Development of Melting-Curve-Based Real-Time PCR for Differentiating Medically Important Candida Species
Abstract
1. Introduction
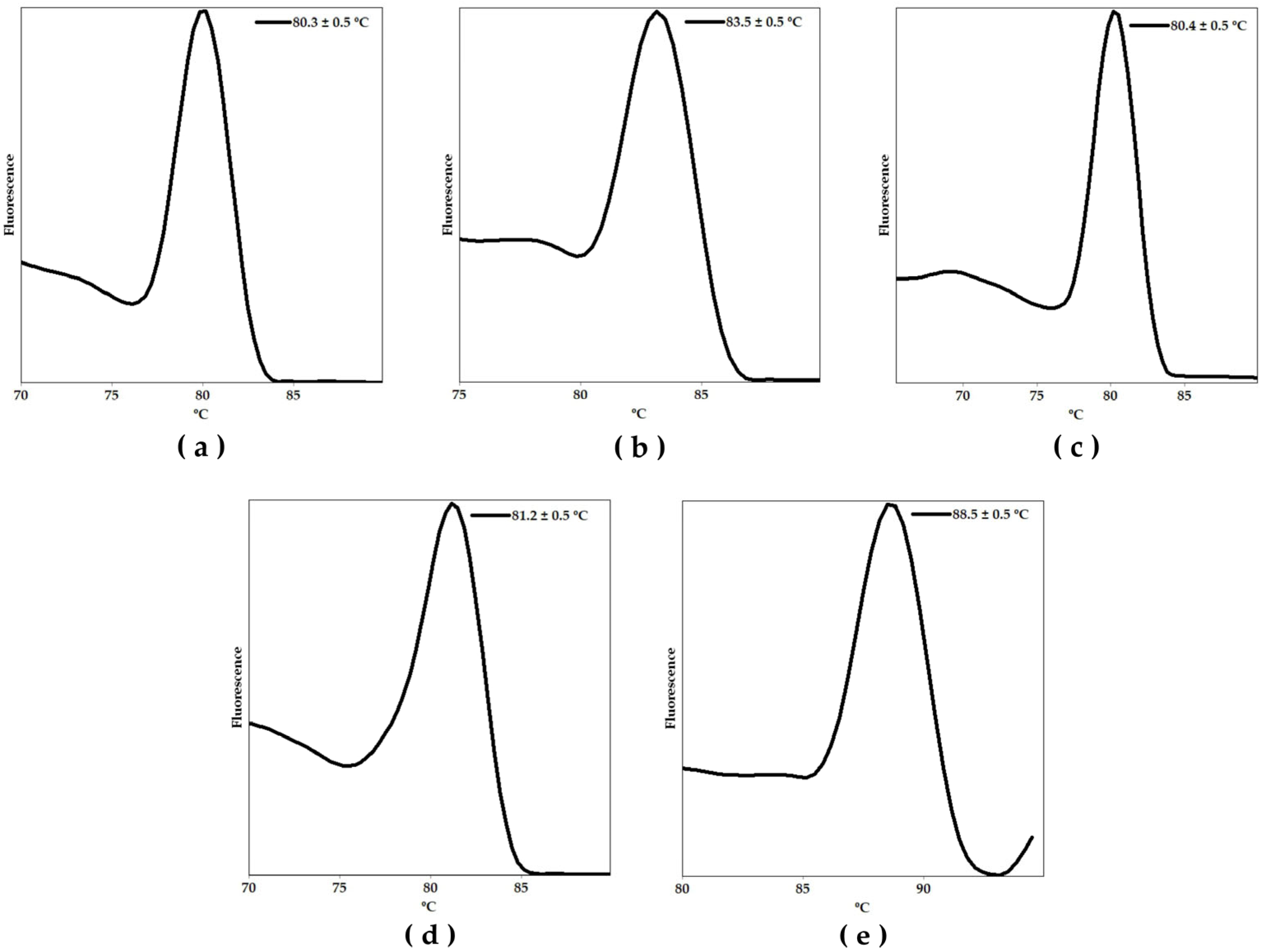
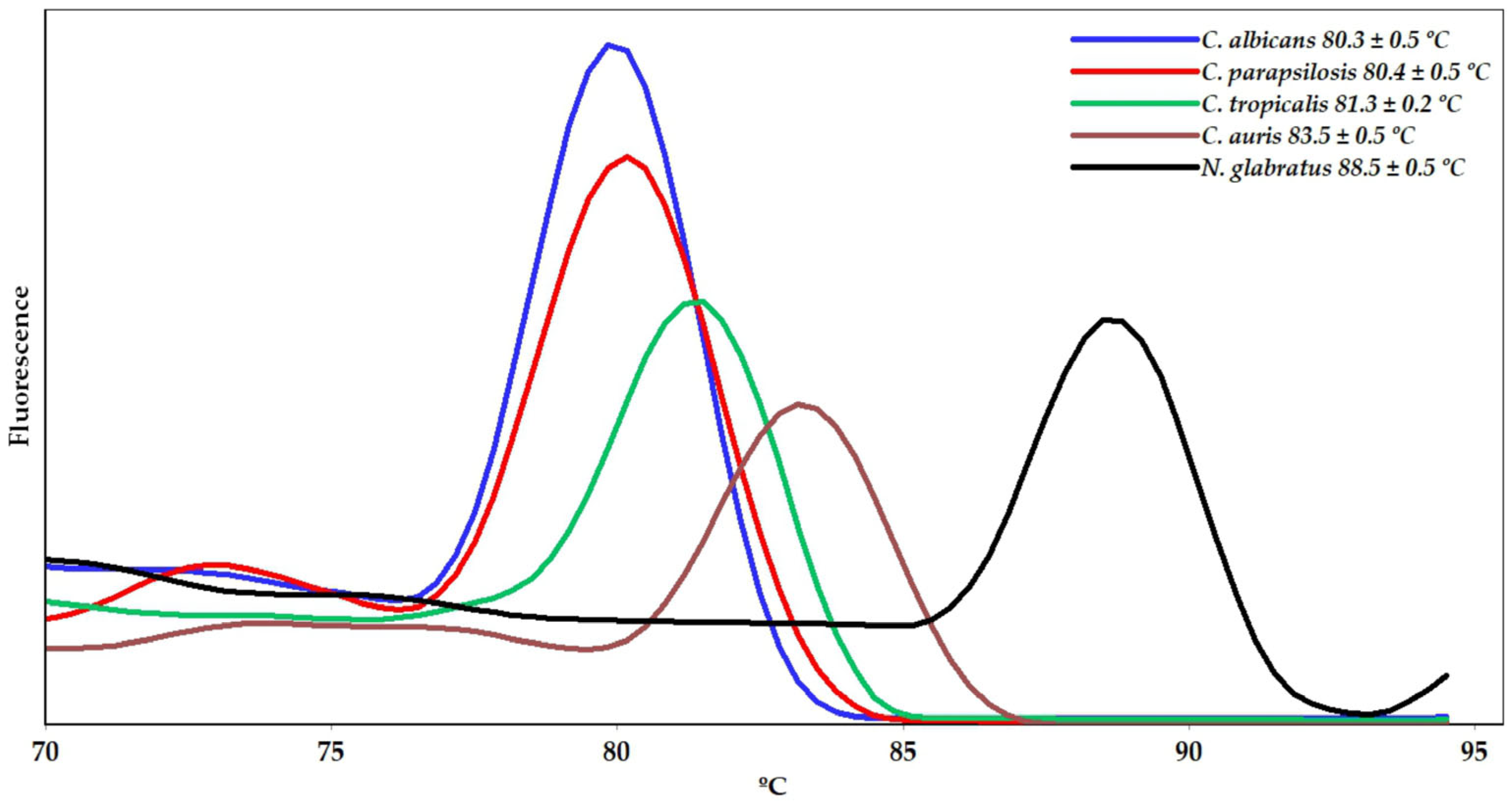
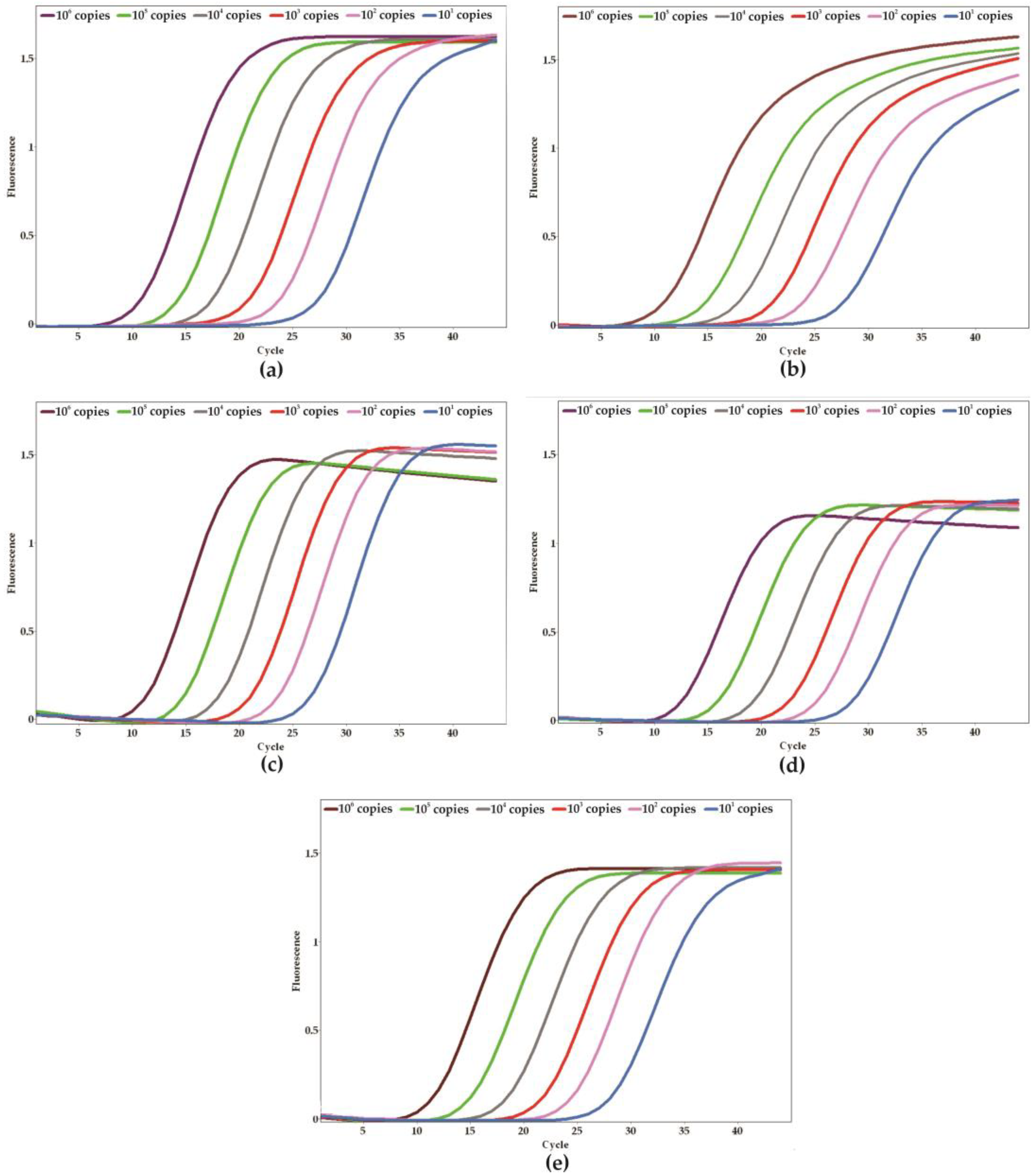
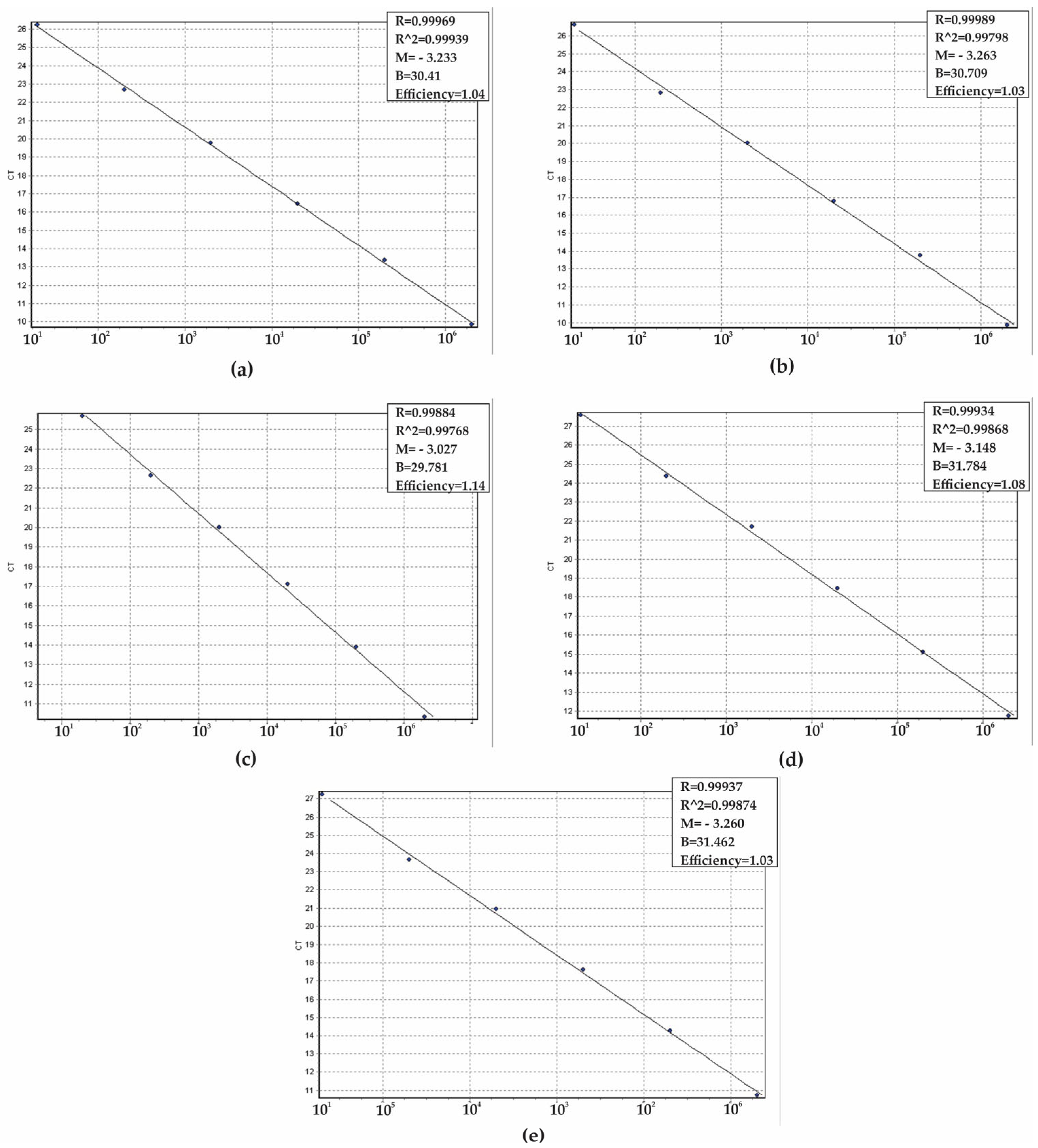
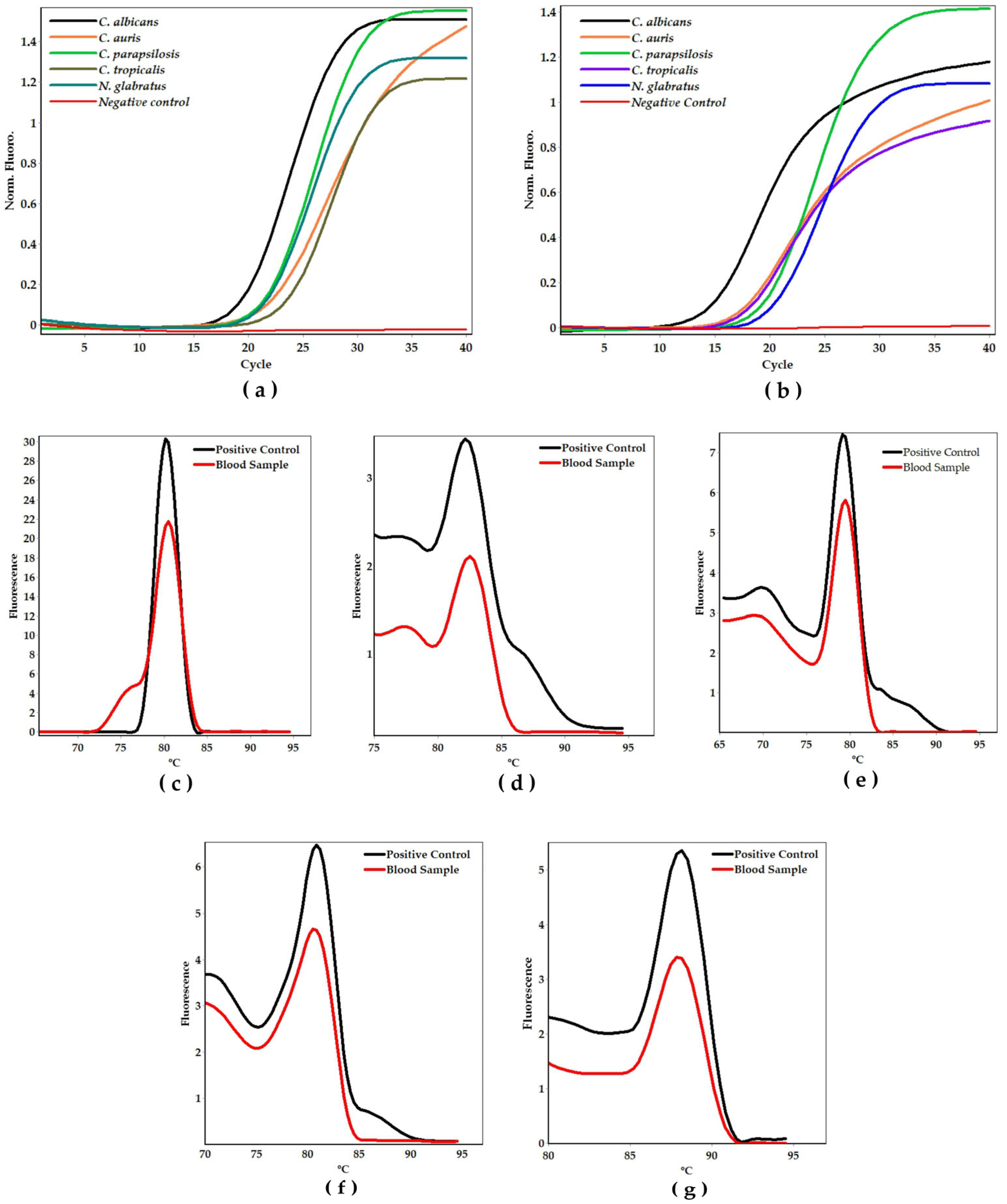
2. Results
3. Discussion
4. Materials and Methods
4.1. PCR Primers and Positive Control Design
4.2. Microorganisms and Culture Conditions
4.3. DNA Extraction
4.4. PCR Design
4.5. Analytical Specificity, Sensitivity, and Validation
4.6. Analytical Validation with DNA-Boiling Purification, Culture-qPCR and Blood Samples
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: A multi-institutional study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Braga, P.R.; Cruz, I.L.; Ortiz, I.; Barreiros, G.; Nouér, S.A.; Nucci, M. Secular trends of candidemia at a Brazilian tertiary care teaching hospital. Braz. J. Infect. Dis. 2018, 22, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Motta, F.A.; Dalla-Costa, L.M.; Muro, M.D.; Cardoso, M.N.; Picharski, G.L.; Jaeger, G.; Burger, M. Risk factors for candidemia mortality in hospitalized children. J. Pediatr. 2017, 93, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Vieira de Melo, A.P.; Zuza-Alves, D.L.; da Silva-Rocha, W.P.; Ferreira, L.B.C.S.; Francisco, E.C.; Salles de Azevedo Melo, A.; Maranhão Chaves, G. Virulence factors of Candida spp. obtained from blood cultures of patients with candidemia attended at tertiary hospitals in Northeast Brazil. J. Mycol. Med. 2019, 29, 132–139. [Google Scholar] [CrossRef]
