The Role of CRABS CLAW Transcription Factor in Floral Organ Development in Plants
Abstract
1. Introduction
2. Structural Features of the CRC Protein
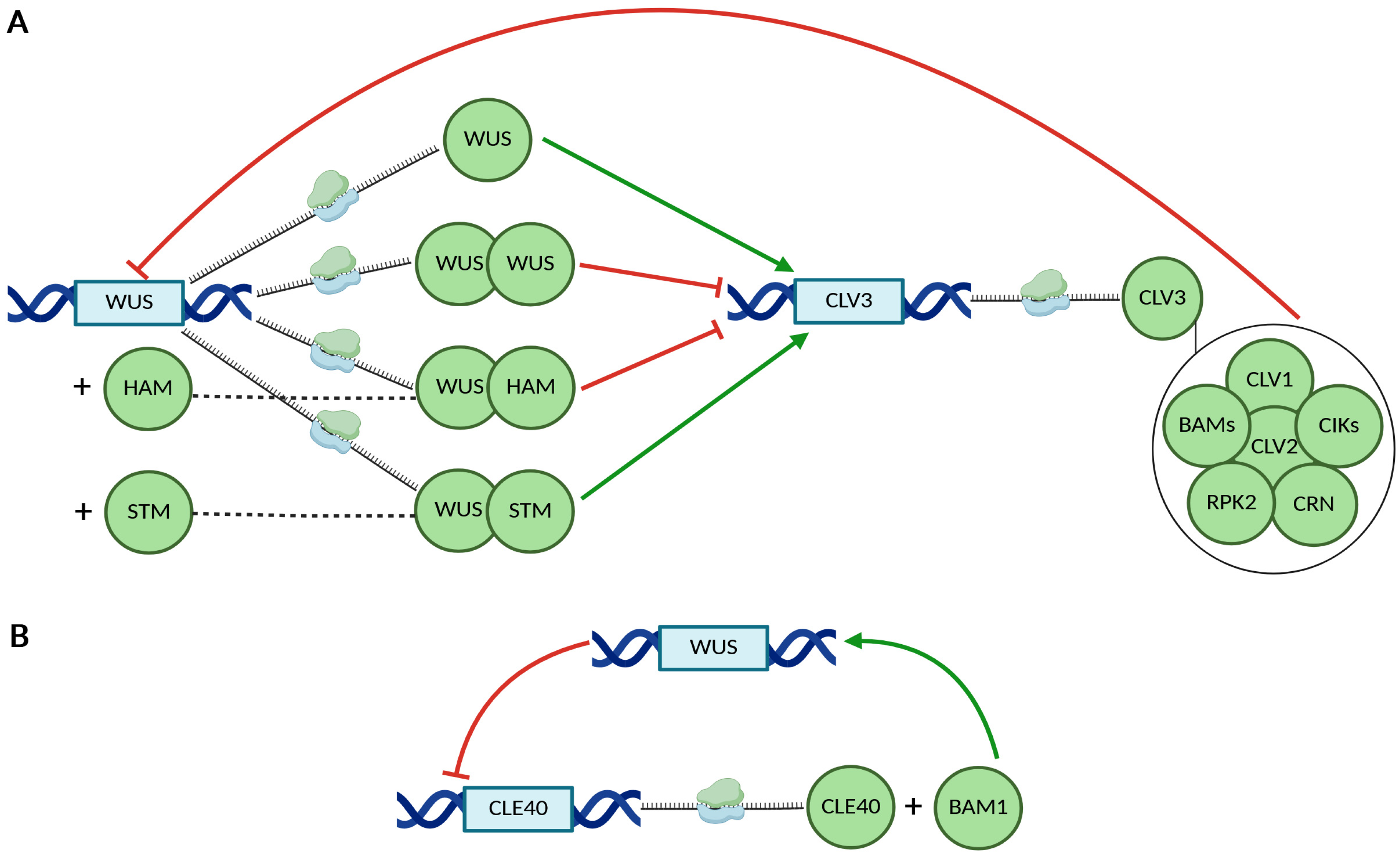
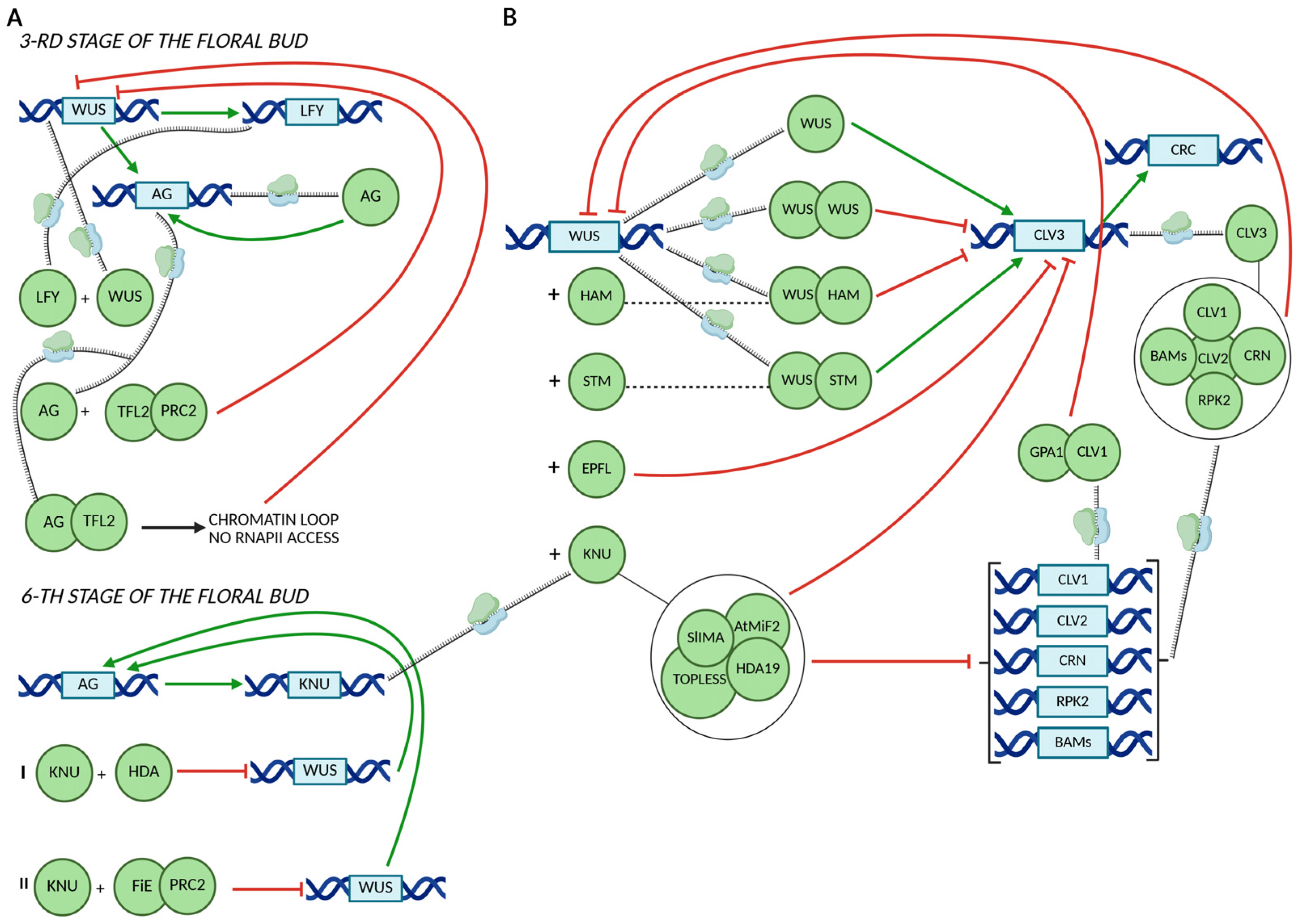
3. CRC in Floral Meristem Termination
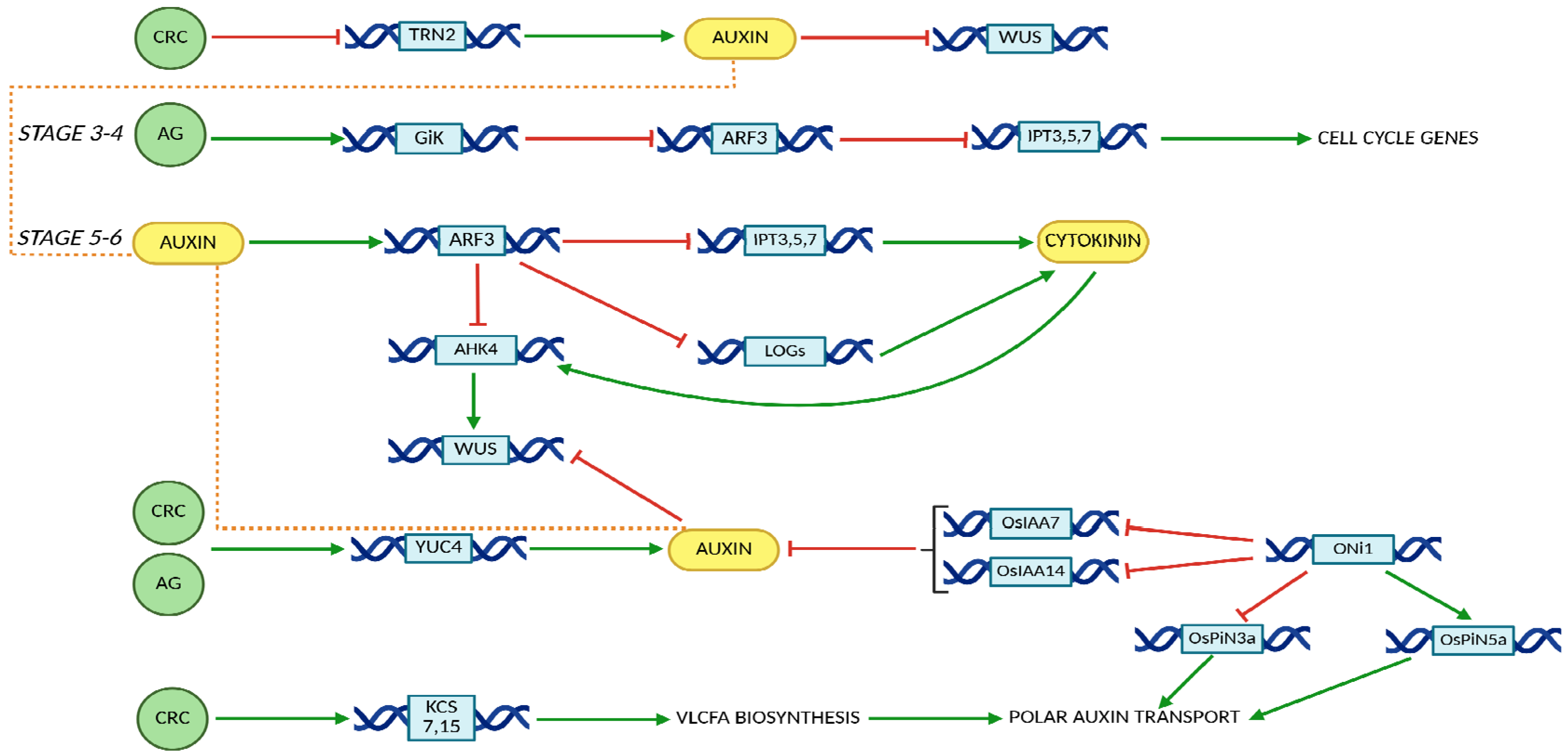
4. CRC in Auxin/Cytokinin Crosstalk
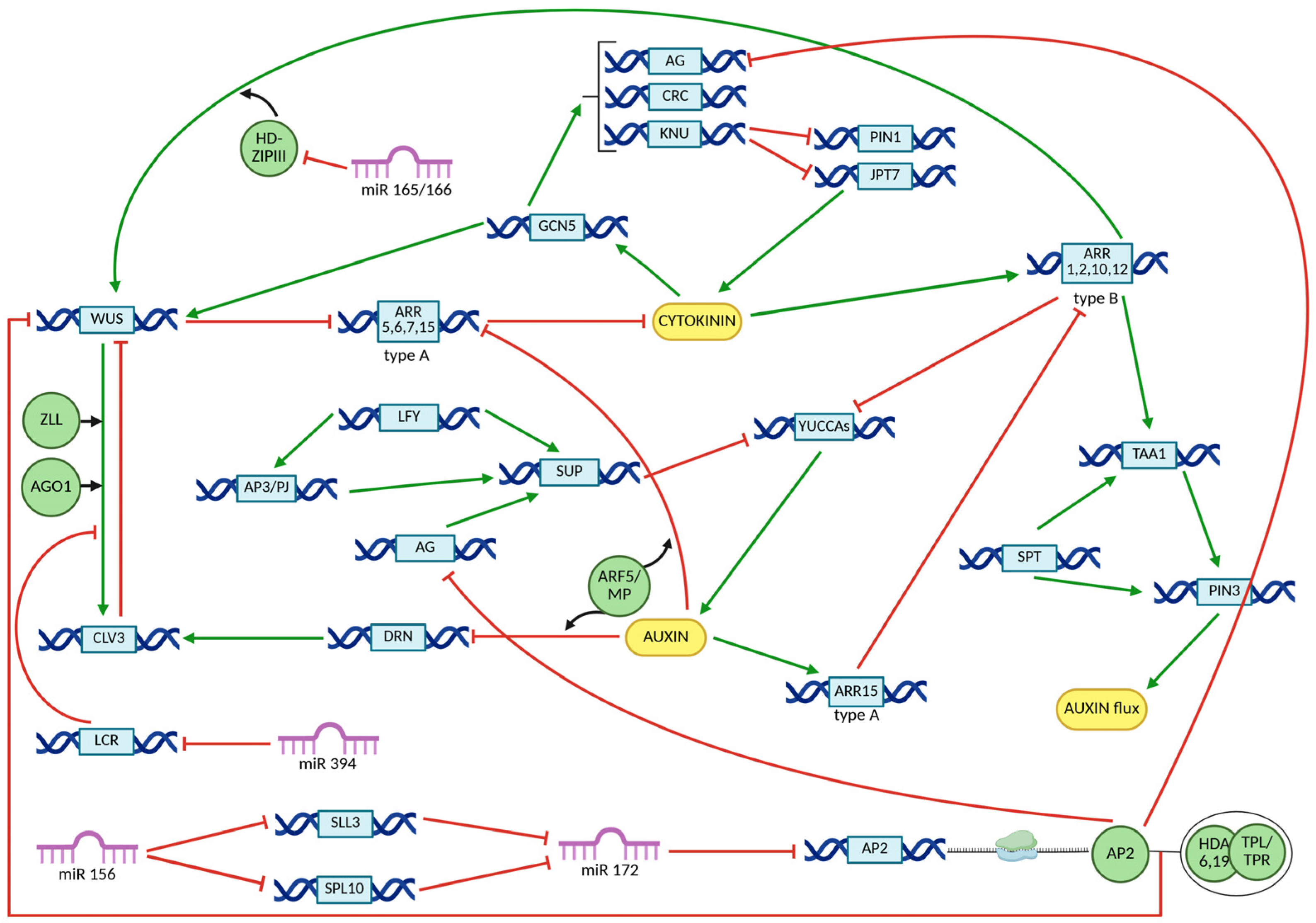
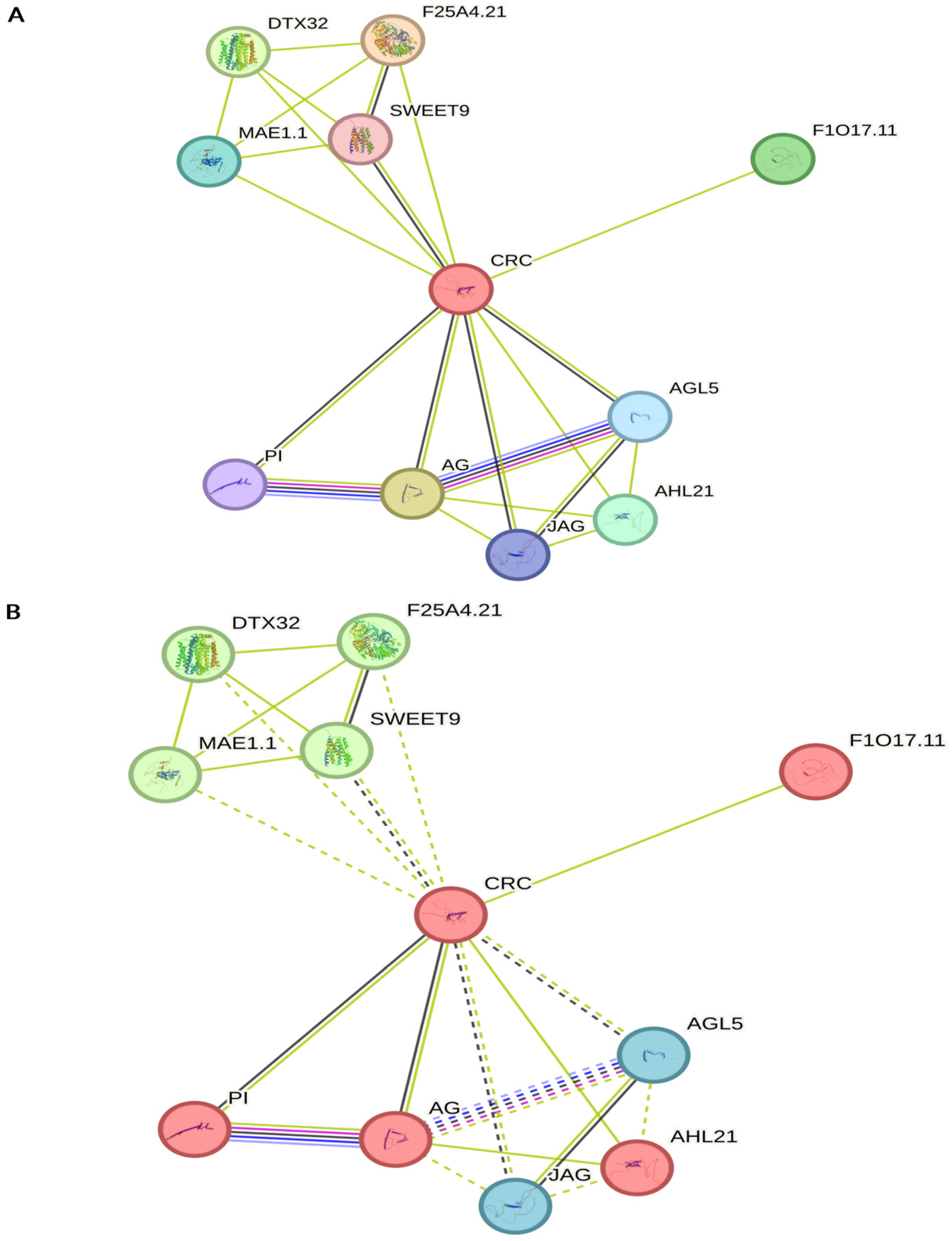
5. CRC Interaction Networks
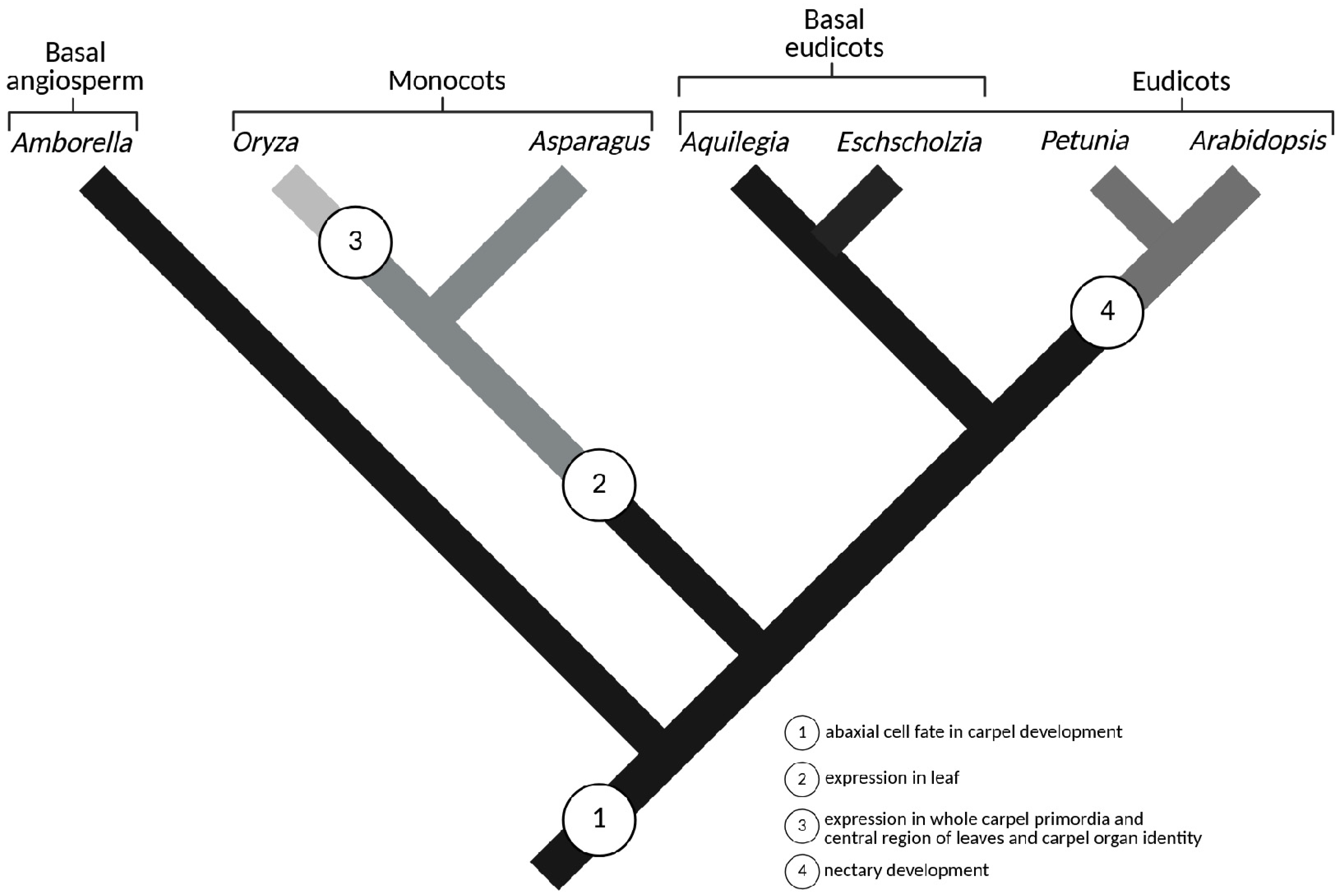
6. Evolutionary Diversification of CRC Gene
7. Polymorphisms and Natural Variation of the CRC Gene
8. Outlook
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Orashakova, S.; Lange, M.; Lange, S.; Wege, S.; Becker, A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 2009, 58, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Thong, Z.; Yu, H. Coming into bloom: The specification of floral meristems. Development 2009, 136, 3379–3391. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, L.; Giménez, E.; Pineda, B.; García-Sogo, B.; Ortiz-Atienza, A.; Micol-Ponce, R.; Angosto, T.; Capel, J.; Moreno, V.; Yuste-Lisbona, F.J.; et al. Tomato CRABS CLAW paralogues interact with chromatin remodelling factors to mediate carpel development and floral determinacy. New Phytol. 2022, 234, 1059–1074. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef] [PubMed]
- Golz, J.F.; Hudson, A. Plant development: YABBYs claw to the fore. Curr. Biol. 1999, 9, R861–R863. [Google Scholar] [CrossRef]
- Han, K.; Lai, M.; Zhao, T.; Yang, X.; An, X.; Chen, Z. Plant YABBY transcription factors: A review of gene expression, biological functions, and prospects. Crit. Rev. Biotechnol. 2025, 45, 214–235. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef]
- Ohmori, Y.; Toriba, T.; Nakamura, H.; Ichikawa, H.; Hirano, H.Y. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J. 2011, 65, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Abiko, M.; Ohmori, Y.; Hirano, H.Y. Genome-wide expression profiling and identification of genes under the control of the DROOPING LEAF gene during midrib development in rice. Genes Genet. Syst. 2008, 83, 237–244. [Google Scholar] [CrossRef]
- Duan, X.; Xie, W.; Chen, X.; Zhang, H.; Zhao, T.; Huang, J.; Zhang, R.; Li, X. Morphological and molecular mechanisms of floral nectary development in Chinese Jujube. BMC Plant Biol. 2024, 24, 1041. [Google Scholar] [CrossRef]
- Lee, J.Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLAW in the Nectaries and Carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef]
- Gross, T.; Becker, A. Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis. Genes 2021, 12, 1663. [Google Scholar] [CrossRef] [PubMed]
- Shang, E.; Wang, X.; Li, T.; Guo, F.; Ito, T.; Sun, B. Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc. Natl. Acad. Sci. USA 2021, 118, e2102826118. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhou, Y.; Cai, J.; Shang, E.; Yamaguchi, N.; Xiao, J.; Looi, L.S.; Wee, W.Y.; Gao, X.; Wagner, D.; et al. Integration of Transcriptional Repression and Polycomb-Mediated Silencing of WUSCHEL in Floral Meristems. Plant Cell 2019, 31, 1488–1505. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Huang, J.; Tatsumi, Y.; Abe, M.; Sugano, S.S.; Kojima, M.; Takebayashi, Y.; Kiba, T.; Yokoyama, R.; Nishitani, K.; et al. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 2018, 9, 5290. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Huang, J.; Xu, Y.; Tanoi, K.; Ito, T. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Herrera-Ubaldo, H.; Campos, S.E.; López-Gómez, P.; Luna-García, V.; Zúñiga-Mayo, V.M.; Armas-Caballero, G.E.; González-Aguilera, K.L.; DeLuna, A.; Marsch-Martínez, N.; Espinosa-Soto, C.; et al. The protein-protein interaction landscape of transcription factors during gynoecium development in Arabidopsis. Mol. Plant 2023, 16, 260–278. [Google Scholar] [CrossRef]
