Circulating MicroRNA Profiles in Pregnant South African Women with Different Types of Diabetes Mellitus
Abstract
1. Introduction
2. Results
2.1. Participant Clinical and Metabolic Characteristics
2.2. MiRNA Expression Profiling
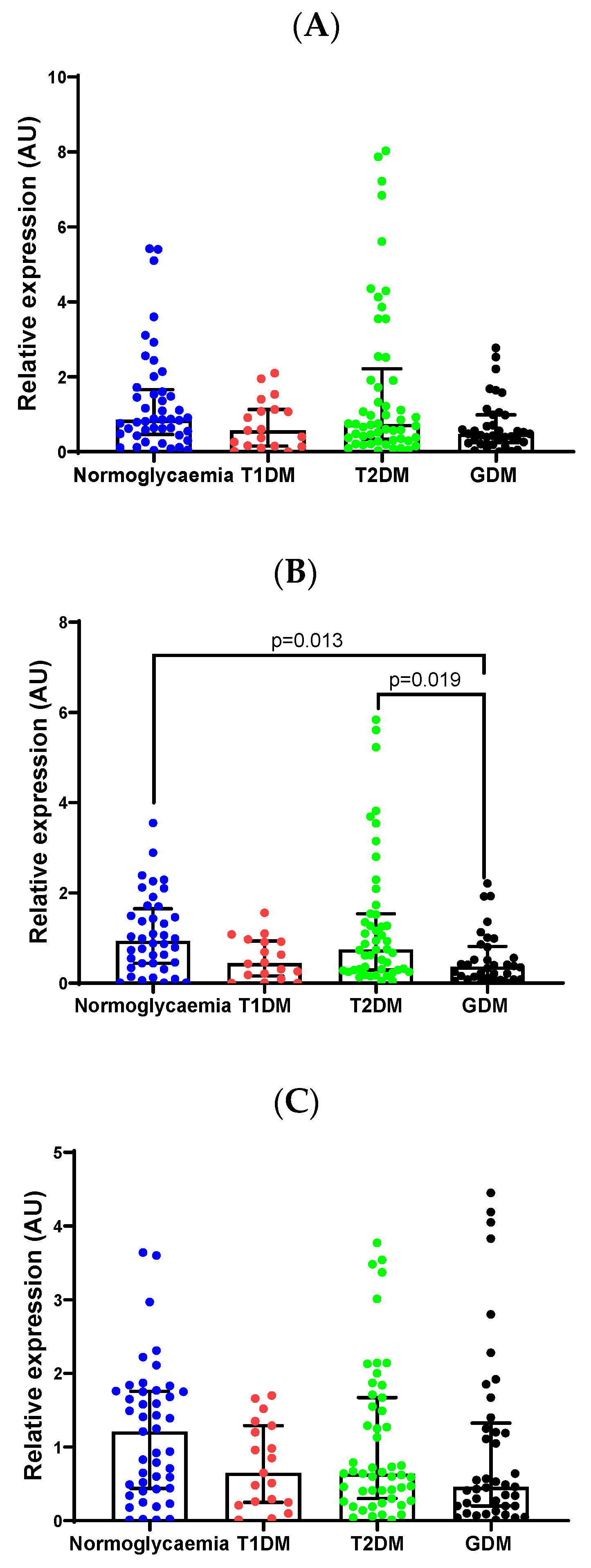
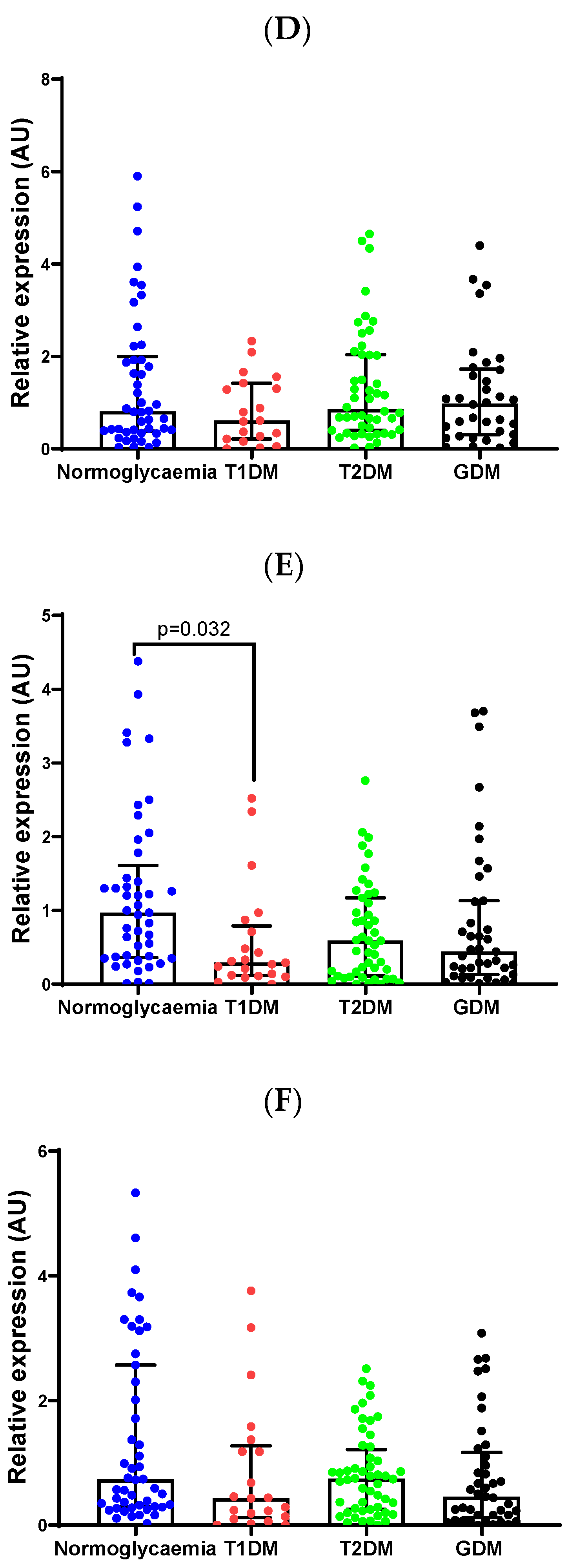
2.3. Validation of Differentially Expressed miRNAs
2.4. Association Between Clinical and Metabolic Parameters
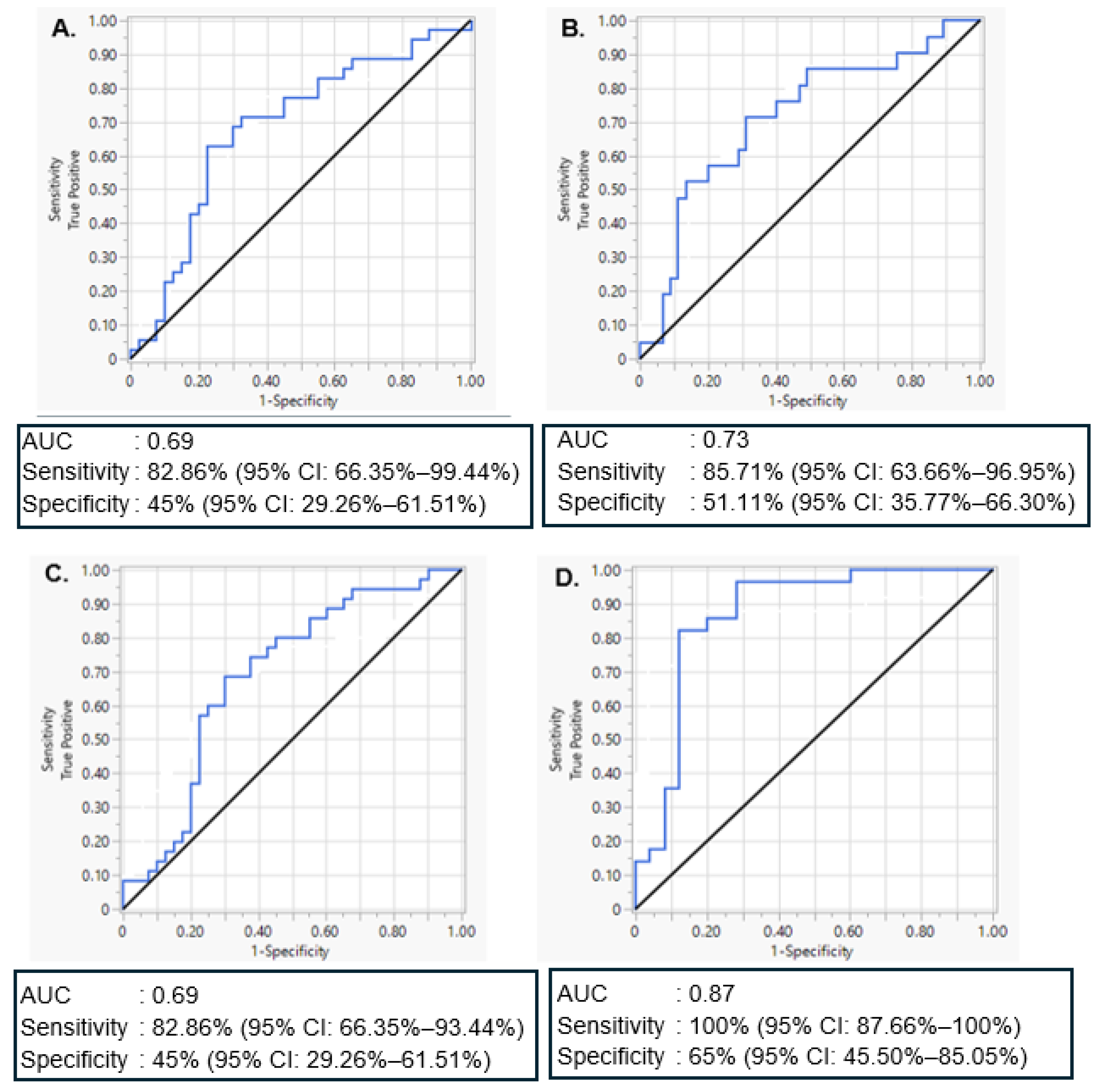
2.5. Evaluation of miR-20a-5p and miR-30d-5p Discriminatory Ability to Predict GDM and T1DM
2.6. MiRNA Gene Targets and Their Enriched Biological Pathways
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Biochemical Parameters
4.3. MiRNA Isolation
4.4. Human Serum/Plasma miScript miRNA PCR Arrays
4.5. MiRCURY LNA Individual PCR Assays
4.6. Bioinformatic Analysis
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | area under the curve |
| BMI | body mass index |
| DNA | deoxyribonucleic acid |
| ELISA | enzyme-linked immunosorbent assay |
| GDM | gestational diabetes mellitus |
| HbA1c | glycated haemoglobin |
| HIV | human immunodeficiency virus |
| IADPSG | International Association of Diabetes and Pregnancy Study Groups Consensus Panel |
| MiRNAs | MicroRNAs |
| OGTT | oral glucose tolerance test |
| PCR | polymerase chain reaction |
| P53 | tumour protein P53 |
| RNA | ribonucleic acid |
| ROC | receiver operating characteristic |
| T1DM | type 1 diabetes mellitus |
| T2DM | type 2 diabetes mellitus |
| WHO | World Health Organization |
| Wnt | Wingless-related integration site |
References
- International Diabetes Federation. IDF Diabetes Atlas 10th edition. 2021. Available online: https://diabetesatlas.org/resources/idf-diabetes-atlas-reports/ (accessed on 3 March 2022).
- Capobianco, G.; Gulotta, A.; Tupponi, G.; Dessole, F.; Pola, M.; Virdis, G.; Petrillo, M.; Mais, V.; Olzai, G.; Antonucci, R.; et al. Materno-Fetal and Neonatal Complications of Diabetes in Pregnancy: A Retrospective Study. J. Clin. Med. 2020, 9, 2707. [Google Scholar] [CrossRef] [PubMed]
- Malaza, N.; Masete, M.; Adam, S.; Dias, S.; Nyawo, T.; Pheiffer, C. A Systematic Review to Compare Adverse Pregnancy Outcomes in Women with Pregestational Diabetes and Gestational Diabetes. Int. J. Environ. Res. Public Health 2022, 19, 10846. [Google Scholar] [CrossRef] [PubMed]
- Bianco, M.E.; Josefson, J.L. Hyperglycemia During Pregnancy and Long-Term Offspring Outcomes. Curr. Diab. Rep. 2019, 19, 143. [Google Scholar] [CrossRef] [PubMed]
