Bacterial Systematic Genetics and Integrated Multi-Omics: Beyond Static Genomics Toward Predictive Models
Abstract
1. Introduction
2. Recent Studies of Bacterial GWAS, Intermediate Molecular Omics, and Multi-Omics Integration
2.1. Genome-Wide Association Studies
2.2. Transcriptome
2.3. Proteome
2.4. Interactome
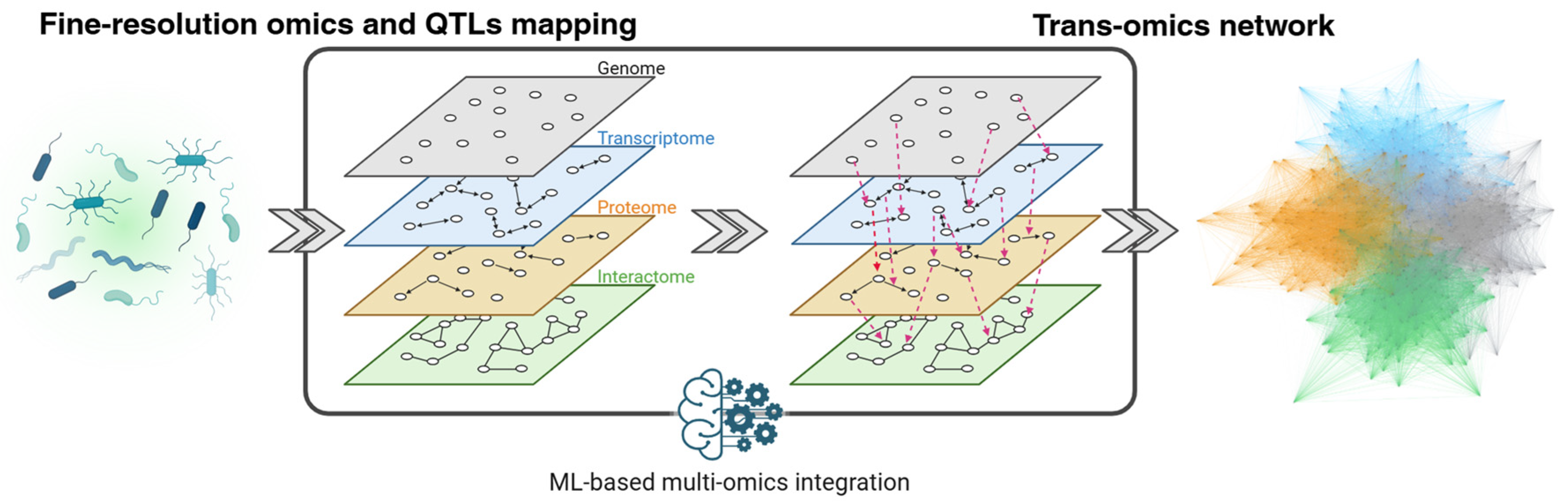
2.5. Bacterial Multi-Omics and QTL Analysis
3. Current Challenges and Promising Technologies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| AP-MS | Affinity purification–mass spectrometry |
| B2H | Bacterial two-hybrid |
| DBG | de Bruijn graph |
| DDA | Data-dependent acquisition |
| DIA | Data-independent acquisition |
| DRS | Direct RNA sequencing |
| FACS | Fluorescence-activated cell sorting |
| GWAS | Genome-wide association studies |
| LD | Linkage disequilibrium |
| LMM | Linear mixed models |
| MDS | Multidimensional scaling |
| ONT | Oxford nanopore technology |
| PCA | Principal component analysis |
| PL | Proximity labeling |
| PPI | Protein–protein interaction |
| PTM | Post-translational modification |
| QTL | Quantitative trait locus |
| SBP | Single-bacterium proteomics |
| SNP | Single-nucleotide polymorphism |
| SVM | Support vector machine |
| TDP | Top-down proteomics |
| TRIPs | Transcription–replication interaction profiles |
| XL-MS | Cross-linking mass spectrometry |
| Y2H | Yeast two-hybrid |
References
- Casjens, S. The Diverse and Dynamic Structure of Bacterial Genomes. Annu. Rev. Genet. 1998, 32, 339–377. [Google Scholar] [CrossRef]
- Wei, W.; Ho, W.-C.; Behringer, M.G.; Miller, S.F.; Bcharah, G.; Lynch, M. Rapid Evolution of Mutation Rate and Spectrum in Response to Environmental and Population-Genetic Challenges. Nat. Commun. 2022, 13, 4752. [Google Scholar] [CrossRef]
- Mosquera-Rendón, J.; Moreno-Herrera, C.X.; Robledo, J.; Hurtado-Páez, U. Genome-Wide Association Studies (GWAS) Approaches for the Detection of Genetic Variants Associated with Antibiotic Resistance: A Systematic Review. Microorganisms 2023, 11, 2866. [Google Scholar] [CrossRef]
- Power, R.A.; Parkhill, J.; de Oliveira, T. Microbial Genome-Wide Association Studies: Lessons from Human GWAS. Nat. Rev. Genet. 2017, 18, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, X.; Han, M.; Sheng, H.; Sun, Y.; Su, L.; Lu, W.; Li, M.; Wang, S.; Chen, J.; et al. Bacterial Genome-Wide Association Studies: Exploring the Genetic Variation Underlying Bacterial Phenotypes. Appl. Environ. Microbiol. 2025, 91, e0251224. [Google Scholar] [CrossRef] [PubMed]
- Saber, M.M.; Shapiro, B.J. Benchmarking Bacterial Genome-Wide Association Study Methods Using Simulated Genomes and Phenotypes. Microb. Genom. 2020, 6, e000337. [Google Scholar] [CrossRef]
- Filiatrault, M.J. Progress in Prokaryotic Transcriptomics. Curr. Opin. Microbiol. 2011, 14, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Shi, Z.J.; Pollard, K.S. Pitfalls of Genotyping Microbial Communities with Rapidly Growing Genome Collections. Cell Syst. 2023, 14, 160–176.e3. [Google Scholar] [CrossRef]
- Pinto, Y.; Bhatt, A.S. Sequencing-Based Analysis of Microbiomes. Nat. Rev. Genet. 2024, 25, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.D.; Lappalainen, T. Functional Characterization of Genetic Variant Effects on Expression. Annu. Rev. Biomed. Data Sci. 2022, 5, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E.; et al. Genomic Analysis Reveals That Pseudomonas aeruginosa Virulence Is Combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef] [PubMed]
- James, T.; Williamson, B.; Tino, P.; Wheeler, N. Whole-Genome Phenotype Prediction with Machine Learning: Open Problems in Bacterial Genomics. Bioinformatics 2025, 41, btaf206. [Google Scholar] [CrossRef] [PubMed]
- Koblitz, J.; Reimer, L.C.; Pukall, R.; Overmann, J. Predicting Bacterial Phenotypic Traits through Improved Machine Learning Using High-Quality, Curated Datasets. Commun. Biol. 2025, 8, 897. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Thomson, N.R. Studying Bacterial Transcriptomes Using RNA-Seq. Curr. Opin. Microbiol. 2010, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Civelek, M.; Lusis, A.J. Systems Genetics Approaches to Understand Complex Traits. Nat. Rev. Genet. 2014, 15, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Callahan, N.; Tullman, J.; Kelman, Z.; Marino, J. Strategies for Development of a next-Generation Protein Sequencing Platform. Trends Biochem. Sci. 2020, 45, 76–89. [Google Scholar] [CrossRef]
- Abele, M.; Soleymaniniya, A.; Bayer, F.P.; Lomp, N.; Doll, E.; Meng, C.; Neuhaus, K.; Scherer, S.; Wenning, M.; Wantia, N.; et al. Proteomic Diversity in Bacteria: Insights and Implications for Bacterial Identification. Mol. Cell. Proteom. 2025, 24, 100917. [Google Scholar] [CrossRef] [PubMed]
- Tsakou, F.; Jersie-Christensen, R.; Jenssen, H.; Mojsoska, B. The Role of Proteomics in Bacterial Response to Antibiotics. Pharmaceuticals 2020, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Nuss, A.M.; Beckstette, M.; Pimenova, M.; Schmühl, C.; Opitz, W.; Pisano, F.; Heroven, A.K.; Dersch, P. Tissue Dual RNA-Seq Allows Fast Discovery of Infection-Specific Functions and Riboregulators Shaping Host-Pathogen Transcriptomes. Proc. Natl. Acad. Sci. USA 2017, 114, E791–E800. [Google Scholar] [CrossRef]