- Müller, C.J.; Larsson, E.; Spíchal, L.; Sundberg, E. Cytokinin-Auxin Crosstalk in the Gynoecial Primordium Ensures Correct Domain Patterning. Plant Physiol. 2017, 175, 1144–1157. [Google Scholar] [CrossRef]
- Kidokoro, S.; Konoura, I.; Soma, F.; Suzuki, T.; Miyakawa, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in Arabidopsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2216183120. [Google Scholar] [CrossRef]
- Pfannebecker, K.C.; Lange, M.; Rupp, O.; Becker, A. Seed Plant-Specific Gene Lineages Involved in Carpel Development. Mol. Biol. Evol. 2017, 34, 925–942. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Gómez-Felipe, A.; Herrera-Ubaldo, H.; de Folter, S. Gynoecium development: Networks in Arabidopsis and beyond. J. Exp. Bot. 2019, 70, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Scutt, C.P.; Vinauger-Douard, M.; Fourquin, C.; Finet, C.; Dumas, C. An evolutionary perspective on the regulation of carpel development. J. Exp. Bot. 2006, 57, 2143–2152. [Google Scholar] [CrossRef]
- Yamada, T.; Yokota, S.; Hirayama, Y.; Imaichi, R.; Kato, M.; Gasser, C.S. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 2011, 67, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Yamaguchi, T.; Tsukaya, H. Expression patterns of AaDL, a CRABS CLAW ortholog in Asparagus asparagoides (Asparagaceae), demonstrate a stepwise evolution of CRC/DL subfamily of YABBY genes. Am. J. Bot. 2010, 97, 591–600. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW Acts as a Bifunctional Transcription Factor in Flower Development. Front. Plant Sci. 2018, 9, 835. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Dumas, R.; Turchi, L.; Lucas, J.; Parcy, F. Plant-TFClass: A structural classification for plant transcription factors. Trends Plant Sci. 2024, 29, 40–51. [Google Scholar] [CrossRef]
- Bobb, A.J.; Eiben, H.G.; Bustos, M.M. PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. Plant J. 1995, 8, 331–343. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 1989, 245, 371–378. [Google Scholar] [CrossRef]
- Stemmer, C.; Ritt, C.; Igloi, G.L.; Grimm, R.; Grasser, K.D. Variability in Arabidopsis thaliana chromosomal high-mobility-group-1-like proteins. Eur. J. Biochem. 1997, 250, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Romanova, M.A.; Maksimova, A.I.; Pawlowski, K.; Voitsekhovskaja, O.V. YABBY Genes in the Development and Evolution of Land Plants. Int. J. Mol. Sci. 2021, 22, 4139. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef]
- Cross, E.M.; Akbari, N.; Ghassabian, H.; Hoad, M.; Pavan, S.; Ariawan, D.; Donnelly, C.M.; Lavezzo, E.; Petersen, G.F.; Forwood, J.K.; et al. A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin α-dependent nuclear localization signals. Protein Sci. 2024, 33, e4876. [Google Scholar] [CrossRef] [PubMed]
