Transcriptomic Analysis of TDP1-Knockout HEK293A Cells Treated with the TDP1 Inhibitor (Usnic Acid Derivative)
Abstract
1. Introduction
2. Results
2.1. The Effect of OL9-119 on the Transcriptome of HEK293A Cells
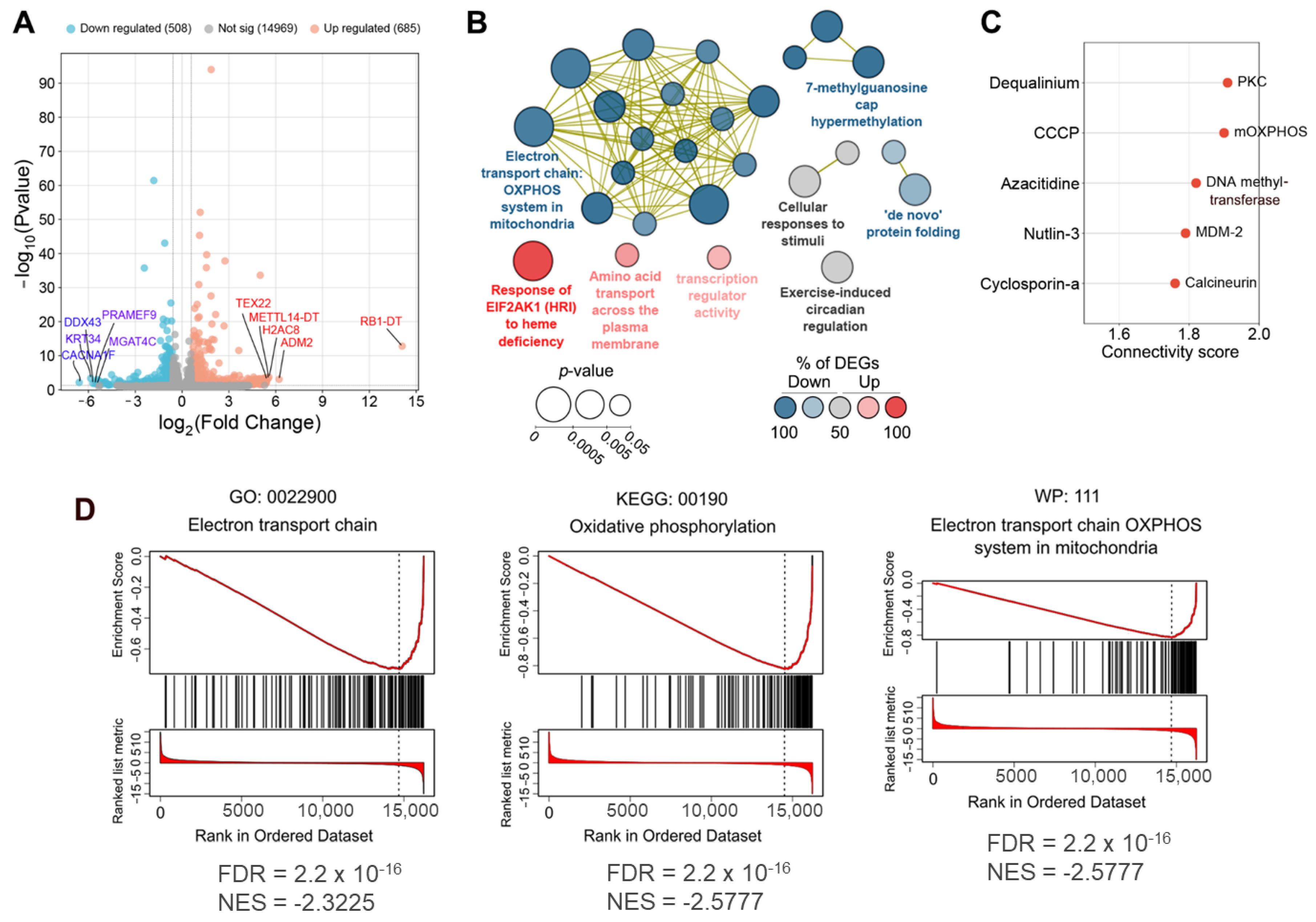
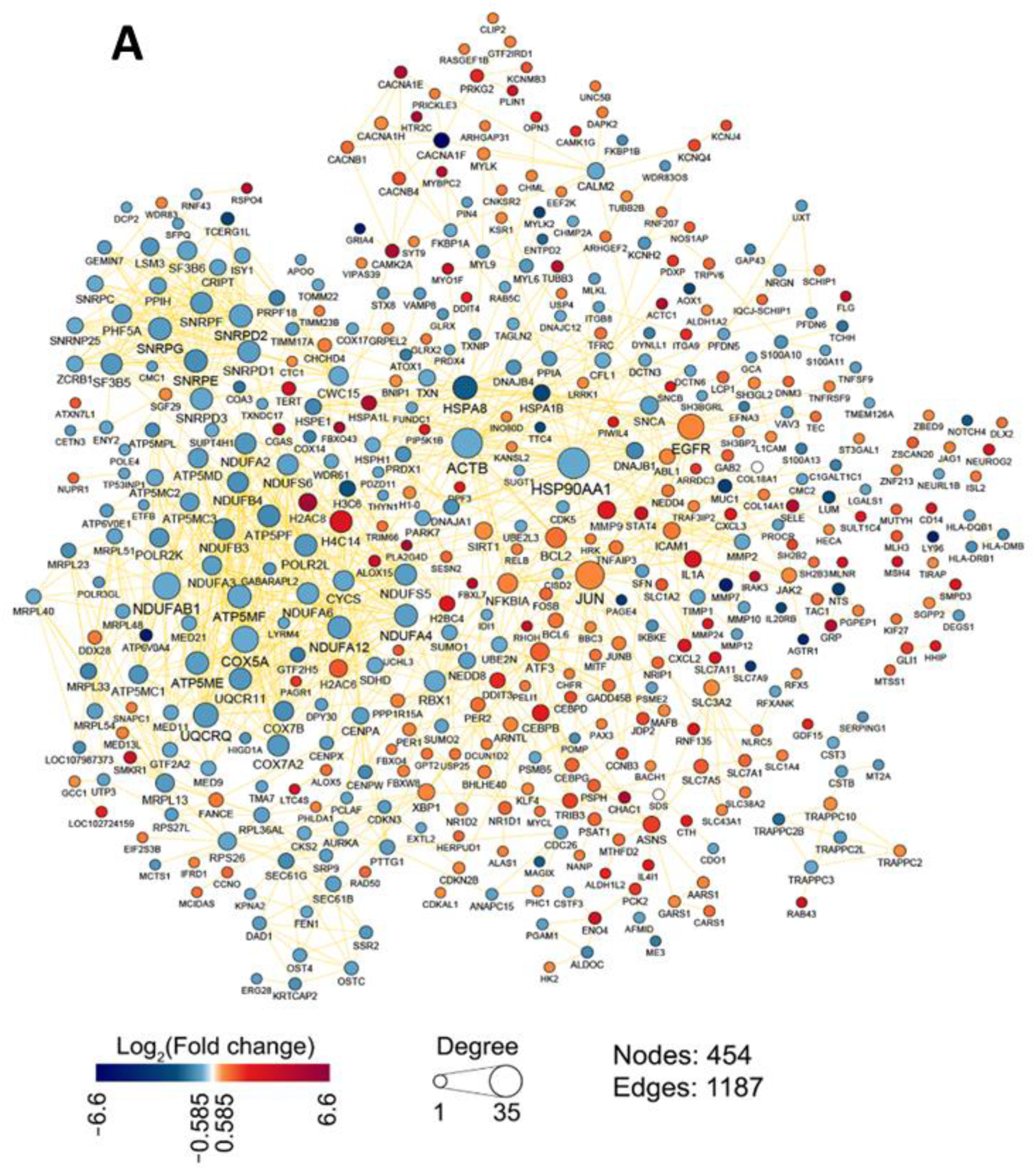