- Khan, S.; Cai, L.; Bilal, H.; Khan, M.N.; Fang, W.; Zhang, D.; Yao, F.; Wang, X.; Wang, Q.; Hou, B.; et al. An 11-year retrospective analysis of candidiasis epidemiology, risk factors, and antifungal susceptibility in a tertiary care hospital in China. Sci. Rep. 2025, 15, 7240. [Google Scholar] [CrossRef]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The changing epidemiology of invasive fungal infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar]
- Pfaller, M.A.; Castanheira, M. Nosocomial candidiasis: Antifungal stewardship and the importance of rapid diagnosis. Med. Mycol. 2016, 54, 1–22. [Google Scholar] [CrossRef]
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Opin. Microbiol. 2020, 56, 7–15. [Google Scholar] [CrossRef]
- Romo, J.A.; Kumamoto, C.A. On commensalism of Candida. J. Fungi 2020, 6, 16. [Google Scholar] [CrossRef]
- Doi, A.M.; Pignatari, A.C.; Edmond, M.B.; Marra, A.R.; Camargo, L.F.; Siqueira, R.A.; da Mota, V.P.; Colombo, A.L. Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian National Surveillance Program. PLoS ONE 2016, 11, e0146909. [Google Scholar] [CrossRef]
- Ghrenassia, E.; Mokart, D.; Mayaux, J.; Demoule, A.; Rezine, I.; Kerhuel, L.; Calvet, L.; De Jong, A.; Azoulay, E.; Darmon, M. Candidemia in critically ill immunocompromised patients: Report of a retrospective multicenter cohort study. Ann. Intensive Care 2019, 9, 62. [Google Scholar] [CrossRef]
- Wu, P.F.; Liu, W.L.; Hsieh, M.H.; Hii, I.M.; Lee, Y.L.; Lin, Y.T.; Ho, M.W.; Liu, C.E.; Chen, Y.H.; Wang, F.D. Epidemiology and antifungal susceptibility of candidemia isolates of non-albicans Candida species from cancer patients. Emerg. Microbes Infect. 2017, 6, e87. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Kanj, S.S.; Govender, N.P.; Thompson, G.R., 3rd; Ostrosky-Zeichner, L.; Govrins, M.A. Invasive candidiasis. Nat. Rev. Dis. Primers 2024, 10, 20. [Google Scholar] [CrossRef]