- Kereliuk, S.M.; Dolinsky, V.W. Recent Experimental Studies of Maternal Obesity, Diabetes during Pregnancy and the Developmental Origins of Cardiovascular Disease. Int. J. Mol. Sci. 2022, 23, 4467. [Google Scholar] [CrossRef]
- Ali, D.S.; Davern, R.; Rutter, E.; Coveney, C.; Devine, H.; Walsh, J.M.; Higgins, M.; Hatunic, M. Pre-Gestational Diabetes and Pregnancy Outcomes. Diabetes Ther. 2020, 11, 2873–2885. [Google Scholar] [CrossRef]
- Murphy, H.R.; Howgate, C.; O’Keefe, J.; Myers, J.; Morgan, M.; Coleman, M.A.; Jolly, M.; Valabhji, J.; Scott, E.M.; Knighton, P.; et al. Characteristics and Outcomes of Pregnant Women with Type 1 or Type 2 Diabetes: A 5-Year National Population-Based Cohort Study. Lancet Diabetes Endocrinol. 2021, 9, 153–164. [Google Scholar] [CrossRef]
- Yan, Y.-S.; Feng, C.; Yu, D.-Q.; Tian, S.; Zhou, Y.; Huang, Y.-T.; Cai, Y.-T.; Chen, J.; Zhu, M.-M.; Jin, M. Long-Term Outcomes and Potential Mechanisms of Offspring Exposed to Intrauterine Hyperglycemia. Front. Nutr. 2023, 10, 1067282. [Google Scholar] [CrossRef]
- Heydarzadeh, S.; Ranjbar, M.; Karimi, F.; Seif, F.; Alivand, M.R. Overview of Host miRNA Properties and Their Association with Epigenetics, Long Non-Coding RNAs, and Xeno-Infectious Factors. Cell Biosci. 2021, 11, 43. [Google Scholar] [CrossRef]
- Jang, J.H.; Lee, T.-J. The Role of microRNAs in Cell Death Pathways. Yeungnam Univ. J. Med. 2021, 38, 107–117. [Google Scholar] [CrossRef]
- Abdel Mageed, S.S.; Doghish, A.S.; Ismail, A.; El-Husseiny, A.A.; Fawzi, S.F.; Mahmoud, A.M.A.; El-Mahdy, H.A. The Role of miRNAs in Insulin Resistance and Diabetic Macrovascular Complications—A Review. Int. J. Biol. Macromol. 2023, 230, 123189. [Google Scholar] [CrossRef]
- Légaré, C.; Clément, A.-A.; Desgagné, V.; Thibeault, K.; White, F.; Guay, S.-P.; Arsenault, B.J.; Scott, M.S.; Jacques, P.-É.; Perron, P.; et al. Human Plasma Pregnancy-Associated miRNAs and Their Temporal Variation within the First Trimester of Pregnancy. Reprod. Biol. Endocrinol. 2022, 20, 14. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell Communication: microRNAs as Hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef]
- Matias-Garcia, P.R.; Wilson, R.; Mussack, V.; Reischl, E.; Waldenberger, M.; Gieger, C.; Anton, G.; Peters, A.; Kuehn-Steven, A. Impact of Long-Term Storage and Freeze-Thawing on Eight Circulating microRNAs in Plasma Samples. PLoS ONE 2020, 15, e0227648. [Google Scholar] [CrossRef] [PubMed]