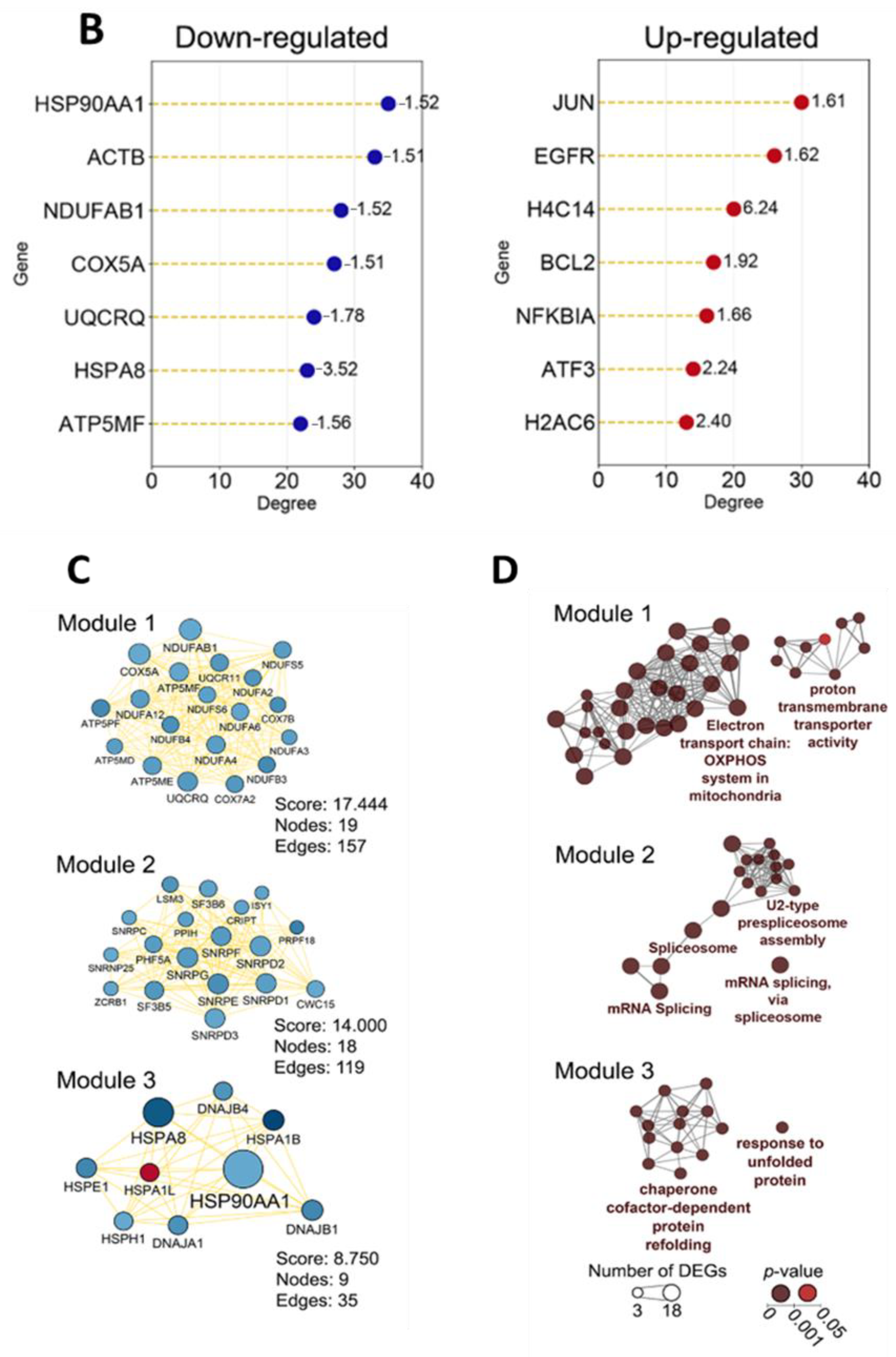
2.1.1. Topological Analysis of OL9-119-Sensitive Regulome
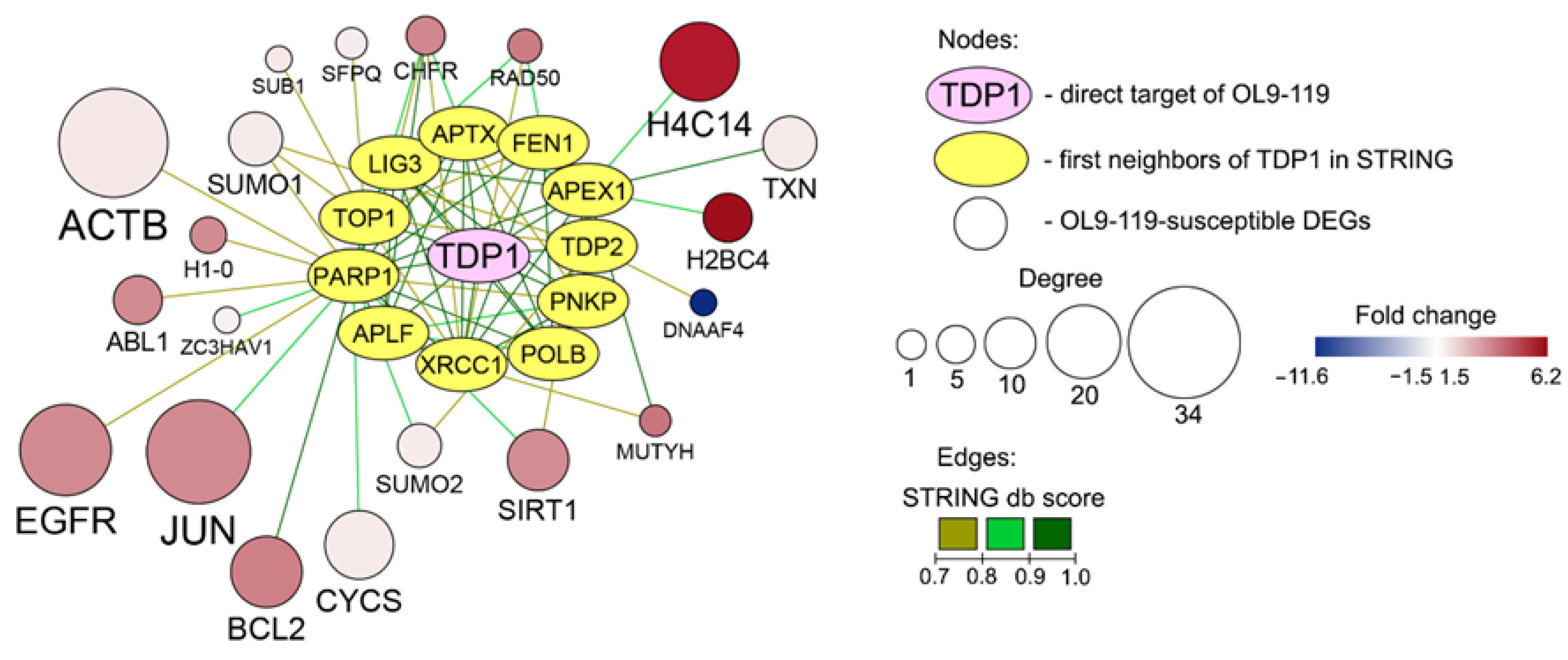
2.1.2. The Relation of TDP1-Inhibiting Activity of OL9-119 with OL9-119-Susceptible Hub Genes
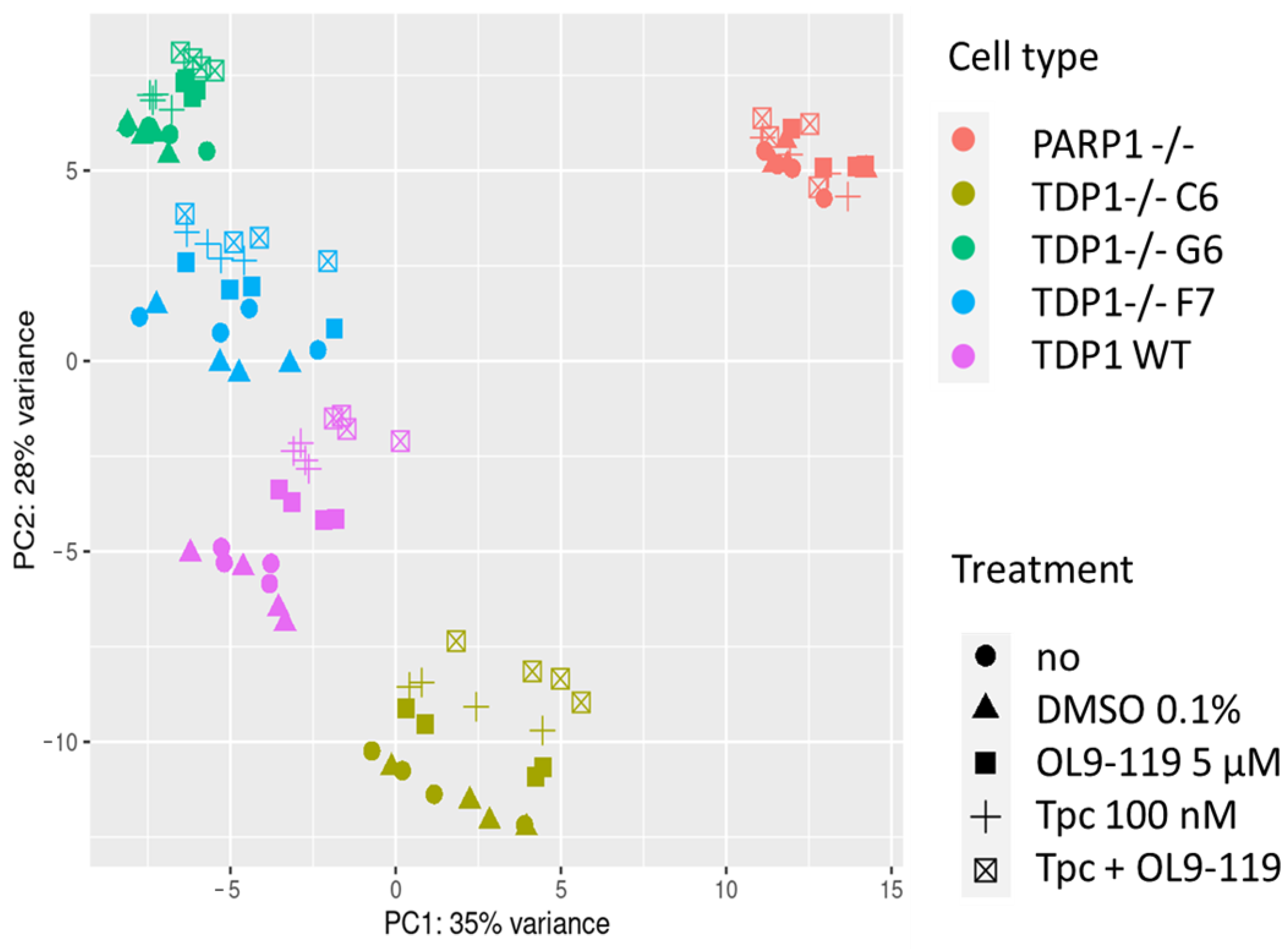