- Denning, D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 24, e428–e438. [Google Scholar] [CrossRef]
- Agnelli, C.; Guimarães, T.; Sukiennik, T.; Lima, P.R.P.; Salles, M.J.; Breda, G.L.; Queiroz-Telles, F.; Chaves Magri, M.M.; Mendes, A.V.; Camargo, L.F.A.; et al. Prognostic trends and current challenges in candidemia: A comparative analysis of two multicenter cohorts within the past decade. J. Fungi 2023, 9, 468. [Google Scholar] [CrossRef]
- Keighley, C.; Kim, H.Y.; Kidd, S.; Chen, S.C.; Alastruey, A.; Dao, A.; Bongomin, F.; Chiller, T.; Wahyuningsih, R.; Forastiero, A.; et al. Candida tropicalis-A systematic review to inform the World Health Organization of a fungal priority pathogens list. Med. Mycol. 2024, 62, myae040. [Google Scholar] [CrossRef]
- de Oliveira, C.S.; Colombo, A.L.; Francisco, E.C.; de Lima, B.; Gandra, R.F.; de Carvalho, M.C.P.; Carrilho, C.M.D.M.; Petinelli, R.; Pelison, M.; Helbel, C.; et al. Clinical and epidemiological aspects of candidemia in eight medical centers in the state of Parana, Brazil: Parana Candidemia Network. Braz. J. Infect. Dis. 2021, 25, 101041. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.Z.; Alam, Q.; Jiman-Fatani, A.; Kamal, M.A.; Abuzenadah, A.M.; Chaudhary, A.G.; Akram, M.; Haque, A. Candida identification: A journey from conventional to molecular methods in medical mycology. World J. Microbiol. Biotechnol. 2014, 30, 1437–1451. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Nguyen, M.H. Diagnosing invasive candidiasis. J. Clin. Microbiol. 2018, 56, e01909-17. [Google Scholar] [CrossRef] [PubMed]