- Collares, C.V.; Evangelista, A.F.; Xavier, D.J.; Rassi, D.M.; Arns, T.; Foss-Freitas, M.C.; Foss, M.C.; Puthier, D.; Sakamoto-Hojo, E.T.; Passos, G.A.; et al. Identifying Common and Specific microRNAs Expressed in Peripheral Blood Mononuclear Cell of Type 1, Type 2, and Gestational Diabetes Mellitus Patients. BMC Res. Notes 2013, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Athira, S.V.; Bhaskar, A.; Misra, P.; Sibin, M.K. Circulatory miR-126 Expression as an Epigenetic Marker in Diabetes Mellitus; a Systematic Review & Meta-Analysis. Gene Rep. 2022, 26, 101502. [Google Scholar] [CrossRef]
- Zhang, L.; Li, K.; Tian, S.; Wang, X.-Q.; Li, J.-H.; Dong, Y.-C.; Xia, H.-F.; Ma, X. Down-Regulation of microRNA-30d-5p Is Associated with Gestational Diabetes Mellitus by Targeting RAB8A. J. Diabetes Complicat. 2021, 35, 107959. [Google Scholar] [CrossRef] [PubMed]
- Ghoreishi, E.; Shahrokhi, S.Z.; Kazerouni, F.; Rahimipour, A. Circulating miR-148b-3p and miR-27a-3p Can Be Potential Biomarkers for Diagnosis of Pre-Diabetes and Type 2 Diabetes: Integrating Experimental and in-Silico Approaches. BMC Endocr. Disord. 2022, 22, 207. [Google Scholar] [CrossRef]
- Ibarra, A.; Vega-Guedes, B.; Brito-Casillas, Y.; Wägner, A.M. Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application. Noncoding RNA 2018, 4, 32. [Google Scholar] [CrossRef]
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352. [Google Scholar] [CrossRef]
- Valverde Tercedor, M.D.C.; Perdomo Ugarte, N.; García Delgado, Y.; Nóvoa Medina, Y.; Expósito Montesdeoca, A.; González Lleó, A.M.; Brito Casillas, Y.; Rodríguez González, G.; Vega Guedes, B.; Wägner, A.M. Wägner Pregestational Diabetes and the Offspring: Comparing the Effects According to the Type of Maternal Diabetes and with Paternal Type 1 Diabetes. Diabetologia 2020, 63 (Suppl. S1), S466. [Google Scholar]
- Masete, M.; Dias, S.; Malaza, N.; Adam, S.; Pheiffer, C. A Big Role for microRNAs in Gestational Diabetes Mellitus. Front. Endocrinol 2022, 13, 892587. [Google Scholar] [CrossRef]
- Valverde Tercedor, C.; Ibarra, A.; Perdomo Ugarte, N.; Davila Batista, V.; Novoa Medina, Y.; Garcia Delgado, Y.; Hernandez-Baraza, L.; Jimenez Monzon, R.; Barreiro Bautista, M.; Gonzalez Lleo, A. Placental microRNAs in Pregestational Diabetes: Effects on Offspring and Functional Study. Diabetologia 2023, 66 (Suppl. S1), S1–S536. [Google Scholar]
- Floh, A.A.; Manlhiot, C.; Redington, A.N.; McCrindle, B.W.; Clarizia, N.A.; Caldarone, C.A.; Schwartz, S.M. Insulin Resistance and Inflammation Are a Cause of Hyperglycemia after Pediatric Cardiopulmonary Bypass Surgery. J. Thorac. Cardiovasc. Surg. 2015, 150, 498–504.e1. [Google Scholar] [CrossRef] [PubMed]