2.1.3. OL9-119 Effects on Transcriptome in the Context of TDP1 Knockout
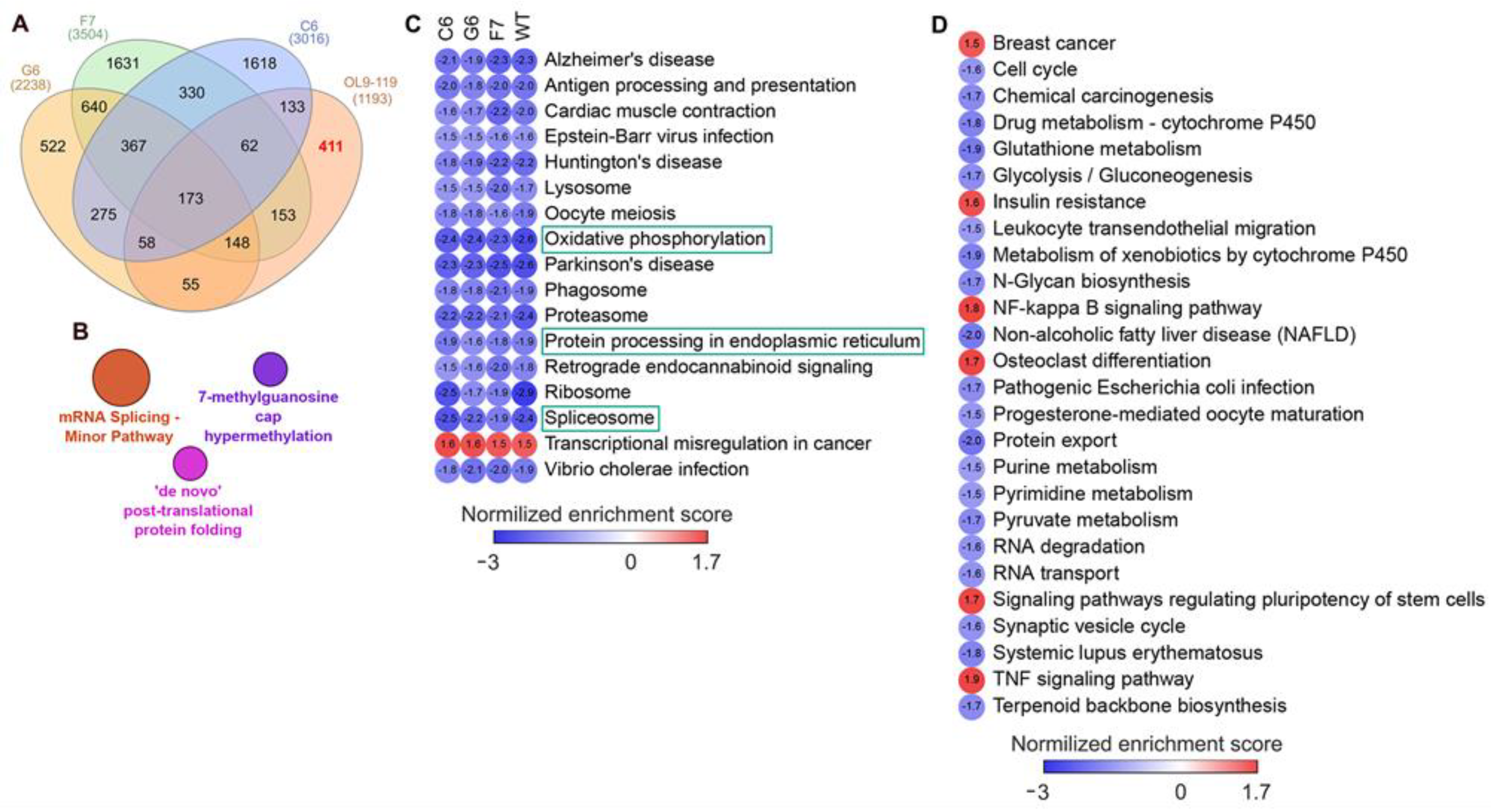
- First, we overlaid DEGs determined in OL9-119-treated wild-type cells with DEGs specific to each TDP1 knockout cell strain (C6, F7, and G6; knockout versus wild-type cells). As shown in the Venn diagram (Figure 5A), the effect of OL9-119 on the HEK293A cell transcriptome is quite similar to the TDP1 knockout effect. Only 34% (411 genes) of all OL9-119-susceptible DEGs are unique to the compound under study. Considering that these genes may determine the off-target effects of OL9-119, we performed a functional annotation of these genes. As shown in Figure 5B, the unique DEGs not associated with the TDP1-targeting effect of OL9-119 were related to RNA processing and protein folding, as mentioned earlier in Section 2.1 (Figure 2B).