- Neppelenbroek, K.H.; Seó, R.S.; Urban, V.M.; Silva, S.; Dovigo, L.N.; Jorge, J.H.; Campanha, N.H. Identification of Candida species in the clinical laboratory: A review of conventional, commercial, and molecular techniques. Oral Dis. 2014, 20, 329–344. [Google Scholar] [CrossRef]
- Higashi, Y.; Niimi, H.; Sakamaki, I.; Yamamoto, Y.; Kitajima, I. Rapid identification of Candida species in candidemia directly from blood samples using imperfect match probes. Sci. Rep. 2020, 10, 5828. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Rapid and accurate identification of Candida albicans and Candida dubliniensis by Real-Time PCR and melting curve analysis. Med. Princ. Pract. 2018, 27, 543–548. [Google Scholar] [CrossRef]
- Alkhars, N.; Al Jallad, N.; Wu, T.T.; Xiao, J. Multilocus sequence typing of Candida albicans oral isolates reveals high genetic relatedness of mother-child dyads in early life. PLoS ONE 2024, 19, e0290938. [Google Scholar] [CrossRef]
- Wengenack, N.L.; Binnicker, M.J. Fungal molecular diagnostics. Clin. Chest Med. 2009, 30, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Torres-Machorro, A.L.; Hernández, R.; Cevallos, A.M.; López-Villaseñor, I. Ribosomal RNA genes in eukaryotic microorganisms: Witnesses of phylogeny? FEMS Microbiol. Rev. 2010, 34, 59–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Eisner, J.D.; Kattar, M.M.; Rassoulian-Barrett, S.L.; LaFe, K.; Yarfitz, S.L.; Limaye, A.P.; Cookson, B.T. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 2000, 38, 2302–2310. [Google Scholar] [CrossRef]
- Turenne, C.Y.; Sanche, S.E.; Hoban, D.J.; Karlowsky, J.A.; Kabani, A.M. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 1999, 37, 1846–1851, Erratum in: J. Clin. Microbiol. 2000, 38, 944.. [Google Scholar] [CrossRef]