- Tsalamandris, S.; Antonopoulos, A.S.; Oikonomou, E.; Papamikroulis, G.-A.; Vogiatzi, G.; Papaioannou, S.; Deftereos, S.; Tousoulis, D. The Role of Inflammation in Diabetes: Current Concepts and Future Perspectives. Eur. Cardiol. 2019, 14, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Wolosowicz, M.; Lukaszuk, B.; Chabowski, A. The Causes of Insulin Resistance in Type 1 Diabetes Mellitus: Is There a Place for Quaternary Prevention? Int. J. Environ. Res. Public Health 2020, 17, 8651. [Google Scholar] [CrossRef]
- Yahaya, T.O.; Salisu, T.; Abdulrahman, Y.B.; Umar, A.K. Update on the Genetic and Epigenetic Etiology of Gestational Diabetes Mellitus: A Review. Egypt. J. Med. Hum. Genet. 2020, 21, 13. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, Z.; Li, H. The Role of Lipid Dysregulation in Gestational Diabetes Mellitus: Early Prediction and Postpartum Prognosis. J. Diabetes Investig. 2023, 15, 15–25. [Google Scholar] [CrossRef]
- Elliott, A.; Walters, R.K.; Pirinen, M.; Kurki, M.; Junna, N.; Goldstein, J.I.; Reeve, M.P.; Siirtola, H.; Lemmelä, S.M.; Turley, P.; et al. Distinct and Shared Genetic Architectures of Gestational Diabetes Mellitus and Type 2 Diabetes. Nat. Genet. 2024, 56, 377–382. [Google Scholar] [CrossRef]
- Vounzoulaki, E.; Khunti, K.; Abner, S.C.; Tan, B.K.; Davies, M.J.; Gillies, C.L. Progression to Type 2 Diabetes in Women with a Known History of Gestational Diabetes: Systematic Review and Meta-Analysis. BMJ 2020, 369, m1361. [Google Scholar] [CrossRef]
- Incani, M.; Baroni, M.G.; Cossu, E. Testing for Type 1 Diabetes Autoantibodies in Gestational Diabetes Mellitus (GDM): Is It Clinically Useful? BMC Endocr. Disord. 2019, 19, 44. [Google Scholar] [CrossRef]
- Radaelli, T.; Lepercq, J.; Varastehpour, A.; Basu, S.; Catalano, P.; Hauguel-de Mouzon, S. Differential Regulation of Genes for Feto-Placental Lipid Pathways in Pregnancy with Gestational and Type 1 Diabetes. Am. J. Obs. Gynecol. 2009, 201, 209.e1–209.e10. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling Maternal Plasma microRNA Expression in Early Pregnancy to Predict Gestational Diabetes Mellitus. Int. J. Gynaecol. Obs. 2015, 130, 49–53. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Jia, Y.-J.; Xing, B.-H.; Shi, D.-D.; Dong, X.-J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel Diagnostic Biomarkers for Gestational Diabetes Mellitus. J. Obstet. Gynaecol. Res. 2017, 43, 974–981. [Google Scholar] [CrossRef]
- Bovell, L.C.; Shanmugam, C.; Putcha, B.-D.K.; Katkoori, V.R.; Zhang, B.; Bae, S.; Singh, K.P.; Grizzle, W.E.; Manne, U. The Prognostic Value of microRNAs Varies with Patient Race/Ethnicity and Stage of Colorectal Cancer. Clin. Cancer Res. 2013, 19, 3955–3965. [Google Scholar] [CrossRef]