- Nakada, R.; Hirano, H.; Matsuura, Y. Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep. 2015, 5, 15055. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Shang, E.; Ito, T.; Sun, B. Control of floral stem cell activity in Arabidopsis. Plant Signal Behav. 2019, 14, 1659706. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Kumar, H.; Mahajan, M.; Sahu, S.K.; Singh, S.K.; Yadav, R.K. Local auxin biosynthesis promotes shoot patterning and stem cell differentiation in Arabidopsis shoot apex. Development 2023, 150, dev202014. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, P.; Li, M.; Zhu, D.; Ma, H.; Xu, H.; Li, S.; Wei, J.; Bian, X.; Wang, M.; et al. Heat stress impairs floral meristem termination and fruit development by affecting the BR-SlCRCa cascade in tomato. Plant Commun. 2024, 5, 100790. [Google Scholar] [CrossRef]
- Kwaśniewska, K.; Breathnach, C.; Fitzsimons, C.; Goslin, K.; Thomson, B.; Beegan, J.; Finocchio, A.; Prunet, N.; Ó’Maoiléidigh, D.S.; Wellmer, F. Expression of KNUCKLES in the Stem Cell Domain Is Required for Its Function in the Control of Floral Meristem Activity in Arabidopsis. Front. Plant Sci. 2021, 12, 704351. [Google Scholar] [CrossRef]
- Guo, L.; Cao, X.; Liu, Y.; Li, J.; Li, Y.; Li, D.; Zhang, K.; Gao, C.; Dong, A.; Liu, X. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 2018, 94, 1083–1097. [Google Scholar] [CrossRef]
- ÓMaoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwasniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- Liu, Z.; Shpak, E.D.; Hong, T. A mathematical model for understanding synergistic regulations and paradoxical feedbacks in the shoot apical meristem. Comput. Struct. Biotechnol. J. 2020, 18, 3877–3889. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Liu, X.; Zhou, Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 2020, 53, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Rodriguez, K.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, E6298–E6306. [Google Scholar] [CrossRef]
- Rodriguez, K.; Kao, L.; Cerbantez-Bueno, V.E.; Delgadillo, C.; Nguyen, D.; Ullah, S.; Delgadillo, C.; Reddy, G.V. HAIRY MERISTEM proteins regulate the WUSCHEL protein levels in mediating CLAVATA3 expression. Physiol. Plant. 2024, 176, e14505. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Hu, C.; Zhu, Y.; Cui, Y.; Cheng, K.; Liang, W.; Wei, Z.; Zhu, M.; Yin, H.; Zeng, L.; Xiao, Y.; et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants. 2018, 4, 205–211. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Cell signaling in the shoot apical meristem. Plant Physiol. 2023, 193, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Shpak, E.D.; Uzair, M. WUSCHEL: The essential regulator of the Arabidopsis shoot Apical Meristem. Curr. Opin. Plant Biol. 2025, 85, 102739. [Google Scholar] [CrossRef]
- Schlegel, J.; Denay, G.; Wink, R.; Pinto, K.G.; Stahl, Y.; Schmid, J.; Blümke, P.; Simon, R.G. Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. eLife 2021, 10, 70934. [Google Scholar] [CrossRef] [PubMed]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Sieburth, L.E.; Running, M.P.; Meyerowitz, E.M. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 1995, 7, 1249–1258. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Bollier, N.; Sicard, A.; Leblond, J.; Latrasse, D.; Gonzalez, N.; Gévaudant, F.; Benhamed, M.; Raynaud, C.; Lenhard, M.; Chevalier, C.; et al. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a Conserved Missing Link in the Regulation of Floral Meristem Termination in Arabidopsis and Tomato. Plant Cell 2018, 30, 83–100. [Google Scholar] [CrossRef]