- Gharizadeh, B.; Norberg, E.; Löffler, J.; Jalal, S.; Tollemar, J.; Einsele, H.; Klingspor, L.; Nyrén, P. Identification of medically important fungi by the Pyrosequencing technology. Mycoses 2004, 47, 29–33. [Google Scholar] [CrossRef]
- De Llanos Frutos, R.; Fernández-Espinar, M.T.; Querol, A. Identification of species of the genus Candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Antonie Van Leeuwenhoek 2004, 85, 175–185. [Google Scholar] [CrossRef]
- Einsele, H.; Hebart, H.; Roller, G.; Löffler, J.; Rothenhofer, I.; Müller, C.A.; Bowden, R.A.; van Burik, J.; Engelhard, D.; Kanz, L.; et al. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 1997, 35, 1353–1360. [Google Scholar] [CrossRef]
- Cornet, M.; Sendid, B.; Fradin, C.; Gaillardin, C.; Poulain, D.; Nguyen, H.V. Molecular identification of closely related Candida species using two ribosomal intergenic spacer fingerprinting methods. J. Mol. Diagn. 2011, 13, 12–22. [Google Scholar] [CrossRef]
- Cebeci Güler, N.; Tosun, I.; Aydin, F. The identification of Meyerozyma guilliermondii from blood cultures and surveillance samples in a university hospital in Northeast Turkey: A ten-year survey. J. Mycol. Med. 2017, 27, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Gabaldón, T.; Consortium OPATHY. Recent Trends in Molecular Diagnostics of Yeast Infections: From PCR to NGS. FEMS Microbiol. Rev. 2019, 43, 517–547. [Google Scholar] [CrossRef]
- World Health Organization. Chapter 6: Data Analysis. In Measles and Rubella Laboratory Network Manual; WHO: Geneva, Switzerland, 2018; Available online: https://www.who.int/docs/default-source/immunization/vpd_surveillance/lab_networks/measles_rubella/manual/chapter-6.pdf (accessed on 11 July 2025).