- Second, to distinguish more precisely between TDP1-independent and TDP1-dependent processes modulated by OL9-119, RNA sequencing was performed on G6, F7, and G6 knockout cells treated with the tested compound (a table of DEGs for all cell types is provided in Table S1). Further GSEA of obtained DEGs (OL9-119-treated vs. untreated cells) revealed 17 processes common for both knockout clones and wild-type cells, which indicates that OL9-119 modulates them independently of TDP1 status (off-target effect) (Figure 5C). Among these processes, three functional terms—namely, oxidative phosphorylation, protein processing in the endoplasmic reticulum (folding), and the spliceosome—have been previously identified (Figure 2B,D, Figure 3B, and Figure 5B). In turn, TDP1-dependent processes, which OL9-119 modulated in only wild-type cells and which were absent in knockout clones, involved various metabolic processes, cell proliferation, inflammation-related terms, and cell differentiation (Figure 5D).
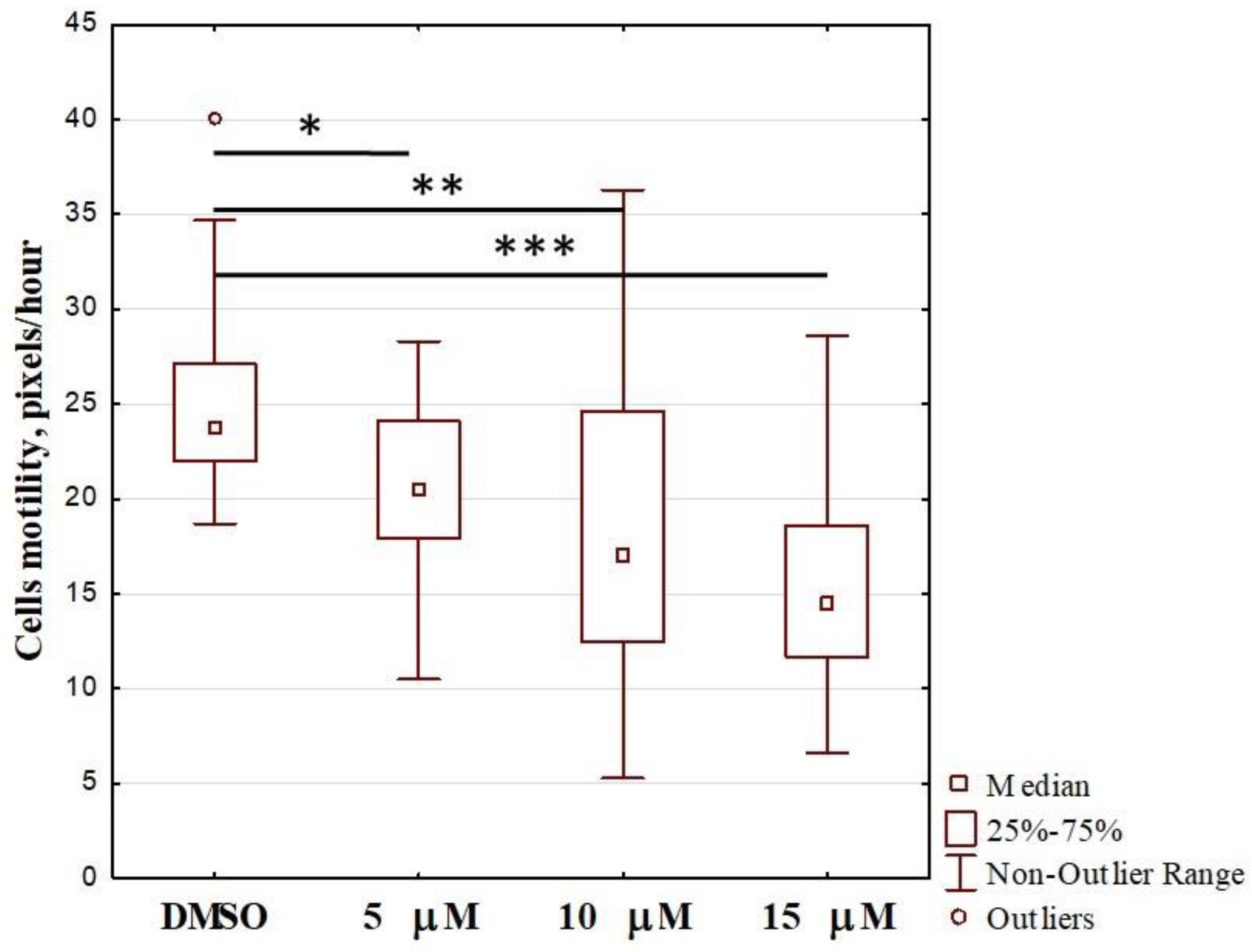
2.2. Effect of OL9-119 on Cell Motility
2.3. The Effect of OL9-119 on Mitochondrial Membrane Potential
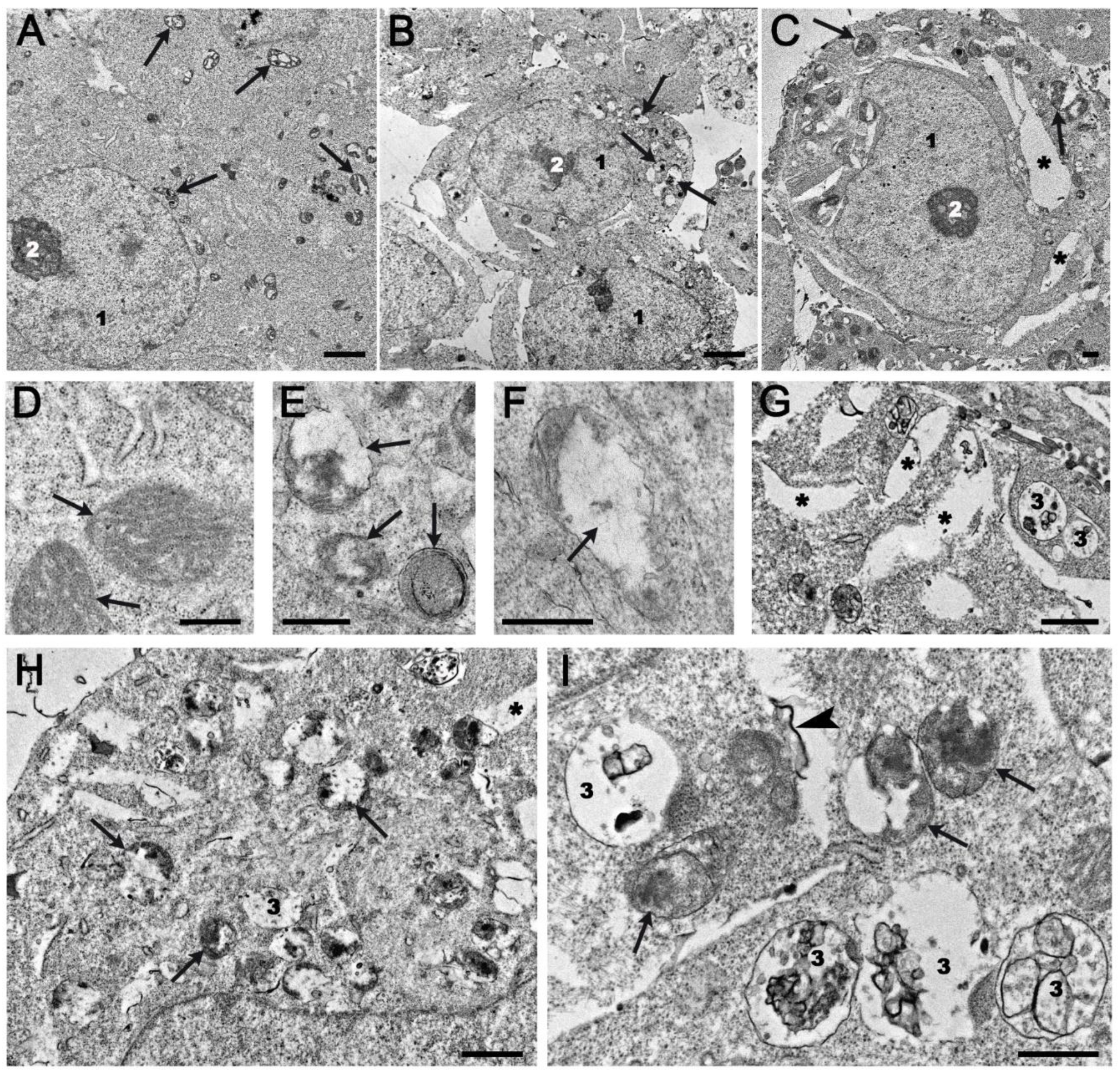
2.4. Ultrastructural Changes in HEK293A Cells Under the Effects of Topotecan and OL9-119
3. Materials and Methods
3.1. Total RNA Preparation and Transcriptome Sequencing
3.2. Bioinformatic Analysis
3.3. Analysis of HEK293A WT Cell Motility Under OL9-119 Treatment
3.4. Analysis of Mitochondrial Membrane Potential