- Sun, B.; Xu, Y.; Ng, K.H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, X.; Zhou, C.; Zhou, Q.; Zhao, Y.; Li, G.; Zhou, D.X. Cooperation between the H3K27me3 Chromatin Mark and Non-CG Methylation in Epigenetic Regulation. Plant Physiol. 2016, 172, 1131–1141. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Han, L.; Huang, Y.; Li, C.; Tian, D.; She, D.; Li, M.; Wang, Z.; Chen, J.; Liu, L.; Wang, S.; et al. Heterotrimeric Gα-subunit regulates flower and fruit development in CLAVATA signaling pathway in cucumber. Hortic. Res. 2024, 11, uhae110. [Google Scholar] [CrossRef]
- Uzair, M.; Urquidi Camacho, R.A.; Liu, Z.; Overholt, A.M.; DeGennaro, D.; Zhang, L.; Herron, B.S.; Hong, T.; Shpak, E.D. An updated model of shoot apical meristem regulation by ERECTA family and CLAVATA3 signaling pathways in Arabidopsis. Development 2024, 151, dev202870. [Google Scholar] [CrossRef]
- Zhang, L.; DeGennaro, D.; Lin, G.; Chai, J.; Shpak, E.D. ERECTA family signaling constrains CLAVATA3 and WUSCHEL to the center of the shoot apical meristem. Development 2021, 148, dev189753. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 1995, 121, 2057–2067. [Google Scholar] [CrossRef]
- Zhou, L.; Iqbal, A.; Yang, M.; Yang, Y. Research Progress on Gene Regulation of Plant Floral Organogenesis. Genes 2025, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mena, C.; de Folter, S.; Costa, M.M.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Engelhorn, J.; Moreau, F.; Fletcher, J.C.; Carles, C.C. ULTRAPETALA1 and LEAFY pathways function independently in specifying identity and determinacy at the Arabidopsis floral meristem. Ann. Bot. 2014, 114, 1497–1505. [Google Scholar] [CrossRef]
- Carles, C.C.; Choffnes-Inada, D.; Reville, K.; Lertpiriyapong, K.; Fletcher, J.C. ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 2005, 132, 897–911. [Google Scholar] [CrossRef]
- Uemura, A.; Yamaguchi, N.; Xu, Y.; Wee, W.; Ichihashi, Y.; Suzuki, T.; Shibata, A.; Shirasu, K.; Ito, T. Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis. Plant Reprod. 2018, 31, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Franks, R.G.; Levin, J.Z.; Liu, Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 2004, 16, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Yan, M.; Liu, C.; Yuan, Z. BELL1 interacts with CRABS CLAW and INNER NO OUTER to regulate ovule and seed development in pomegranate. Plant Physiol. 2023, 191, 1066–1083. [Google Scholar] [CrossRef]
- Pelayo, M.A.; Yamaguchi, N.; Ito, T. One factor, many systems: The floral homeotic protein AGAMOUS and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 2021, 61, 102009. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.J.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef]
- Lai, X.; Stigliani, A.; Lucas, J.; Hugouvieux, V.; Parcy, F.; Zubieta, C. Genome-wide binding of SEPALLATA3 and AGAMOUS complexes determined by sequential DNA-affinity purification sequencing. Nucleic Acids Res. 2020, 48, 9637–9648. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 Regulates Floral Meristem Determinacy by Repressing Cytokinin Biosynthesis and Signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Reyes-Olalde, J.I.; Marsch-Martinez, N.; de Folter, S. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Front. Plant Sci. 2014, 5, 191. [Google Scholar] [CrossRef] [PubMed]