- Jin, Q.; Zhai, Y.; Qiang, R.; Ma, X.; Zhao, C.; Zhong, J.; Li, J.; Chen, Q.; Han, M.; Du, H.; et al. Dual RNA-Seq Reveals the Complement Protein C3-Mediated Host-Pathogen Interaction in the Brain Abscess Caused by Staphylococcus Aureus. mSystems 2025, 10, e0154024. [Google Scholar] [CrossRef] [PubMed]
- Blattman, S.B.; Jiang, W.; Oikonomou, P.; Tavazoie, S. Prokaryotic Single-Cell RNA Sequencing by in Situ Combinatorial Indexing. Nat. Microbiol. 2020, 5, 1192–1201. [Google Scholar] [CrossRef]
- Kuchina, A.; Brettner, L.M.; Paleologu, L.; Roco, C.M.; Rosenberg, A.B.; Carignano, A.; Kibler, R.; Hirano, M.; DePaolo, R.W.; Seelig, G. Microbial Single-Cell RNA Sequencing by Split-Pool Barcoding. Science 2021, 371, eaba5257. [Google Scholar] [CrossRef] [PubMed]
- Homberger, C.; Hayward, R.J.; Barquist, L.; Vogel, J. Improved Bacterial Single-Cell RNA-Seq through Automated MATQ-Seq and Cas9-Based Removal of rRNA Reads. mBio 2023, 14, e03557-22. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Amemiya, H.M.; He, L.L.; Gandhi, S.J.; Nicol, R.; Bhattacharyya, R.P.; Smillie, C.S.; Hung, D.T. Bacterial Droplet-Based Single-Cell RNA-Seq Reveals Antibiotic-Associated Heterogeneous Cellular States. Cell 2023, 186, 877–891.e14. [Google Scholar] [CrossRef] [PubMed]
- Dar, D.; Dar, N.; Cai, L.; Newman, D.K. Spatial Transcriptomics of Planktonic and Sessile Bacterial Populations at Single-Cell Resolution. Science 2021, 373, eabi4882. [Google Scholar] [CrossRef] [PubMed]
- Budnik, B.; Levy, E.; Harmange, G.; Slavov, N. SCoPE-MS: Mass Spectrometry of Single Mammalian Cells Quantifies Proteome Heterogeneity during Cell Differentiation. Genome Biol. 2018, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A Critical Review of Bottom-up Proteomics: The Good, the Bad, and the Future of This Field. Proteomes 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Mount, H.O.; Urbanus, M.L.; Sheykhkarimli, D.; Coté, A.G.; Laval, F.; Coppin, G.; Kishore, N.; Li, R.; Spirohn-Fitzgerald, K.; Petersen, M.O.; et al. A Comprehensive Two-Hybrid Analysis to Explore the Legionella pneumophila Effector-Effector Interactome. mSystems 2024, 9, e0100424. [Google Scholar] [CrossRef] [PubMed]
- Pardo, M.; Choudhary, J.S. Assignment of Protein Interactions from Affinity Purification/mass Spectrometry Data. J. Proteome Res. 2012, 11, 1462–1474. [Google Scholar] [CrossRef]
- Shatsky, M.; Allen, S.; Gold, B.L.; Liu, N.L.; Juba, T.R.; Reveco, S.A.; Elias, D.A.; Prathapam, R.; He, J.; Yang, W.; et al. Bacterial Interactomes: Interacting Protein Partners Share Similar Function and Are Validated in Independent Assays More Frequently than Previously Reported. Mol. Cell. Proteom. 2016, 15, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fan, X.; Hu, Y. In Vivo Interactome Profiling by Enzyme-Catalyzed Proximity Labeling. Cell Biosci. 2021, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Movassagh, M.; Schiff, S.J.; Paulson, J.N. mbQTL: An R/Bioconductor Package for Microbial Quantitative Trait Loci (QTL) Estimation. Bioinformatics 2023, 39, btad565. [Google Scholar] [CrossRef] [PubMed]
- She, R.; Jarosz, D.F. Mapping Causal Variants with Single-Nucleotide Resolution Reveals Biochemical Drivers of Phenotypic Change. Cell 2018, 172, 478–490.e15. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-Wide Efficient Mixed-Model Analysis for Association Studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Mai, T.T.; Galardini, M.; Wheeler, N.E.; Horsfield, S.T.; Parkhill, J.; Corander, J. Improved Prediction of Bacterial Genotype-Phenotype Associations Using Interpretable Pangenome-Spanning Regressions. mBio 2020, 11, e01344-20. [Google Scholar] [CrossRef]
- Earle, S.G.; Wu, C.-H.; Charlesworth, J.; Stoesser, N.; Gordon, N.C.; Walker, T.M.; Spencer, C.C.A.; Iqbal, Z.; Clifton, D.A.; Hopkins, K.L.; et al. Identifying Lineage Effects When Controlling for Population Structure Improves Power in Bacterial Association Studies. Nat. Microbiol. 2016, 1, 16041. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Hallgrímsdóttir, I.; Eisen, M.; Pachter, L. Association Mapping from Sequencing Reads Using K-Mers. eLife 2018, 7, e32920. [Google Scholar] [CrossRef] [PubMed]
- Mehrab, Z.; Mobin, J.; Tahmid, I.A.; Rahman, A. Efficient Association Mapping from K-Mers-An Application in Finding Sex-Specific Sequences. PLoS ONE 2021, 16, e0245058. [Google Scholar] [CrossRef]
- Weisberg, A.J.; Chang, J.H. Mobile Genetic Element Flexibility as an Underlying Principle to Bacterial Evolution. Annu. Rev. Microbiol. 2023, 77, 603–624. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Gu, C.; Kim, H.U.; Lee, S.Y. Current Status of Pan-Genome Analysis for Pathogenic Bacteria. Curr. Opin. Biotechnol. 2020, 63, 54–62. [Google Scholar] [CrossRef]
- Lees, J.A.; Vehkala, M.; Välimäki, N.; Harris, S.R.; Chewapreecha, C.; Croucher, N.J.; Marttinen, P.; Davies, M.R.; Steer, A.C.; Tong, S.Y.C.; et al. Sequence Element Enrichment Analysis to Determine the Genetic Basis of Bacterial Phenotypes. Nat. Commun. 2016, 7, 12797. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, S.T.; Rau, M.H.; Sánchez, B.J.; Jensen, K.; Zeidan, A.A. From Genotype to Phenotype: Computational Approaches for Inferring Microbial Traits Relevant to the Food Industry. FEMS Microbiol. Rev. 2023, 47, fuad030. [Google Scholar] [CrossRef] [PubMed]