3.5. Transmission Electron Microscope (TEM) Examinations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, D.R.; Interthal, H.; Champoux, J.J.; Hol, W.G. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure 2002, 10, 237–248. [Google Scholar] [CrossRef]
- Yang, S.W.; Burgin, A.B., Jr.; Huizenga, B.N.; Robertson, C.A.; Yao, K.C.; Nash, H.A. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA 1996, 93, 11534–11539. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.L.; Luzina, O.A.; Chepanova, A.A.; Dyrkheeva, N.S.; Salakhutdinov, N.F.; Lavrik, O.I. Natural Products and Their Derivatives as Inhibitors of the DNA Repair Enzyme Tyrosyl-DNA Phosphodiesterase 1. Int. J. Mol. Sci. 2023, 24, 5781. [Google Scholar] [CrossRef] [PubMed]
- Roca, J. The mechanisms of DNA topoisomerases. Trends Biochem. Sci. 1995, 20, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Huang, S.N.; Gao, R.; Das, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 2014, 19, 114–129. [Google Scholar] [CrossRef]
- Comeaux, E.Q.; van Waardenburg, R.C. Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target. Drug Metab. Rev. 2014, 46, 494–507. [Google Scholar] [CrossRef]
- Lebedeva, N.A.; Rechkunova, N.I.; El-Khamisy, S.F.; Lavrik, O.I. Tyrosyl-DNA phosphodiesterase 1 initiates repair of apurinic/apyrimidinic sites. Biochimie 2012, 94, 1749–1753. [Google Scholar] [CrossRef]
- Dyrkheeva, N.; Anarbaev, R.; Lebedeva, N.; Kuprushkin, M.; Kuznetsova, A.; Kuznetsov, N.; Rechkunova, N.; Lavrik, O. Human Tyrosyl-DNA phosphodiesterase 1 possesses transphosphooligonucleotidation activity with primary alcohols. Front. Cell Dev. Biol. 2020, 8, 604732. [Google Scholar] [CrossRef]