- Sud, N.; Zhang, H.; Pan, K.; Cheng, X.; Cui, J.; Su, Q. Aberrant Expression of microRNA Induced by High-Fructose Diet: Implications in the Pathogenesis of Hyperlipidemia and Hepatic Insulin Resistance. J. Nutr. Biochem. 2017, 43, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.C.; Magenta, A.; Beji, S.; Lakatta, E.G.; Capogrossi, M.C. Aging, MicroRNAs, and Heart Failure. Curr. Probl. Cardiol. 2020, 45, 100406. [Google Scholar] [CrossRef] [PubMed]
- Karere, G.M.; Cox, L.A.; Bishop, A.C.; South, A.M.; Shaltout, H.A.; Mercado-Deane, M.-G.; Cuda, S. Sex Differences in miRNA Expression and Cardiometabolic Risk Factors in Hispanic Adolescents With Obesity. J. Pediatr. 2021, 235, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Ma, L.; Zhou, J.; Jiang, M.; Rao, E.; Zhao, Y.; Guo, F. miR-17-92 Plays an Oncogenic Role and Conveys Chemo-Resistance to Cisplatin in Human Prostate Cancer Cells. Int. J. Oncol. 2016, 48, 1737–1748. [Google Scholar] [CrossRef]
- Wan, S.; Zhang, J.; Chen, X.; Lang, J.; Li, L.; Chen, F.; Tian, L.; Meng, Y.; Yu, X. MicroRNA-17-92 Regulates Beta-Cell Restoration After Streptozotocin Treatment. Front. Endocrinol. 2020, 11, 9. [Google Scholar] [CrossRef]
- Tang, X.; Muniappan, L.; Tang, G.; Özcan, S. Identification of Glucose-Regulated miRNAs from Pancreatic β Cells Reveals a Role for miR-30d in Insulin Transcription. RNA 2009, 15, 287–293. [Google Scholar] [CrossRef]
- Zhao, X.; Mohan, R.; Özcan, S.; Tang, X. MicroRNA-30d Induces Insulin Transcription Factor MafA and Insulin Production by Targeting Mitogen-Activated Protein 4 Kinase 4 (MAP4K4) in Pancreatic β-Cells. J. Biol. Chem. 2012, 287, 31155–31164. [Google Scholar] [CrossRef]
- Gómez Muñoz, L.; Perna-Barrull, D.; Murillo-Vallés, M.; Armengol, M.P.; Alcalde, M.; Català, M.; Rodríguez-Fernández, S.; Sunye, S.; Valls, A.; Pérez Sánchez, J.; et al. Immunoregulatory Biomarkers of the Remission Phase in Type 1 Diabetes: miR-30d-5p Modulates PD-1 Expression and Regulatory T Cell Expansion. Non-Coding RNA 2023, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Petersen, K.F.; Shulman, G.I. Lipid-Induced Insulin Resistance: Unravelling the Mechanism. Lancet 2010, 375, 2267–2277. [Google Scholar] [CrossRef] [PubMed]