- Pinhata, J.M.; Cergole-Novella, M.C.; Carmo, A.M.S.; Silva, R.R.F.; Ferrazoli, L.; Sacchi, C.T.; Oliveira, R.S. Rapid Detection of Mycobacterium tuberculosis Complex by Real-Time PCR in Sputum Samples and Its Use in the Routine Diagnosis of a Reference Laboratory. J. Med. Microbiol. 2015, 64, 1040–1045. [Google Scholar] [CrossRef]
- Liu, W.; Lee, L.P. Toward rapid and accurate molecular diagnostics at home. Adv. Mater. 2023, 35, e2206525. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Wickes, B. Fungal diagnostics. Cold Spring Harb. Perspect. Med. 2014, 4, a019299. [Google Scholar] [CrossRef] [PubMed]
- Sidiq, F.; Hoostal, M.; Rogers, S.O. Rapid identification of fungi in culture-negative clinical blood and respiratory samples by DNA sequence analyses. BMC Res. Notes 2016, 9, 293. [Google Scholar] [CrossRef]
- Garcia Garces, H.; Hrycyk, M.F.; Giacobino, J.; Capela Machado, G.; Domingos Arantes, T.; Theodoro, R.C.; Bosco, S.M.G.; Bagagli, E. Molecular identification and phylogenetical analysis of dermatophyte fungi from Latin America. Mycoses 2016, 59, 787–797. [Google Scholar] [CrossRef]
- Trabelsi, H.; Neji, S.; Hadrich, I.; Khemakhem, N.; Sellami, H.; Makni, F.; Ayadi, A. Contribution of the internal transcribed spacer regions to the detection and identification of human fungal pathogens. Curr. Res. Transl. Med. 2019, 67, 100–106. [Google Scholar] [CrossRef]
- Wickes, B.L.; Wiederhold, N.P. Molecular diagnostics in medical mycology. Nat. Commun. 2018, 9, 5135. [Google Scholar] [CrossRef]
- Tavares, E.R.; Azevedo, C.S.; Panagio, L.A.; Pelisson, M.; Pinge-Filho, P.; Venancio, E.J.; Barros, T.F.; Yamada-Ogatta, S.F.; Yamauchi, L.M. Accurate and sensitive real-time PCR assays using intergenic spacer 1 region to differentiate Cryptococcus gattii sensu lato and Cryptococcus neoformans sensu lato. Med. Mycol. 2016, 54, 89–96. [Google Scholar]
- Bidaud, A.L.; Chowdhary, A.; Dannaoui, E. Candida auris: An emerging drug resistant yeast—A mini-review. J. Mycol. Med. 2018, 28, 568–573. [Google Scholar] [CrossRef]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Tsay, S.; Kallen, A.; Jackson, B.R.; Chiller, T.M.; Vallabhaneni, S. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin. Infect. Dis. 2018, 66, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Granada, M.; Cook, E.; Sherlock, G.; Rosenzweig, F. Microbe profile: Candida glabrata—A master of deception. Microbiology 2024, 170, 001518. [Google Scholar] [CrossRef]
- Dai, Z.; Lan, X.; Cai, M.; Liao, Y.; Zhang, J.; Ye, N.; Lu, X.; Wang, J.; Xiao, Y.; Zhang, Y.; et al. Nineteen years retrospective analysis of epidemiology, antifungal resistance and a nomogram model for 30-day mortality in nosocomial candidemia patients. Front. Cell. Infect. Microbiol. 2025, 15, 1504866. [Google Scholar] [CrossRef]
- Peixoto, P.H.; Silva, M.L.; Portela, F.V.; da Silva, B.; Milanez, E.; de Oliveira, D.; Ribeiro, A.; de Almeida, H.; Lima-Neto, R.; Guedes, G.M.; et al. Clinical, epidemiological and laboratory features of invasive Candida parapsilosis complex infections in a brazilian pediatric reference hospital during the COVID-19 pandemic. J. Fungi 2023, 9, 844. [Google Scholar] [CrossRef]
- Lima, R.; Ribeiro, F.C.; Colombo, A.L.; de Almeida, J.N., Jr. The emerging threat antifungal-resistant Candida tropicalis in humans, animals, and environment. Front. Fungal Biol. 2022, 3, 957021. [Google Scholar] [CrossRef]
- Otaguiri, E.S.; Morguette, A.E.B.; Morey, A.T.; Tavares, E.R.; Kerbauy, G.; de Almeida Torres, R.S.L.; Chaves Júnior, M.; Tognim, M.C.B.; Góes, V.M.; Krieger, M.A.; et al. Development of a melting-curve based multiplex real-time PCR assay for simultaneous detection of Streptococcus agalactiae and genes encoding resistance to macrolides and lincosamides. BMC Pregn. Childbirth 2018, 18, 126. [Google Scholar] [CrossRef]
- Van Bergen, K.J.M.; Stuitje, A.R.; Akkers, R.C.; Vermeer, H.J.; Castel, R.; Mank, T.G. Performance of a novel melting curve-based qPCR assay for malaria parasites in routine clinical practice in non-endemic setting. Malar. J. 2023, 22, 191. [Google Scholar] [CrossRef]