- Jaillard, M.; Lima, L.; Tournoud, M.; Mahé, P.; van Belkum, A.; Lacroix, V.; Jacob, L. A Fast and Agnostic Method for Bacterial Genome-Wide Association Studies: Bridging the Gap between K-Mers and Genetic Events. PLoS Genet. 2018, 14, e1007758. [Google Scholar] [CrossRef] [PubMed]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid Scoring of Genes in Microbial Pan-Genome-Wide Association Studies with Scoary. Genome Biol. 2016, 17, 238, Erratum in Genome Biol. 2016, 17, 262. [Google Scholar] [CrossRef]
- Roder, T.; Pimentel, G.; Fuchsmann, P.; Stern, M.T.; von Ah, U.; Vergères, G.; Peischl, S.; Brynildsrud, O.; Bruggmann, R.; Bär, C. Scoary2: Rapid Association of Phenotypic Multi-Omics Data with Microbial Pan-Genomes. Genome Biol. 2024, 25, 93. [Google Scholar] [CrossRef] [PubMed]
- Carrara, A.; Bertelli, C.; Gardiol, C.; Marquis, B.; Andrey, D.O.; Schrenzel, J.; Pillonel, T.; Greub, G. Association of Pathogenic Determinants of Fusobacterium necrophorum with Bacteremia, and Lemierre’s Syndrome. Sci. Rep. 2024, 14, 19804. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, Q.; Peierdun, M.; Takiff, H.E.; Gao, Q. The Mutational Signatures of Poor Treatment Outcomes on the Drug-Susceptible Mycobacterium Tuberculosis Genome. eLife 2023, 12, e84815. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.H.F.; Ma, K.C.; Westervelt, K.A.; Hullahalli, K.; Waldor, M.K.; Grad, Y.H. CanB Is a Metabolic Mediator of Antibiotic Resistance in Neisseria Gonorrhoeae. Nat. Microbiol. 2023, 8, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiang, D.; Yao, X.; Luo, Y.; Yang, Z.; Ren, M.; Zhang, G.; Yu, Y.; Lu, A.; Wang, Y. Pan-Genome Wide Association Study of Glaesserella parasuis Highlights Genes Associated with Virulence and Biofilm Formation. Front. Microbiol. 2023, 14, 1160433. [Google Scholar] [CrossRef]
- Monteith, W.; Pascoe, B.; Mourkas, E.; Clark, J.; Hakim, M.; Hitchings, M.D.; McCarthy, N.; Yahara, K.; Asakura, H.; Sheppard, S.K. Contrasting Genes Conferring Short- and Long-Term Biofilm Adaptation in Listeria. Microb. Genom. 2023, 9, 001114. [Google Scholar] [CrossRef]
- Zheng, L.; Yang, C.; Chen, P.; Zhu, L.; Wen, H.; Liu, M.; Guan, J.; Lu, G.; Jing, J.; Sun, S.; et al. Vibrio Parahaemolyticus from Migratory Birds in China Carries an Extra Copy of tRNA-Gly and Plasmid-Mediated Quinolone Resistance Gene qnrD. Microbiol. Spectr. 2023, 11, e0217022. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Chen, S.; Jiang, R.; Zhou, H.; Li, Y.; Ouyang, D.; Gong, Y.; Yao, Z.; Ye, X. Inferring Staphylococcus aureus Host Species and Cross-Species Transmission from a Genome-Based Model. BMC Genom. 2025, 26, 149. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.K.; van der Putten, B.C.L.; Fuchs, T.M.; Vinh, T.N.; Bootsma, M.; Oldenkamp, R.; La Ragione, R.; Matamoros, S.; Hoa, N.T.; Berens, C.; et al. Genome-Wide Association Reveals Host-Specific Genomic Traits in Escherichia coli. BMC Biol. 2023, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Boeck, L.; Burbaud, S.; Skwark, M.; Pearson, W.H.; Sangen, J.; Wuest, A.W.; Marshall, E.K.P.; Weimann, A.; Everall, I.; Bryant, J.M.; et al. Mycobacterium abscessus Pathogenesis Identified by Phenogenomic Analyses. Nat. Microbiol. 2022, 7, 1431–1441. [Google Scholar] [CrossRef]
- CRyPTIC Consortium and the 100,000 Genomes Project; Allix-Béguec, C.; Arandjelovic, I.; Bi, L.; Beckert, P.; Bonnet, M.; Bradley, P.; Cabibbe, A.M.; Cancino-Muñoz, I.; Caulfield, M.J.; et al. Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing. N. Engl. J. Med. 2018, 379, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- The CRyPTIC Consortium. Genome-Wide Association Studies of Global Mycobacterium Tuberculosis Resistance to 13 Antimicrobials in 10,228 Genomes Identify New Resistance Mechanisms. PLoS Biol. 2022, 20, e3001755. [Google Scholar]
- Perez-Sepulveda, B.M.; Heavens, D.; Pulford, C.V.; Predeus, A.V.; Low, R.; Webster, H.; Dykes, G.F.; Schudoma, C.; Rowe, W.; Lipscombe, J.; et al. An Accessible, Efficient and Global Approach for the Large-Scale Sequencing of Bacterial Genomes. Genome Biol. 2021, 22, 349. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.C.; Monk, J.M.; Szubin, R.; Hefner, Y.; Palsson, B.O. Global Pathogenomic Analysis Identifies Known and Candidate Genetic Antimicrobial Resistance Determinants in Twelve Species. Nat. Commun. 2023, 14, 7690. [Google Scholar] [CrossRef]
- Achtman, M.; Zhou, Z.; Alikhan, N.-F.; Tyne, W.; Parkhill, J.; Cormican, M.; Chiou, C.-S.; Torpdahl, M.; Litrup, E.; Prendergast, D.M.; et al. Genomic Diversity of Salmonella enterica—The UoWUCC 10K Genomes Project. Wellcome Open Res. 2020, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- Schober, I.; Koblitz, J.; Sardà Carbasse, J.; Ebeling, C.; Schmidt, M.L.; Podstawka, A.; Gupta, R.; Ilangovan, V.; Chamanara, J.; Overmann, J.; et al. BacDive in 2025: The Core Database for Prokaryotic Strain Data. Nucleic Acids Res. 2025, 53, D748–D756. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A One-Stop Repository for Bacterial Whole-Genome Sequence Typing and Source Tracking. Nucleic Acids Res. 2021, 49, D644–D650. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Brettin, T.; Long, S.W.; Musser, J.M.; Olsen, R.J.; Olson, R.; Shukla, M.; Stevens, R.L.; Xia, F.; Yoo, H.; et al. Developing an in Silico Minimum Inhibitory Concentration Panel Test for Klebsiella pneumoniae. Sci. Rep. 2018, 8, 421. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4765–4774. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016. [Google Scholar]