- Cerbantez-Bueno, V.E.; Zúñiga-Mayo, V.M.; Reyes-Olalde, J.I.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Marsch-Martinez, N.; de Folter, S. Redundant and Non-redundant Functions of the AHK Cytokinin Receptors During Gynoecium Development. Front. Plant Sci. 2020, 11, 568277. [Google Scholar] [CrossRef]
- Knauer, S.; Holt, A.L.; Rubio-Somoza, I.; Tucker, E.J.; Hinze, A.; Pisch, M.; Javelle, M.; Timmermans, M.C.; Tucker, M.R.; Laux, T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 2013, 24, 125–132. [Google Scholar] [CrossRef]
- Han, N.; Zhu, H.; Li, F.; Wang, M.; Tian, Z.; Wei, J.; Zhang, Z. Genome-wide identification of YABBY genes and functional characterization of CRABS CLAW (AktCRC) in flower development of Akebia trifoliata. Int. J. Biol. Macromol. 2025, 311, 143892. [Google Scholar] [CrossRef]
- Wang, G.; Wu, Z.; Sun, B. KNUCKLES regulates floral meristem termination by controlling auxin distribution and cytokinin activity. Plant Cell 2024, 37, koae312. [Google Scholar] [CrossRef] [PubMed]
- Hawar, A.; Chen, W.; Zhu, T.; Wang, X.; Liu, J.; Xiong, S.; Ito, T.; Chen, D.; Sun, B. The histone acetyltransferase GCN5 regulates floral meristem activity and flower development in Arabidopsis. Plant Cell 2025, 37, koaf135. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.Q.; Lian, H.; Zhou, C.M.; Xu, L.; Jiao, Y.; Wang, J.W. A Two-Step Model for de Novo Activation of WUSCHEL during Plant Shoot Regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef]
- Bomblies, K.; Dagenais, N.; Weigel, D. Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 1999, 216, 260–264. [Google Scholar] [CrossRef]
- Fouracre, J.P.; Poethig, R.S. Role for the shoot apical meristem in the specification of juvenile leaf identity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 10168–10177. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, T.; Zheng, B.; Yumul, R.E.; Liu, X.; You, C.; Gao, Z.; Xiao, L.; Chen, X. APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 2017, 215, 1197–1209. [Google Scholar] [CrossRef]
- Landau, U.; Asis, L.; Eshed Williams, L. The ERECTA, CLAVATA and class III HD-ZIP Pathways Display Synergistic Interactions in Regulating Floral Meristem Activities. PLoS ONE 2015, 10, e0125408. [Google Scholar] [CrossRef]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs Specify the Shoot Stem Cell Niche by Dual Regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef]
- Nowak, K.; Morończyk, J.; Grzyb, M.; Szczygieł-Sommer, A.; Gaj, M.D. miR172 Regulates WUS during Somatic Embryogenesis in Arabidopsis via AP2. Cells 2022, 11, 718. [Google Scholar] [CrossRef]
- Sakai, H.; Krizek, B.A.; Jacobsen, S.E.; Meyerowitz, E.M. Regulation of SUP expression identifies multiple regulators involved in arabidopsis floral meristem development. Plant Cell 2000, 12, 1607–1618. [Google Scholar] [CrossRef]
- Tucker, M.R.; Hinze, A.; Tucker, E.J.; Takada, S.; Jürgens, G.; Laux, T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development 2008, 135, 2839–2843. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Takasugi, T.; Ito, Y. Altered expression of auxin-related genes in the fatty acid elongase mutant oni1 of rice. Plant Signal Behav. 2011, 6, 887–888. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.H.; Yu, H.; Ito, T. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 2009, 7, e1000251. [Google Scholar] [CrossRef] [PubMed]