- Boxhammer, E.; Paar, V.; Wernly, B.; Kiss, A.; Mirna, M.; Aigner, A.; Acar, E.; Watzinger, S.; Podesser, B.K.; Zauner, R.; et al. MicroRNA-30d-5p—A Potential New Therapeutic Target for Prevention of Ischemic Cardiomyopathy after Myocardial Infarction. Cells 2023, 12, 2369. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Zhou, F.; Li, J.; Lu, G.; Zhao, Y. MiR-20a-5p Regulates MPP+-Induced Oxidative Stress and Neuroinflammation in HT22 Cells by Targeting IRF9/NF-κB Axis. Evid. Based Complement. Altern. Med. 2021, 2021, 6621206. [Google Scholar] [CrossRef]
- Sarfstein, R.; Werner, H. Tumor Suppressor P53 Regulates Insulin Receptor (INSR) Gene Expression via Direct Binding to the INSR Promoter. Oncotarget 2020, 11, 2424–2437. [Google Scholar] [CrossRef]
- Chen, J.; Ning, C.; Mu, J.; Li, D.; Ma, Y.; Meng, X. Role of Wnt Signaling Pathways in Type 2 Diabetes Mellitus. Mol. Cell Biochem. 2021, 476, 2219–2232. [Google Scholar] [CrossRef]
- He, W.; Tang, M.; Gu, R.; Wu, X.; Mu, X.; Nie, X. The Role of P53 in Regulating Chronic Inflammation and PANoptosis in Diabetic Wounds. Aging Dis. 2024, 16, 373–393. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization Strategies Differently Affect Circulating miRNA Profile Associated with the Training Status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Zendjabil, M. Preanalytical, Analytical and Postanalytical Considerations in Circulating microRNAs Measurement. Biochem. Med. 2024, 34, 020501. [Google Scholar] [CrossRef]
- Becker, N.; Lockwood, C. Pre-Analytical Variables in miRNA Analysis. Clin. Biochem. 2013, 46, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Torres, J.A.; Baccarelli, A.; Bollati, V. Epigenetics and Lifestyle. Epigenomics 2011, 3, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Juan, J.; Kang, X.; Yao, M.; Chen, X.; Wei, Z.; Kong, L.; Chen, H.; Cui, S.; Gao, F.; et al. Comparative Analysis of Perinatal Outcomes in Pregnant Women with Pregestational Diabetes Mellitus Based on Diagnostic Timing. Sci. Rep. 2025, 15, 9613. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Bian, D.; Hao, L.; Huang, F.; Xu, M.; Qin, J.; Liu, Y. microRNA-503 Contribute to Pancreatic Beta Cell Dysfunction by Targeting the mTOR Pathway in Gestational Diabetes Mellitus. EXCLI J. 2017, 16, 1177–1187. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Li, H.; Wan, J.; Zhou, Q.; Zhou, Y.; Zhang, C. microRNA-96 Protects Pancreatic β-Cell Function by Targeting PAK1 in Gestational Diabetes Mellitus. BioFactors 2018, 44, 539–547. [Google Scholar] [CrossRef]
- Kupec, T.; Bleilevens, A.; Iborra, S.; Najjari, L.; Wittenborn, J.; Maurer, J.; Stickeler, E. Stability of Circulating microRNAs in Serum. PLoS ONE 2022, 17, e0268958. [Google Scholar] [CrossRef]