- Sy-Janairo, M.L.L.; Janairo, J.I.B. Non-Endoscopic Applications of Machine Learning in Gastric Cancer: A Systematic Review. J. Gastrointest. Cancer 2024, 55, 47–64. [Google Scholar] [CrossRef]
- Creecy, J.P.; Conway, T. Quantitative Bacterial Transcriptomics with RNA-Seq. Curr. Opin. Microbiol. 2015, 23, 133–140. [Google Scholar] [CrossRef]
- Pountain, A.W.; Jiang, P.; Yao, T.; Homaee, E.; Guan, Y.; McDonald, K.J.C.; Podkowik, M.; Shopsin, B.; Torres, V.J.; Golding, I.; et al. Transcription-Replication Interactions Reveal Bacterial Genome Regulation. Nature 2024, 626, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, F.; Ferreira-Cerca, S.; Grohmann, D. Nanopore Sequencing of RNA and cDNA Molecules in Escherichia coli. RNA 2022, 28, 400–417. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Guo, Z.; Shao, Y.; Ye, L.; Wang, M.; Deng, X.; Chen, S.; Li, R. Analysis of Bacterial Transcriptome and Epitranscriptome Using Nanopore Direct RNA Sequencing. Nucleic Acids Res. 2024, 52, 8746–8762. [Google Scholar] [CrossRef]
- McNulty, R.; Sritharan, D.; Pahng, S.H.; Meisch, J.P.; Liu, S.; Brennan, M.A.; Saxer, G.; Hormoz, S.; Rosenthal, A.Z. Probe-Based Bacterial Single-Cell RNA Sequencing Predicts Toxin Regulation. Nat. Microbiol. 2023, 8, 934–945. [Google Scholar] [CrossRef]
- Espinoza Miranda, S.S.; Abbaszade, G.; Hess, W.R.; Drescher, K.; Saliba, A.-E.; Zaburdaev, V.; Chai, L.; Dreisewerd, K.; Grünberger, A.; Westendorf, C.; et al. Resolving Spatiotemporal Dynamics in Bacterial Multicellular Populations: Approaches and Challenges. Microbiol. Mol. Biol. Rev. 2025, 89, e0013824. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.-E.; Santos, S.C.; Vogel, J. New RNA-Seq Approaches for the Study of Bacterial Pathogens. Curr. Opin. Microbiol. 2017, 35, 78–87. [Google Scholar] [CrossRef]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-Seq of Pathogen and Host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef]
- Ryan, D.; Bornet, E.; Prezza, G.; Alampalli, S.V.; Franco de Carvalho, T.; Felchle, H.; Ebbecke, T.; Hayward, R.J.; Deutschbauer, A.M.; Barquist, L.; et al. An Expanded Transcriptome Atlas for Bacteroides Thetaiotaomicron Reveals a Small RNA That Modulates Tetracycline Sensitivity. Nat. Microbiol. 2024, 9, 1130–1144. [Google Scholar] [CrossRef]
- Deng, X.; Chen, K.; Luo, G.-Z.; Weng, X.; Ji, Q.; Zhou, T.; He, C. Widespread Occurrence of N6-Methyladenosine in Bacterial mRNA. Nucleic Acids Res. 2015, 43, 6557–6567. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, F.; Jüttner, M.; Knüppel, R.; Ferreira-Cerca, S.; Grohmann, D. Nanopore-Based RNA Sequencing Deciphers the Formation, Processing, and Modification Steps of rRNA Intermediates in Archaea. RNA 2023, 29, 1255–1273. [Google Scholar] [CrossRef]
- Riquelme-Barrios, S.; Vásquez-Camus, L.; Cusack, S.A.; Burdack, K.; Petrov, D.P.; Yeşiltaç-Tosun, G.N.; Kaiser, S.; Giehr, P.; Jung, K. Direct RNA Sequencing of the Escherichia coli Epitranscriptome Uncovers Alterations under Heat Stress. Nucleic Acids Res. 2025, 53, gkaf175. [Google Scholar] [CrossRef] [PubMed]
- Liu-Wei, W.; van der Toorn, W.; Bohn, P.; Hölzer, M.; Smyth, R.P.; von Kleist, M. Sequencing Accuracy and Systematic Errors of Nanopore Direct RNA Sequencing. BMC Genom. 2024, 25, 528. [Google Scholar] [CrossRef]
- Calvo-Roitberg, E.; Daniels, R.F.; Pai, A.A. Challenges in Identifying mRNA Transcript Starts and Ends from Long-Read Sequencing Data. Genome Res. 2024, 34, 1719–1734. [Google Scholar] [CrossRef]
- Ackermann, M. A Functional Perspective on Phenotypic Heterogeneity in Microorganisms. Nat. Rev. Microbiol. 2015, 13, 497–508. [Google Scholar] [CrossRef]
- Walls, A.W.; Rosenthal, A.Z. Bacterial Phenotypic Heterogeneity through the Lens of Single-Cell RNA Sequencing. Transcription 2024, 15, 48–62. [Google Scholar] [CrossRef]
- Imdahl, F.; Vafadarnejad, E.; Homberger, C.; Saliba, A.-E.; Vogel, J. Single-Cell RNA-Sequencing Reports Growth-Condition-Specific Global Transcriptomes of Individual Bacteria. Nat. Microbiol. 2020, 5, 1202–1206. [Google Scholar] [CrossRef]
- Chong, T.N.; Shapiro, L. Bacterial Cell Differentiation Enables Population Level Survival Strategies. mBio 2024, 15, e0075824. [Google Scholar] [CrossRef]
- Zhu, M.; Dai, X. Shaping of Microbial Phenotypes by Trade-Offs. Nat. Commun. 2024, 15, 4238. [Google Scholar] [CrossRef]
- Wang, B.; Lin, A.E.; Yuan, J.; Novak, K.E.; Koch, M.D.; Wingreen, N.S.; Adamson, B.; Gitai, Z. Single-Cell Massively-Parallel Multiplexed Microbial Sequencing (M3-Seq) Identifies Rare Bacterial Populations and Profiles Phage Infection. Nat. Microbiol. 2023, 8, 1846–1862. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, Y.; Sheng, K.; Rosenthal, R.; Liu, N.; Hua, X.; Zhang, T.; Chen, J.; Song, M.; Lv, Y.; et al. Droplet-Based High-Throughput Single Microbe RNA Sequencing by smRandom-Seq. Nat. Commun. 2023, 14, 5130. [Google Scholar] [CrossRef] [PubMed]
- Sarfatis, A.; Wang, Y.; Twumasi-Ankrah, N.; Moffitt, J.R. Highly Multiplexed Spatial Transcriptomics in Bacteria. Science 2025, 387, eadr0932, Erratum in Science 2025, 387, eadx0881. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of Protein Stability by Post-Translational Modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein Post-Translational Modifications in Bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Zubarev, R.A. The Challenge of the Proteome Dynamic Range and Its Implications for in-Depth Proteomics. Proteomics 2013, 13, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Park, A.J.; Krieger, J.R.; Khursigara, C.M. Survival Proteomes: The Emerging Proteotype of Antimicrobial Resistance. FEMS Microbiol. Rev. 2016, 40, 323–342. [Google Scholar] [CrossRef]