- Snipes, S.A.; Rodriguez, K.; DeVries, A.E.; Miyawaki, K.N.; Perales, M.; Xie, M.; Reddy, G.V. Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLoS Genet. 2018, 14, e1007351. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Gissot, L.; Beaudoin, F.; Haslam, R.; Michaelson, L.; Marion, J.; Molino, D.; Lima, A.; Bach, L.; Morin, H.; et al. Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 2010, 22, 364–375. [Google Scholar] [CrossRef]
- Han, X.; Yin, L.; Xue, H. Co-expression analysis identifies CRC and AP1 the regulator of Arabidopsis fatty acid biosynthesis. J. Integr. Plant Biol. 2012, 54, 486–499. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, A.; Liu, C.; Saeed, M.; Li, J.; Wu, Y.; Wu, Y.; Gu, H.; Yuan, J.; Wang, B.; et al. A Comprehensive Analysis In Silico of KCS Genes in Maize Revealed Their Potential Role in Response to Abiotic Stress. Plants 2024, 13, 3507. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Shamimuzzaman, M.; Vodkin, L. Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genom. 2013, 14, 477. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Carabelli, M.; Turchi, L.; Morelli, G.; Østergaard, L.; Ruberti, I.; Moubayidin, L. Coordination of biradial-to-radial symmetry and tissue polarity by HD-ZIP II proteins. Nat. Commun. 2021, 12, 4321. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Bowman, J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 1999, 99, 199–209. [Google Scholar] [CrossRef]
- Azhakanandam, S.; Nole-Wilson, S.; Bao, F.; Franks, R.G. SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 2008, 146, 1165–1181. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Mayo, V.M.; Marsch-Martínez, N.; de Folter, S. JAIBA, a class-II HD-ZIP transcription factor involved in the regulation of meristematic activity, and important for correct gynoecium and fruit development in Arabidopsis. Plant J. 2012, 71, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, S.; Sohlberg, J.J.; Long, J.A.; Fridborg, I.; Sundberg, E. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development. 2002, 129, 4707–4717. [Google Scholar] [CrossRef]
- Gong, P.; Song, C.; Liu, H.; Li, P.; Zhang, M.; Zhang, J.; Zhang, S.; He, C. Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. J. Exp. Bot. 2021, 72, 6882–6903. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Zhou, X.; Wan, Y.; Song, X.; Li, W.; Guo, W. Suppressing a β-1,3-glucanase gene expression increases the seed and fibre yield in cotton. Plant J. 2024, 120, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Meister, R.J.; Oldenhof, H.; Bowman, J.L.; Gasser, C.S. Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiol. 2005, 137, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, N.; Lan, J.; Pan, Y.; Jiang, Y.; Wu, Y.; Chen, X.; Feng, X.; Qin, G. Arabidopsis transcription factor TCP4 controls the identity of the apical gynoecium. Plant Cell 2024, 36, 2668–2688. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J.; Chavez Montes, R.A.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Gonzalez-Aguilera, K.L.; Ballester, P.; Ripoll, J.J.; Ezquer, I.; et al. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Jamil, I.; Giri, A.; Moubayidin, L. Organ symmetry establishment during gynoecium development. Curr. Opin. Plant Biol. 2025, 85, 102732. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zuñiga-Mayo, V.M.; Chávez Montes, R.A.; Marsch-Martínez, N.; de Folter, S. Inside the gynoecium: At the carpel margin. Trends Plant Sci. 2013, 18, 644–655. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Marsch-Martínez, N.; de Folter, S. The class II HD-ZIP JAIBA gene is involved in meristematic activity and important for gynoecium and fruit development in Arabidopsis. Plant Signal Behav. 2012, 11, 1501–1503. [Google Scholar] [CrossRef] [PubMed]