- International Association of Diabetes and Pregnancy Study Groups Consensus Panel; Metzger, B.E.; Gabbe, S.G.; Persson, B.; Buchanan, T.A.; Catalano, P.A.; Damm, P.; Dyer, A.R.; de Leiva, A.; Hod, M.; et al. International Association of Diabetes and Pregnancy Study Groups Recommendations on the Diagnosis and Classification of Hyperglycemia in Pregnancy. Diabetes Care 2010, 33, 676–682. [Google Scholar] [CrossRef]
- World Health Organization. Diagnostic Criteria and Classification of Hyperglycaemia First Detected in Pregnancy, WHO/NMH/MND/13.2 ed.; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Grubbs, F.E. Procedures for Detecting Outlying Observations in Samples. Technometrics 1969, 11, 1–21. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.-L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing More than Half a Million Experimentally Supported miRNA:mRNA Interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Armstrong, R.A. When to Use the Bonferroni Correction. Ophthalmic Physiol. Opt. 2014, 34, 502–508. [Google Scholar] [CrossRef]
- Youden, W.J. Index for Rating Diagnostic Tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
| Variable | Normoglycemia (n = 47) | T1DM (n = 23) | T2DM (n = 54) * | GDM (n = 43) |
|---|---|---|---|---|
| Age (years) | 31.0 (27.0–36.0) c | 29.0 (27.0–32.0) d,e | 35.0 (30.0–37.0) d | 36.0 (33.0–37.0) c,e |
| Gestational age at recruitment (weeks) | 22.0 (20.0–25.0) a,b | 17.0 (14.0–21.0) a,d | 21.0 (17.0–25.0) e | 26.0 (24.0–26.0) b,d,e |
| Body mass index (kg/m2) | 30.1 (26.1–38.5) d | 31.2 (23.6–34.0) e | 31.9 (28.8–37.7) f | 39.7 (33.7–46.5) d,e,f |
| Glycated haemoglobin (%) | 5.1 (4.9–5.5) c,d | 9.2 (7.6–10.7) c,e | 7.5 (6.3–8.9) d,f | 5.7 (5.3–6.1) f,e |
| 0-h blood glucose OGTT (mmol/L) | 4.0 (3.8–4.6) d,e | ND | 7.6 (7.0–9.2) d,f | 5.5 (5.2–6.0) e,f |
| 2-h blood glucose OGTT (mmol/L) | 5.0 (4.3–6.2) d,e | ND | 12.9 (10.7) d,f | 8.2 (6.5–9.5) e,f |
| # History of hypertension (%): Yes | 4.3% d,e | 13.0% | 31.5% e | 25.5% d |
| Triglycerides (mmol/L) | 1.5 (0.5–2.1) a | 0.9 (0–2.5) b | 2.1 (0.4–3.9) | 2.4 (1.5–3.8) a,b |
| C-peptide (ng/mL) | 1.4 (0–3.2) a | 0.5 (0–1.2) b,d | 1.8 (0.9–2.7) b | 2.2 (1.0–4.3) a,d |
| MiRNA | T1DM | T2DM | GDM | |||
|---|---|---|---|---|---|---|
| Fold Regulation | p Value | Fold Regulation | p Value | Fold Regulation | p Value | |
| miR-19b-3p | ↓ 5.3 | 0.500 | ↓ 2.0 | 0.180 | ↓ 9.8 | 0.033 |
| miR-20a-5p | ↓ 4.6 | 0.047 | ↓ 1.7 | 0.520 | ↓ 9.8 | 0.110 |
| miR-29a-3p | ↑ 2.5 | 0.180 | ↑ 1.9 | 0.002 | ↑ 2.9 | 0.200 |
| KEGG Pathway | p-Value | Genes | MiRNAs |
|---|---|---|---|
| Oocyte meiosis | 1.53 × 10−8 | 42 | miR-20a-5p |
| miR-30d-5p | |||