- Wang, D.-Z.; Kong, L.-F.; Li, Y.-Y.; Xie, Z.-X. Environmental Microbial Community Proteomics: Status, Challenges and Perspectives. Int. J. Mol. Sci. 2016, 17, 1275. [Google Scholar] [CrossRef] [PubMed]
- Rajczewski, A.T.; Blakeley-Ruiz, J.A.; Meyer, A.; Vintila, S.; McIlvin, M.R.; Van Den Bossche, T.; Searle, B.C.; Griffin, T.J.; Saito, M.A.; Kleiner, M.; et al. Data-Independent Acquisition Mass Spectrometry as a Tool for Metaproteomics: Interlaboratory Comparison Using a Model Microbiome. Proteomics 2025, 25, e202400187. [Google Scholar] [CrossRef]
- Balakrishnan, R.; Mori, M.; Segota, I.; Zhang, Z.; Aebersold, R.; Ludwig, C.; Hwa, T. Principles of Gene Regulation Quantitatively Connect DNA to RNA and Proteins in Bacteria. Science 2022, 378, eabk2066. [Google Scholar] [CrossRef]
- Gregorich, Z.R.; Ge, Y. Top-down Proteomics in Health and Disease: Challenges and Opportunities. Proteomics 2014, 14, 1195–1210. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L. Consortium for Top Down Proteomics Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Dupré, M.; Duchateau, M.; Malosse, C.; Borges-Lima, D.; Calvaresi, V.; Podglajen, I.; Clermont, D.; Rey, M.; Chamot-Rooke, J. Optimization of a Top-Down Proteomics Platform for Closely Related Pathogenic Bacterial Discrimination. J. Proteome. Res. 2021, 20, 202–211. [Google Scholar] [CrossRef]
- Zhao, Z.; Guo, Y.; Chowdhury, T.; Anjum, S.; Li, J.; Huang, L.; Cupp-Sutton, K.A.; Burgett, A.; Shi, D.; Wu, S. Top-down Proteomics Analysis of Picogram-Level Complex Samples Using Spray-Capillary-Based Capillary Electrophoresis-Mass Spectrometry. Anal. Chem. 2024, 96, 8763–8771. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Steen, J.A.; Mann, M. Mass-Spectrometry-Based Proteomics: From Single Cells to Clinical Applications. Nature 2025, 638, 901–911. [Google Scholar] [CrossRef]
- Végvári, Á.; Zhang, X.; Zubarev, R.A. Toward Single Bacterium Proteomics. J. Am. Soc. Mass Spectrom. 2023, 34, 2098–2106. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. In Virulence Mechanisms of Bacterial Pathogens, 5th ed.; American Society of Microbiology: Washington, DC, USA, 2016; pp. 215–239. ISBN 9781555819279. [Google Scholar]
- Sauvage, S.; Hardouin, J. Exoproteomics for Better Understanding Pseudomonas aeruginosa Virulence. Toxins 2020, 12, 571. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Khan, F.A.; Menghwar, H.; Faisal, M.; Ashraf, M.; Rasheed, M.A.; Marawan, M.A.; Dawood, A.; Chen, Y.; Chen, H.; et al. Progresses on Bacterial Secretomes Enlighten Research on Mycoplasma Secretome. Microb. Pathog. 2020, 144, 104160. [Google Scholar] [CrossRef]
- Russo, D.A.; Oliinyk, D.; Pohnert, G.; Meier, F.; Zedler, J.A.Z. EXCRETE Workflow Enables Deep Proteomics of the Microbial Extracellular Environment. Commun. Biol. 2024, 7, 1189. [Google Scholar] [CrossRef]
- Joung, J.K.; Ramm, E.I.; Pabo, C.O. A Bacterial Two-Hybrid Selection System for Studying Protein-DNA and Protein-Protein Interactions. Proc. Natl. Acad. Sci. USA 2000, 97, 7382–7387. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, S.; Liu, C.-M.; Fernie, A.R.; Yan, S. Recent Advances in Mass Spectrometry-Based Protein Interactome Studies. Mol. Cell. Proteom. 2025, 24, 100887. [Google Scholar] [CrossRef]
- Typas, A.; Sourjik, V. Bacterial Protein Networks: Properties and Functions. Nat. Rev. Microbiol. 2015, 13, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.; Herman, R.; Mancini, L.; Capitanchik, C.; Davey, K.; Dawson, C.S.; Ule, J.; Thomas, G.H.; Willis, A.E.; Lilley, K.S.; et al. Interrogation of RNA-Protein Interaction Dynamics in Bacterial Growth. Mol. Syst. Biol. 2024, 20, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Gómez Borrego, J.; Torrent Burgas, M. Structural Assembly of the Bacterial Essential Interactome. eLife 2024, 13, e94919. [Google Scholar] [CrossRef]
- Caufield, J.H.; Abreu, M.; Wimble, C.; Uetz, P. Protein Complexes in Bacteria. PLoS Comput. Biol. 2015, 11, e1004107. [Google Scholar] [CrossRef]
- Babu, M.; Bundalovic-Torma, C.; Calmettes, C.; Phanse, S.; Zhang, Q.; Jiang, Y.; Minic, Z.; Kim, S.; Mehla, J.; Gagarinova, A.; et al. Global Landscape of Cell Envelope Protein Complexes in Escherichia coli. Nat. Biotechnol. 2018, 36, 103–112. [Google Scholar] [CrossRef]
- Gómez Borrego, J.; Torrent Burgas, M. Analysis of Host-Bacteria Protein Interactions Reveals Conserved Domains and Motifs That Mediate Fundamental Infection Pathways. Int. J. Mol. Sci. 2022, 23, 11489. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; McClendon, C.L. Reaching for High-Hanging Fruit in Drug Discovery at Protein-Protein Interfaces. Nature 2007, 450, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kan, C.H.; Yang, X.; Ma, C. Inhibition of Bacterial RNA Polymerase Function and Protein-Protein Interactions: A Promising Approach for next-Generation Antibacterial Therapeutics. RSC Med. Chem. 2024, 15, 1471–1487. [Google Scholar] [CrossRef]