- Sohlberg, J.J.; Myrenås, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—Being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Robertson, D.L.; Van de Peer, Y.; Oliver, S.G. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 2008, 33, 220–229. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING database in 2025: Protein networks with directionality of regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef]
- Pfannebecker, K.C.; Lange, M.; Rupp, O.; Becker, A. An Evolutionary Framework for Carpel Developmental Control Genes. Mol. Biol. Evol. 2017, 34, 330–348. [Google Scholar] [CrossRef]
- Dreni, L.; Kater, M.M. MADS reloaded: Evolution of the AGAMOUS subfamily genes. New Phytol. 2014, 201, 717–732. [Google Scholar] [CrossRef]
- Prunet, N.; Morel, P.; Thierry, A.M.; Eshed, Y.; Bowman, J.L.; Negrutiu, I.; Trehin, C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 2008, 20, 901–919. [Google Scholar] [CrossRef]
- Phukela, B.; Geeta, R.; Das, S.; Tandon, R. Ancestral segmental duplication in Solanaceae is responsible for the origin of CRCa-CRCb paralogues in the family. Mol. Genet. Genom. 2020, 295, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Fourquin, C.; Primo, A.; Martínez-Fernández, I.; Huet-Trujillo, E.; Ferrándiz, C. The CRC orthologue from Pisum sativum shows conserved functions in carpel morphogenesis and vascular development. Ann. Bot. 2014, 114, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Ohmori, Y.; Tanaka, W.; Hirabayashi, C.; Murai, K.; Ogihara, Y.; Yamaguchi, T.; Hirano, H.Y. The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes. Genet. Syst. 2009, 84, 137–146. [Google Scholar] [CrossRef]
- Fourquin, C.; Vinauger-Douard, M.; Fogliani, B.; Dumas, C.; Scutt, C.P. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 2005, 102, 4649–4654. [Google Scholar] [CrossRef] [PubMed]
- Fourquin, C.; Vinauger-Douard, M.; Chambrier, P.; Berne-Dedieu, A.; Scutt, C.P. Functional conservation between CRABS CLAW orthologues from widely diverged angiosperms. Ann. Bot. 2007, 100, 651–657. [Google Scholar] [CrossRef]
- Lee, J.Y.; Baum, S.F.; Oh, S.H.; Jiang, C.Z.; Chen, J.C.; Bowman, J.L. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 2005, 132, 5021–5032. [Google Scholar] [CrossRef]
- Endress, P.K. The flowers in extant basal angiosperms and inferences on ancestral flowers. Int. J. Plant Sci. 2001, 162, 1111–1140. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Hsiao, Y.Y.; Li, C.I.; Yeh, C.M.; Mitsuda, N.; Yang, H.X.; Chiu, C.C.; Chang, S.B.; Liu, Z.J.; Tsai, W.C. The ancestral duplicated DL/CRC orthologs, PeDL1 and PeDL2, function in orchid reproductive organ innovation. J. Exp. Bot. 2021, 72, 5442–5461. [Google Scholar] [CrossRef]
- Morel, P.; Heijmans, K.; Ament, K.; Chopy, M.; Trehin, C.; Chambrier, P.; Rodrigues Bento, S.; Bimbo, A.; Vandenbussche, M. The Floral C-Lineage Genes Trigger Nectary Development in Petunia and Arabidopsis. Plant Cell 2018, 30, 2020–2037. [Google Scholar] [CrossRef]
