with HPV Infection and Host Gene Expression. Cancers 2021, 13, 3513. [Google Scholar] [CrossRef]
- Sen, S.; Mandal, P.; Bhattacharya, A.; Kundu, S.; Roy Chowdhury, R.; Ranjan Mondal, N.; Chatterjee, T.; Chakravarty, B.; Roy, S.; Sengupta, S. Impact of Viral and Host DNA Methylations on HPV16-Related Cervical Cancer Pathogenesis. Tumor Biol. 2017, 39, 5. [Google Scholar] [CrossRef]
- McCabe, M.T.; Powell, D.R.; Zhou, W.; Vertino, P.M. Homozygous Deletion of the STK11/LKB1 Locus and the Generation of Novel Fusion Transcripts in Cervical Cancer Cells. Cancer Genet. Cytogenet. 2010, 197, 130–141. [Google Scholar] [CrossRef]
- Nguyen, N.P.D.; Deshpande, V.; Luebeck, J.; Mischel, P.S.; Bafna, V. ViFi: Accurate Detection of Viral Integration and MRNA Fusion Reveals Indiscriminate and Unregulated Transcription in Proximal Genomic Regions in Cervical Cancer. Nucleic Acids Res. 2018, 46, 3309–3325. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Transposable Elements in Cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Madakashira, B.P.; Sadler, K.C. DNA Methylation, Nuclear Organization, and Cancer. Front. Genet. 2017, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]


| Search Query in Google Scholar | Epigenetic Modification |
|---|---|
| “HPV” “methylation” AND (“cervical cancer” OR “cervix”) AND (“mtDNA” or “mitochondrial DNA”) AND (“EPIC” OR “450K” OR “450 platform” OR “27K” OR “27 platform” OR “bisulfite sequencing” OR “pyrosequencing”). | Mitochondrial DNA methylation |
| [intitle:cervical cancer] “HPV” “histone modification” “epigenetic” AND (“cervical cancer” OR “cervix”) AND (“CHIP” OR “Chromatin Immunoprecipitation” OR “Mass spectrometry”) | Histone modifications |
| [intitle:cervical cancer] [intitle:RNA] “HPV” “RNA” “biomarker” “epigenetic” AND (“cervical cancer” OR “cervix”) | Non-coding RNA expression |
| [intitle: “cervical cancer”] “methylation” AND (“Transposable element” OR “Transposons” OR “Retrotransposons” OR “Alu” OR “LINE-1” OR “SINE” OR “LTR” OR “Non-LTR” OR repetitive elements”) | Repetitive elements methylation |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Primary research paper | Review |
| Manuscript published in English | Book or book chapter |
| Study reported association between cervical cancer and epigenetic modification | Thesis or dissertation |
| Proceedings from conferences, symposia or workshops | |
| Article in blog or newsletter | |
| Retracted study |
| Epigenetic Modification Related to Cervical Cancer | Number of Manuscripts | Publication Period |
|---|---|---|
| Mitochondrial DNA methylation | 2 | [2011–2017] |
| Histone modifications | 12 | [2013–2024] |
| Non-coding RNA expression | 31 | [2015–2025] |
| Repetitive elements methylation | 4 | [2010–2021] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladoukakis, E.; Andriamiadana, G.; Hajizadah, F.; James, L.G.E.; Nedjai, B. Epigenetic Biomarkers for Cervical Cancer Progression: A Scoping Review. Int. J. Mol. Sci. 2025, 26, 9423. https://doi.org/10.3390/ijms26199423
Ladoukakis E, Andriamiadana G, Hajizadah F, James LGE, Nedjai B. Epigenetic Biomarkers for Cervical Cancer Progression: A Scoping Review. International Journal of Molecular Sciences. 2025; 26(19):9423. https://doi.org/10.3390/ijms26199423
Chicago/Turabian StyleLadoukakis, Efthymios, Gracia Andriamiadana, Fatema Hajizadah, Lewis G. E. James, and Belinda Nedjai. 2025. "Epigenetic Biomarkers for Cervical Cancer Progression: A Scoping Review" International Journal of Molecular Sciences 26, no. 19: 9423. https://doi.org/10.3390/ijms26199423
APA StyleLadoukakis, E., Andriamiadana, G., Hajizadah, F., James, L. G. E., & Nedjai, B. (2025). Epigenetic Biomarkers for Cervical Cancer Progression: A Scoping Review. International Journal of Molecular Sciences, 26(19), 9423. https://doi.org/10.3390/ijms26199423