- Battesti, A.; Bouveret, E. The Bacterial Two-Hybrid System Based on Adenylate Cyclase Reconstitution in Escherichia coli. Methods 2012, 58, 325–334. [Google Scholar] [CrossRef]
- Fields, S.; Song, O. A Novel Genetic System to Detect Protein-Protein Interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Karimova, G.; Pidoux, J.; Ullmann, A.; Ladant, D. A Bacterial Two-Hybrid System Based on a Reconstituted Signal Transduction Pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5752–5756. [Google Scholar] [CrossRef]
- Mehla, J.; Caufield, J.H.; Sakhawalkar, N.; Uetz, P. A Comparison of Two-Hybrid Approaches for Detecting Protein-Protein Interactions. Methods Enzymol. 2017, 586, 333–358. [Google Scholar]
- Carlson, M.L.; Stacey, R.G.; Young, J.W.; Wason, I.S.; Zhao, Z.; Rattray, D.G.; Scott, N.; Kerr, C.H.; Babu, M.; Foster, L.J.; et al. Profiling the Escherichia coli Membrane Protein Interactome Captured in Peptidisc Libraries. eLife 2019, 8, e46615. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Larsen, B.; Lin, Z.-Y.; Breitkreutz, A.; Mellacheruvu, D.; Fermin, D.; Qin, Z.S.; Tyers, M.; Gingras, A.-C.; Nesvizhskii, A.I. SAINT: Probabilistic Scoring of Affinity Purification-Mass Spectrometry Data. Nat. Methods 2011, 8, 70–73. [Google Scholar] [CrossRef]
- Mann, M. Functional and Quantitative Proteomics Using SILAC. Nat. Rev. Mol. Cell Biol. 2006, 7, 952–958. [Google Scholar] [CrossRef]
- Penn, B.H.; Netter, Z.; Johnson, J.R.; Von Dollen, J.; Jang, G.M.; Johnson, T.; Ohol, Y.M.; Maher, C.; Bell, S.L.; Geiger, K.; et al. An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses. Mol. Cell 2018, 71, 637–648.e5. [Google Scholar] [CrossRef]
- Richards, A.L.; Eckhardt, M.; Krogan, N.J. Mass Spectrometry-Based Protein-Protein Interaction Networks for the Study of Human Diseases. Mol. Syst. Biol. 2021, 17, e8792. [Google Scholar] [CrossRef]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed Evolution of APEX2 for Electron Microscopy and Proximity Labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef]
- Choi-Rhee, E.; Schulman, H.; Cronan, J.E. Promiscuous Protein Biotinylation by Escherichia coli Biotin Protein Ligase. Protein Sci. 2004, 13, 3043–3050. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Cho, K.F.; Cavanagh, P.E.; Ting, A.Y. Deciphering Molecular Interactions by Proximity Labeling. Nat. Methods 2021, 18, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Herfurth, M.; Müller, F.; Søgaard-Andersen, L.; Glatter, T. A miniTurbo-Based Proximity Labeling Protocol to Identify Conditional Protein Interactomes in Vivo in Myxococcus xanthus. STAR Protoc. 2023, 4, 102657. [Google Scholar] [CrossRef]
- Yu, C.; Huang, L. New Advances in Cross-Linking Mass Spectrometry toward Structural Systems Biology. Curr. Opin. Chem. Biol. 2023, 76, 102357. [Google Scholar] [CrossRef]
- Nouchikian, L.; Fernandez-Martinez, D.; Renard, P.-Y.; Sabot, C.; Duménil, G.; Rey, M.; Chamot-Rooke, J. Do Not Waste Time—Ensure Success in Your Cross-Linking Mass Spectrometry Experiments before You Begin. Anal. Chem. 2024, 96, 2506–2513. [Google Scholar] [CrossRef] [PubMed]
- Khakzad, H.; Happonen, L.; Tran Van Nhieu, G.; Malmström, J.; Malmström, L. In Vivo Cross-Linking MS of the Complement System MAC Assembled on Live Gram-Positive Bacteria. Front. Genet. 2020, 11, 612475. [Google Scholar] [CrossRef] [PubMed]
- Schweppe, D.K.; Harding, C.; Chavez, J.D.; Wu, X.; Ramage, E.; Singh, P.K.; Manoil, C.; Bruce, J.E. Host-Microbe Protein Interactions during Bacterial Infection. Chem. Biol. 2015, 22, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Bellinzona, G.; Sassera, D.; Bonvin, A.M.J.J. Accelerating Protein-Protein Interaction Screens with Reduced AlphaFold-Multimer Sampling. Bioinform. Adv. 2024, 4, vbae153. [Google Scholar] [CrossRef]
- Lee, C.Y.; Hubrich, D.; Varga, J.K.; Schäfer, C.; Welzel, M.; Schumbera, E.; Djokic, M.; Strom, J.M.; Schönfeld, J.; Geist, J.L.; et al. Systematic Discovery of Protein Interaction Interfaces Using AlphaFold and Experimental Validation. Mol. Syst. Biol. 2024, 20, 75–97. [Google Scholar] [CrossRef]
- Abulude, I.J.; Luna, I.C.R.; Varela, A.S.; Camilli, A.; Kadouri, D.E.; Guo, X. Using AlphaFold-Multimer to Study Novel Protein-Protein Interactions of Predation Essential Hypothetical Proteins in Bdellovibrio. Front. Bioinform. 2025, 5, 1566486. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING Database in 2025: Protein Networks with Directionality of Regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.-J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Petrey, D.; Zhao, H.; Trudeau, S.J.; Murray, D.; Honig, B. PrePPI: A Structure Informed Proteome-Wide Database of Protein-Protein Interactions. J. Mol. Biol. 2023, 435, 168052. [Google Scholar] [CrossRef]
- Yang, X.; Lian, X.; Fu, C.; Wuchty, S.; Yang, S.; Zhang, Z. HVIDB: A Comprehensive Database for Human-Virus Protein-Protein Interactions. Brief. Bioinform. 2021, 22, 832–844. [Google Scholar] [CrossRef]
- Torchet, R.; Druart, K.; Ruano, L.C.; Moine-Franel, A.; Borges, H.; Doppelt-Azeroual, O.; Brancotte, B.; Mareuil, F.; Nilges, M.; Ménager, H.; et al. The iPPI-DB Initiative: A Community-Centered Database of Protein-Protein Interaction Modulators. Bioinformatics 2021, 37, 89–96. [Google Scholar] [CrossRef]
- Michnick, S.W. Three Decades of Protein-Fragment Complementation. Nat. Rev. Mol. Cell Biol. 2024, 26, 3–4. [Google Scholar] [CrossRef]