- Wan, Z.; Zhang, Y.; He, Z.; Liu, J.; Lan, K.; Hu, Y.; Zhang, C. A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four Human Coronaviruses. Int. J. Mol. Sci. 2016, 17, 1880. [Google Scholar] [CrossRef] [PubMed]
- Tavares, E.R.; de Lima, T.F.; Bartolomeu-Gonçalves, G.; de Castro, I.M.; de Lima, D.G.; Borges, P.H.G.; Nakazato, G.; Kobayashi, R.K.T.; Venancio, E.J.; Tarley, C.R.T.; et al. Development of a melting-curve-based multiplex real-time PCR assay for the simultaneous detection of viruses causing respiratory infection. Microorganisms 2023, 11, 2692. [Google Scholar] [CrossRef] [PubMed]
- Dubey, D.; Rath, S.; Subhadarshini, S.S.; Purohit, G.K.; Tripathy, D.; Panigrahi, R.; Palai, S.; Singhsamanta, D. Exploring rapid molecular methods for diagnosing Candida species infecting humans: A narrative review. Med. Infect. Dis. 2023, 5, 336–346. [Google Scholar] [CrossRef]
- Maffezzoli, P.; Kestler, M.; Burillo, A.; Corcione, S.; De Rosa, F.G.; Muñoz, P.; Bouza, E. Diagnostic and prognostic value of time to positivity in blood cultures. An opinion paper. Rev. Esp. Quimioter. 2025, 38, 8–20. [Google Scholar] [CrossRef]
- Zacharioudakis, I.M.; Zervou, F.N.; Mylonakis, E. T2 Magnetic Resonance Assay: Overview of Available Data and Clinical Implications. J. Fungi 2018, 4, 45. [Google Scholar] [CrossRef]
- Zervou, F.N.; Zacharioudakis, I.M.; Kurpewski, J.; Mylonakis, E. T2 Magnetic Resonance for Fungal Diagnosis. Methods Mol. Biol. 2017, 1508, 305–319. [Google Scholar]
- Thermo Fisher Scientific. Creating Standard Curves with Genomic DNA or Plasmid DNA Templates for Use in Quantitative PCR; Application Note; Thermo Fisher Scientific: Waltham, MA, USA, 2008; Available online: https://assets.thermofisher.com/TFS-Assets/LSG/Application-Notes/cms_042486.pdf (accessed on 10 July 2025).
| Species | Target | Primers (5′ → 3′) | Amplicon Size (pb) | Annealing Temperature (°C) | |
|---|---|---|---|---|---|
| C. albicans | IGS2 | CaNTS2-F | GCCTCAATTTGAACGTGGTACTGC | 302 | 62 |
| CaNTS2-R | TAGCCAAACCAACCATTACGGGTG | ||||
| C. auris | ITS1 | CauITS1-F | GGATTTTAAAACTAACCCAACG | 240 | 62 |
| CauITS1-R | TTTTGTGAATGCAACGCC | ||||
| C. parapsilosis | IGS2 | CpNTS2-F | CCCTGATGCCACCAACACC | 218 | 62 |
| CpNTS2-R | GCTAGAGCGTCGTTGTAAGAAG | ||||
| C. tropicalis | IGS2 | CtNTS2-F | GGGCGTAGAATTCGATGGGAGTGA | 302 | 62 |
| CtNTS2-R | GACACTTGGGAGGGGCTTACTAGAG | ||||
| N. glabratus | IGS2 | NgNTS2-F | AGTACCCCCGGACCGAGCTT | 295 | 62 |
| NgNTS2-R | CGTGCGACGGCACACGTTTT | ||||
| Ribonuclease P [34,35] | RNaseP-F | AGATTTGGACCTGCGAGCG | 65 | 62 | |
| RNaseP-R | GAGCGGCTGTCTCCACAAGT | ||||
| Strains/Isolates | Origin | CaNTS2 | CauNTS2 | CpNTS2 | CtNTS2 | NgNTS2 |
|---|---|---|---|---|---|---|
| Positive control | Plasmid | + | + | + | + | + |
| Candida albicans ATCC 26790 | Fiocruz a | + | − | − | − | − |
| C. albicans ATCC 10231 | Fiocruz a | + | − | − | − | − |
| C. albicans ATCC 14053 | BioMMLab b | + | − | − | − | − |
| C. albicans 4 | BioMMLab b | + | − | − | − | − |
| C. albicans 5 | BioMMLab b | + | − | − | − | − |
| C. albicans 8 | BioMMLab b | + | − | − | − | − |
| Candida auris CBS 10913 | LQA c | − | + | − | − | − |
| C. auris CBS 12766 | LQA c | − | + | − | − | − |
| Candida parapsilosis ATCC 22019 | Fiocruz a | − | − | + | − | − |
| C. parapsilosis ATCC 1975 | Fiocruz a | − | − | + | − | − |
| C. parapsilosis 41 | BioMMLab b | − | − | + | − | − |
| C. parapsilosis 42 | BioMMLab b | − | − | + | − | − |
| C. parapsilosis 43 | BioMMLab b | − | − | + | − | − |
| Candida tropicalis ATCC 28707 | Fiocruz a | − | − | − | + | − |
| C. tropicalis 12 | BioMMLab b | − | − | − | + | − |
| C. tropicalis 13 | BioMMLab b | − | − | − | + | − |
| C. tropicalis 14 | BioMMLab b | − | − | − | + | − |
| Nakaseomyces glabratus * ATCC 2001 | Fiocruz a | − | − | − | − | + |
| N. glabratus ATCC 518 | Fiocruz a | − | − | − | − | + |