| Pathways in cancer | 4.93 × 10−8 | 99 | miR-20a-5p |
| miR-30d-5p | |||
| Ubiquitin mediated proteolysis | 3.69 × 10−7 | 49 | miR-20a-5p |
| miR-30d-5p | |||
| Lysine degradation | 5.10 × 10−7 | 15 | miR-20a-5p |
| miR-30d-5p | |||
| Protein processing in endoplasmic reticulum | 3.18 × 10−6 | 53 | miR-20a-5p |
| miR-30d-5p | |||
| Cell cycle | 7.28 × 10−6 | 46 | miR-20a-5p |
| miR-30d-5p | |||
| Hippo signaling pathway | 1.14 × 10−5 | 42 | miR-20a-5p |
| miR-30d-5p | |||
| mRNA surveillance pathway | 2.00 × 10−4 | 32 | miR-20a-5p |
| miR-30d-5p | |||
| p53 signaling pathway | 2.00 × 10−3 | 26 | miR-20a-5p |
| miR-30d-5p | |||
| Wnt signaling pathway | 2.00 × 10−3 | 42 | miR-20a-5p |
| miR-30d-5p | |||
| Colorectal cancer | 2.00 × 10−3 | 21 | miR-20a-5p |
| miR-30d-5p | |||
| RNA transport | 1.00 × 10−2 | 44 | miR-20a-5p |
| miR-30d-5p |
| Measurement | GDM | T2DM | Normoglycemia |
|---|---|---|---|
| 0-h blood glucose OGTT (mmol/L) | ≥5.1 | ≥7.0 | <5.1 |
| 1-h Glucose (mmol/L) | ≥10 | - | <10 |
| 2-h blood glucose OGTT (mmol/L) | ≥8.5–11 | ≥11.1 | <8.5 |
| Random Glucose (mmol/L) | - | ≥11.1 | - |
| HbA1c (%) | - | ≥6.5 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masete, M.; Dias, S.; Malaza, N.; Adam, S.; Mutavhatsindi, H.; Valverde-Tercedor, C.; Vega-Guedes, B.; Wägner, A.M.; Pheiffer, C. Circulating MicroRNA Profiles in Pregnant South African Women with Different Types of Diabetes Mellitus. Int. J. Mol. Sci. 2025, 26, 9337. https://doi.org/10.3390/ijms26199337
Masete M, Dias S, Malaza N, Adam S, Mutavhatsindi H, Valverde-Tercedor C, Vega-Guedes B, Wägner AM, Pheiffer C. Circulating MicroRNA Profiles in Pregnant South African Women with Different Types of Diabetes Mellitus. International Journal of Molecular Sciences. 2025; 26(19):9337. https://doi.org/10.3390/ijms26199337
Chicago/Turabian StyleMasete, Matladi, Stephanie Dias, Nompumelelo Malaza, Sumaiya Adam, Hygon Mutavhatsindi, Carmen Valverde-Tercedor, Begoña Vega-Guedes, Ana Maria Wägner, and Carmen Pheiffer. 2025. "Circulating MicroRNA Profiles in Pregnant South African Women with Different Types of Diabetes Mellitus" International Journal of Molecular Sciences 26, no. 19: 9337. https://doi.org/10.3390/ijms26199337
APA StyleMasete, M., Dias, S., Malaza, N., Adam, S., Mutavhatsindi, H., Valverde-Tercedor, C., Vega-Guedes, B., Wägner, A. M., & Pheiffer, C. (2025). Circulating MicroRNA Profiles in Pregnant South African Women with Different Types of Diabetes Mellitus. International Journal of Molecular Sciences, 26(19), 9337. https://doi.org/10.3390/ijms26199337