- Yang, D.; Jin, Y.; He, X.; Dong, A.; Wang, J.; Wu, R. Inferring Multilayer Interactome Networks Shaping Phenotypic Plasticity and Evolution. Nat. Commun. 2021, 12, 5304. [Google Scholar] [CrossRef]
- Cui, H.; Tejada-Lapuerta, A.; Brbić, M.; Saez-Rodriguez, J.; Cristea, S.; Goodarzi, H.; Lotfollahi, M.; Theis, F.J.; Wang, B. Towards Multimodal Foundation Models in Molecular Cell Biology. Nature 2025, 640, 623–633. [Google Scholar] [CrossRef]
- Rühling, M.; Schmelz, F.; Kempf, A.; Paprotka, K.; Fraunholz, M.J. Identification of the Staphylococcus aureus Endothelial Cell Surface Interactome by Proximity Labeling. mBio 2025, 16, e0365424. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Han, S.M.; Lee, J.-Y.; Kim, K.S.; Lee, J.-E.; Lee, D.-W. Advancing Gut Microbiome Research: The Shift from Metagenomics to Multi-Omics and Future Perspectives. J. Microbiol. Biotechnol. 2025, 35, e2412001. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Rai, N.; Zorraquino, V.; Tagkopoulos, I. Multi-Omics Integration Accurately Predicts Cellular State in Unexplored Conditions for Escherichia coli. Nat. Commun. 2016, 7, 13090. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Cheng, Y.; Lv, X.; Liu, Y.; Li, J.; Du, G.; Chen, J.; Liu, L. A Multi-Omics, Machine Learning-Aware, Genome-Wide Metabolic Model of Bacillus subtilis Refines the Gene Expression and Cell Growth Prediction. Adv. Sci. 2024, 11, e2408705. [Google Scholar] [CrossRef]
- Georgouli, K.; Yeom, J.-S.; Blake, R.C.; Navid, A. Multi-Scale Models of Whole Cells: Progress and Challenges. Front. Cell Dev. Biol. 2023, 11, 1260507. [Google Scholar] [CrossRef] [PubMed]
- Karr, J.R.; Sanghvi, J.C.; Macklin, D.N.; Gutschow, M.V.; Jacobs, J.M.; Bolival, B., Jr.; Assad-Garcia, N.; Glass, J.I.; Covert, M.W. A Whole-Cell Computational Model Predicts Phenotype from Genotype. Cell 2012, 150, 389–401. [Google Scholar] [CrossRef]
- Macklin, D.N.; Ahn-Horst, T.A.; Choi, H.; Ruggero, N.A.; Carrera, J.; Mason, J.C.; Sun, G.; Agmon, E.; DeFelice, M.M.; Maayan, I.; et al. Simultaneous Cross-Evaluation of Heterogeneous E. coli Datasets via Mechanistic Simulation. Science 2020, 369, eaav3751. [Google Scholar] [CrossRef]
- Choi, H.; Covert, M.W. Whole-Cell Modeling of E. Coli Confirms That in Vitro tRNA Aminoacylation Measurements Are Insufficient to Support Cell Growth and Predicts a Positive Feedback Mechanism Regulating Arginine Biosynthesis. Nucleic Acids Res. 2023, 51, 5911–5930. [Google Scholar] [CrossRef]
- Skalnik, C.J.; Cheah, S.Y.; Yang, M.Y.; Wolff, M.B.; Spangler, R.K.; Talman, L.; Morrison, J.H.; Peirce, S.M.; Agmon, E.; Covert, M.W. Whole-Cell Modeling of E. Coli Colonies Enables Quantification of Single-Cell Heterogeneity in Antibiotic Responses. PLoS Comput. Biol. 2023, 19, e1011232. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Didelot, X.; Meric, G.; Torralbo, A.; Jolley, K.A.; Kelly, D.J.; Bentley, S.D.; Maiden, M.C.J.; Parkhill, J.; Falush, D. Genome-Wide Association Study Identifies Vitamin B5 Biosynthesis as a Host Specificity Factor in Campylobacter. Proc. Natl. Acad. Sci. USA 2013, 110, 11923–11927. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, D.P.; Streich, J.C.; Burdick, L.H.; Klingeman, D.M.; Chhetri, H.B.; Brelsford, C.M.; Ellis, J.C.; Close, D.M.; Jacobson, D.A.; Michener, J.K. Protoplast Fusion in Bacillus Species Produces Frequent, Unbiased, Genome-Wide Homologous Recombination. Nucleic Acids Res. 2022, 50, 6211–6223. [Google Scholar] [CrossRef]
- Fernández-García, G.; Valdés-Chiara, P.; Villazán-Gamonal, P.; Alonso-Fernández, S.; Manteca, A. Essential Genes Discovery in Microorganisms by Transposon-Directed Sequencing (Tn-Seq): Experimental Approaches, Major Goals, and Future Perspectives. Int. J. Mol. Sci. 2024, 25, 11298. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Fenster, J.A.; Fankhauser, R.G.; Kaar, J.L.; Tenaillon, O.; Gill, R.T. CRISPR/Cas9 Recombineering-Mediated Deep Mutational Scanning of Essential Genes in Escherichia Coli. Mol. Syst. Biol. 2020, 16, e9265. [Google Scholar] [CrossRef]
- Ugolini, G.S.; Wang, M.; Secchi, E.; Pioli, R.; Ackermann, M.; Stocker, R. Microfluidic Approaches in Microbial Ecology. Lab Chip 2024, 24, 1394–1418. [Google Scholar] [CrossRef]
- Ripandelli, R.A.A.; van Oijen, A.M.; Robinson, A. Single-Cell Microfluidics: A Primer for Microbiologists. J. Phys. Chem. B 2024, 128, 10311–10328. [Google Scholar] [CrossRef]
- Blaszczak, E.; Lazarewicz, N.; Sudevan, A.; Wysocki, R.; Rabut, G. Protein-Fragment Complementation Assays for Large-Scale Analysis of Protein-Protein Interactions. Biochem. Soc. Trans. 2021, 49, 1337–1348. [Google Scholar] [CrossRef]
- Liu, Z.; Miller, D.; Li, F.; Liu, X.; Levy, S.F. A Large Accessory Protein Interactome Is Rewired across Environments. eLife 2020, 9, e62365. [Google Scholar] [CrossRef]
- Ebrahimi, V.; Hashemi, A. Challenges of in Vitro Genome Editing with CRISPR/Cas9 and Possible Solutions: A Review. Gene 2020, 753, 144813. [Google Scholar] [CrossRef] [PubMed]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using Machine Learning Approaches for Multi-Omics Data Analysis: A Review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef] [PubMed]
- Reed, T.J.; Tyl, M.D.; Tadych, A.; Troyanskaya, O.G.; Cristea, I.M. Tapioca: A Platform for Predicting de Novo Protein-Protein Interactions in Dynamic Contexts. Nat. Methods 2024, 21, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, Q.; Aufschnaiter, A.; Ott, M.; Meyer, J.G. Multi-Omic Integration by Machine Learning (MIMaL). Bioinformatics 2022, 38, 4908–4918, Erratum in Bioinformatics 2023, 39, btad146. [Google Scholar] [CrossRef]