| N. glabratus 23 | BioMMLab b | − | − | − | − | + |
| N. glabratus 24 | BioMMLab b | − | − | − | − | + |
| N. glabratus 26 | BioMMLab b | − | − | − | − | + |
| Candida bracarensis PT1217 | UMinho d | − | − | − | − | − |
| Candida dubliniensis ATCC 974 | Fiocruz a | − | − | − | − | − |
| C. dubliniensis 83C | BioMMLab b | − | − | − | − | − |
| Candida guilliermondii PT822 | UMinho d | − | − | − | − | − |
| Candida kefyr 1 | BioMMLab b | − | − | − | − | − |
| Candida lusitaneae PT1007 | UMinho d | − | − | − | − | − |
| Candida metapsilosis PT2263 | UMinho d | − | − | − | − | − |
| Candida orthopsilosis PT2259 | UMinho d | − | − | − | − | − |
| Aspergillus flavus | CMRP/UEL e | − | − | − | − | − |
| Aspergillus fumigatus | CMRP/UEL e | − | − | − | − | − |
| Aspergillus niger | CMRP/UEL e | − | − | − | − | − |
| Aspergillus terreus | CMRP/UEL e | − | − | − | − | − |
| Cryptococcus gattii ATCC 24065 | Fiocruz a | − | − | − | − | − |
| C. gattii ATCC 56990 | Fiocruz a | − | − | − | − | − |
| C. gattii ATCC 24066 | Fiocruz a | − | − | − | − | − |
| Cryptococcus neoformans ATCC 34872 | Fiocruz a | − | − | − | − | − |
| C. neoformans ATCC 66031 | Fiocruz a | − | − | − | − | − |
| Hismiddlelasma capsulatum | EJV/UEL f | − | − | − | − | − |
| Issatchenkia orientalis ATCC 6258 | BioMMLab b | − | − | − | − | − |
| Paracoccidiodes brasiliensis 18 | EJV/UEL f | − | − | − | − | − |
| Pichia kudriavzevii ** ATCC 34135 | Fiocruz a | − | − | − | − | − |
| P. kudriavzevii ATCC 520 | Fiocruz a | − | − | − | − | − |
| P. kudriavzevii 36 | BioMMLab b | − | − | − | − | − |
| P. kudriavzevii 42 | BioMMLab b | − | − | − | − | − |
| P. kudriavzevii 39 | BioMMLab b | − | − | − | − | − |
| Sporothrix sp. | EJV/UEL f | − | − | − | − | − |
| Trichophyton rubrum | EJV/UEL f | − | − | − | − | − |
| Enterococcus faecium ATCC 6569 | Fiocruz b | − | − | − | − | − |
| Enterococcus faecalis ATCC 29212 | Fiocruz b | − | − | − | − | − |
| Staphylococcus aureus BEC 9393 | Fiocruz b | − | − | − | − | − |
| S. aureus MRSA PSA 598 | BioMMLab b | − | − | − | − | − |
| S. aureus MRSA PSA 149 | BioMMLab b | − | − | − | − | − |
| Staphylococcus epidermidis ATCC 35984 | Fiocruz a | − | − | − | − | − |
| Staphylococcus haemolyticus ATCC 29968 | Fiocruz a | − | − | − | − | − |
| Pseudomonas aeroginosa PA01 | Fiocruz a | − | − | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavares, E.R.; Santos, V.P.; Castro, I.M.d.; Borges, P.H.G.; Silva-Rodrigues, G.; Olak, A.P.S.; Bartolomeu-Gonçalves, G.; Santos, J.P.; Morey, A.T.; Ishida, K.; et al. Development of Melting-Curve-Based Real-Time PCR for Differentiating Medically Important Candida Species. Int. J. Mol. Sci. 2025, 26, 9411. https://doi.org/10.3390/ijms26199411
Tavares ER, Santos VP, Castro IMd, Borges PHG, Silva-Rodrigues G, Olak APS, Bartolomeu-Gonçalves G, Santos JP, Morey AT, Ishida K, et al. Development of Melting-Curve-Based Real-Time PCR for Differentiating Medically Important Candida Species. International Journal of Molecular Sciences. 2025; 26(19):9411. https://doi.org/10.3390/ijms26199411
Chicago/Turabian StyleTavares, Eliandro Reis, Virginia Prezzi Santos, Isabela Madeira de Castro, Paulo Henrique Guilherme Borges, Gislaine Silva-Rodrigues, Anna Paula Silva Olak, Guilherme Bartolomeu-Gonçalves, Jussevania Pereira Santos, Alexandre Tadachi Morey, Kelly Ishida, and et al. 2025. "Development of Melting-Curve-Based Real-Time PCR for Differentiating Medically Important Candida Species" International Journal of Molecular Sciences 26, no. 19: 9411. https://doi.org/10.3390/ijms26199411
APA StyleTavares, E. R., Santos, V. P., Castro, I. M. d., Borges, P. H. G., Silva-Rodrigues, G., Olak, A. P. S., Bartolomeu-Gonçalves, G., Santos, J. P., Morey, A. T., Ishida, K., Yamada-Ogatta, S. F., & Yamauchi, L. M. (2025). Development of Melting-Curve-Based Real-Time PCR for Differentiating Medically Important Candida Species. International Journal of Molecular Sciences, 26(19), 9411. https://doi.org/10.3390/ijms26199411