- Deng, M.H.; Zhao, K.; Lv, J.H.; Huo, J.L.; Zhang, Z.Q.; Zhu, H.S.; Zou, X.X.; Wen, J.F. Flower transcriptome dynamics during nectary development in pepper (Capsicum annuum L.). Genet. Mol. Biol. 2020, 43, e20180267. [Google Scholar] [CrossRef]
- Katzer, A.M.; Wessinger, C.A.; Anaya, B.M.; Hileman, L.C. CRABS CLAW-independent floral nectary development in Penstemon barbatus. Am. J. Bot. 2025, 112, e70058. [Google Scholar] [CrossRef]
- Wei, L.; Li, C.; Duan, Y.; Qu, W.; Wang, H.; Miao, H.; Zhang, H. A SNP Mutation of SiCRC Regulates Seed Number Per Capsule and Capsule Length of cs1 Mutant in Sesame. Int. J. Mol. Sci. 2019, 20, 4056. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Gao, J.; Shen, W.; Wu, Z.; Dai, C.; Wen, J.; Yi, B.; Ma, C.; Shen, J.; Fu, T.; et al. BnaCRCs with domestication preference positively correlate with the seed-setting rate of canola. Plant J. 2022, 111, 1717–1731. [Google Scholar] [CrossRef] [PubMed]
- Shivhare, D.; Musialak-Lange, M.; Julca, I.; Gluza, P.; Mutwil, M. Removing auto-activators from yeast-two-hybrid assays by conditional negative selection. Sci. Rep. 2021, 11, 5477. [Google Scholar] [CrossRef] [PubMed]
- Strickler, S.R.; Bombarely, A.; Mueller, L.A. Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am. J. Bot. 2012, 99, 257–266. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Penin, A.A. Gene Expression Maps in Plants: Current State and Prospects. Plants 2019, 8, 309. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Qian, Q.; Lu, M.; Chen, M.; Fan, Z.; Shang, Y.; Bu, C.; Du, Z.; Song, S.; Zeng, J.; et al. PlantPan: A comprehensive multi-species plant pan-genome database. Plant J. 2025, 122, e70144. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.M.; Velásquez-Zapata, V.; Wise, R.P. Next-Generation Yeast Two-Hybrid Screening to Discover Protein-Protein Interactions. Methods Mol. Biol. 2023, 2690, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Velásquez-Zapata, V.; Elmore, J.M.; Banerjee, S.; Dorman, K.S.; Wise, R.P. Next-generation yeast-two-hybrid analysis with Y2H-SCORES identifies novel interactors of the MLA immune receptor. PLoS Comput. Biol. 2021, 17, e1008890. [Google Scholar] [CrossRef]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, J.X.; Zhu, Y.; Zhu, Y.; Liu, C.Z.; Chen, Y.Y.; Zeng, F.L.; Chen, S.; Wang, Y.C. Building an improved transcription factor-centered yeast one hybrid system to identify DNA motifs bound by protein comprehensively. BMC Plant Biol. 2023, 23, 236. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymczyk, P.; Nowak, J.; Majewska, M. The Role of CRABS CLAW Transcription Factor in Floral Organ Development in Plants. Int. J. Mol. Sci. 2025, 26, 9377. https://doi.org/10.3390/ijms26199377
Szymczyk P, Nowak J, Majewska M. The Role of CRABS CLAW Transcription Factor in Floral Organ Development in Plants. International Journal of Molecular Sciences. 2025; 26(19):9377. https://doi.org/10.3390/ijms26199377
Chicago/Turabian StyleSzymczyk, Piotr, Jadwiga Nowak, and Małgorzata Majewska. 2025. "The Role of CRABS CLAW Transcription Factor in Floral Organ Development in Plants" International Journal of Molecular Sciences 26, no. 19: 9377. https://doi.org/10.3390/ijms26199377
APA StyleSzymczyk, P., Nowak, J., & Majewska, M. (2025). The Role of CRABS CLAW Transcription Factor in Floral Organ Development in Plants. International Journal of Molecular Sciences, 26(19), 9377. https://doi.org/10.3390/ijms26199377