| Technology | Layer | Resolution | Key Features | Advantages | Limitations | Ref. |
|---|---|---|---|---|---|---|
| Bulk RNA-seq | Transcriptome | Population | Short-read sequencing of pooled transcripts | High throughput, broad dynamic range | Averages out cell heterogeneity | [69] |
| Dual RNA-seq | Transcriptome | Population (host + microbe) | Simultaneous sequencing of host and bacterial transcripts | Captures infection dialogue | Complex analysis; host RNA often dominates | [20] |
| Oxford Nanopre Technology (ONT) Long-Read Sequencing | Transcriptome | Population | Direct sequencing of cDNA or native RNA; maps operons & detects RNA modifications | Preserves modifications; resolves full-length transcripts | Lower throughput; base-calling errors | [72] |
| Single-cell RNA-seq | Transcriptome | Single-cell | FACS + random-hexamer priming (MATQ-seq); split-pool barcoding (PETRI-seq, MicroSPLiT); droplet-based platforms (M3-seq, BacDrop, smRandom-seq); droplet + probe (ProBac-seq) | Detects extremely rare subpopulations (<0.1%); reveals heterogeneity within clonal populations | Lower throughput; complex workflows; higher cost | [21,22,23,24,70,73,85,88,89] |
| Spatial transcriptomics (e.g., par-seqFISH) | Transcriptome | Spatial | Sequential hybridization and imaging of marker genes in fixed biofilm | Spatial mapping of expression at micron scale | Limited number of target genes; requires fixed samples | [25] |
| DIA-MS | Proteome | Population | Systematic fragmentation of all detectable precursor ions | High reproducibility; fewer missing values; quantitative | Requires optimized spectral libraries | [96] |
| Top-Down Proteomics (TDP) | Proteome | Proteoform | Intact protein analysis to capture sequence variants and PTMs | Direct identification of proteoforms; PTM mapping | Low throughput; specialized equipment | [101,102] |
| Single-Bacterium Proteomics (SBP) | Proteome | Single-cell | SCOPE-MS with carrier proteome | Detects proteins in individual bacterial cells | Very low protein amounts; method still developing | [104] |
| EXCRETE Workflow | Proteome | Secretome | Bead-based aggregation & digestion | High-yield, high-throughput secretome profiling | Limited to extracellular proteins; may miss low-abundance targets | [108] |
| Method | Interaction Type | In Vivo/ In Vitro | Resolution | Strengths | Limitations | Ref. |
|---|---|---|---|---|---|---|
| Yeast Two-Hybrid (Y2H) | Binary PPIs | In Vivo (yeast) | Protein–protein | High throughput; well-established; cost-effective | Non-native environment for bacterial proteins; may produce false positives/negatives | [120] |
| Bacterial Two-Hybrid (B2H) | Binary PPIs | In Vivo | Protein–protein | Native bacterial environment; effective for membrane proteins; high throughput | May miss transient interactions; exogenous system may alter relative abundance of hybrid proteins | [121] |
| Protein Fragment Complementation Assay (PCA) | Binary PPIs | In Vivo | Protein–protein | Detects interactions under native regulatory control | Requires genome tagging of all target genes; potential labeling bias | [144] |
| Affinity Purification–MS (AP-MS) | Stable complexes | In Vitro | Complex composition | Quantitative (q-AP-MS); adaptable to many proteins | Requires tagged bait; may disrupt physiological interactions; may miss weak/transient interactions | [126] |
| Proximity Labeling (PL) | Stable + transient | In Vivo | Spatial proximity (~10–20 nm) | Captures weak/transient interactions; preserves native state | Labeling bias; requires fusion construct; difficult to distinguish between direct/indirect associations | [131] |
| Cross-Linking MS (XL-MS) | Stable + transient | In Vivo/In Vitro | Residue-level | Provides structural constraints; models large complexes | May miss weak/transient interactions due to cross-linker accessibility; complex workflow | [134,135] |
| AlphaFold-Multimer | Predicted PPIs | In Silico | Structural model | Proteome-scale predictions; structural insight | Requires experimental validation | [138] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakaguchi, T.; Irifune, Y.; Kamada, R.; Sakaguchi, K. Bacterial Systematic Genetics and Integrated Multi-Omics: Beyond Static Genomics Toward Predictive Models. Int. J. Mol. Sci. 2025, 26, 9326. https://doi.org/10.3390/ijms26199326
Sakaguchi T, Irifune Y, Kamada R, Sakaguchi K. Bacterial Systematic Genetics and Integrated Multi-Omics: Beyond Static Genomics Toward Predictive Models. International Journal of Molecular Sciences. 2025; 26(19):9326. https://doi.org/10.3390/ijms26199326
Chicago/Turabian StyleSakaguchi, Tatsuya, Yuta Irifune, Rui Kamada, and Kazuyasu Sakaguchi. 2025. "Bacterial Systematic Genetics and Integrated Multi-Omics: Beyond Static Genomics Toward Predictive Models" International Journal of Molecular Sciences 26, no. 19: 9326. https://doi.org/10.3390/ijms26199326
APA StyleSakaguchi, T., Irifune, Y., Kamada, R., & Sakaguchi, K. (2025). Bacterial Systematic Genetics and Integrated Multi-Omics: Beyond Static Genomics Toward Predictive Models. International Journal of Molecular Sciences, 26(19), 9326. https://doi.org/10.3390/ijms26199326