- Li, J.; Summerlin, M.; Nitiss, K.C.; Nitiss, J.L.; Hanakahi, L.A. TDP1 is required for efficient non-homologous end joining in human cells. DNA Repair 2017, 60, 40–49. [Google Scholar] [CrossRef]
- Chiang, S.C.; Meagher, M.; Kassouf, N.; Hafezparast, M.; McKinnon, P.J.; Haywood, R.; El-Khamisy, S.F. Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical-induced DNA damage. Sci. Adv. 2017, 3, e1602506. [Google Scholar] [CrossRef]
- Chowdhuri, S.P.; Das, B.B. Top1-PARP1 association and beyond: From DNA topology to break repair. NAR Cancer 2021, 3, zcab003. [Google Scholar] [CrossRef] [PubMed]
- Das, B.B.; Antony, S.; Gupta, S.; Dexheimer, T.S.; Redon, C.E.; Garfield, S.; Shiloh, Y.; Pommier, Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009, 28, 3667–3680. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.C.; Carroll, J.; El-Khamisy, S.F. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle 2010, 9, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.J.; Chiang, S.C.; Wells, O.S.; Rookyard, C.; El-Khamisy, S.F. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat. Commun. 2012, 3, 733. [Google Scholar] [CrossRef]
- Das, B.B.; Ghosh, A.; Bhattacharjee, S.; Bhattacharyya, A. Trapped topoisomerase-DNA covalent complexes in the mitochondria and their role in human diseases. Mitochondrion 2021, 60, 234–244. [Google Scholar] [CrossRef]
- El-Khamisy, S.F.; Caldecott, K.W. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis 2006, 21, 219–224. [Google Scholar] [CrossRef]
- Zakharenko, A.; Dyrkheeva, N.; Lavrik, O. Dual DNA topoisomerase 1 and tyrosyl-DNA phosphodiesterase 1 inhibition for improved anticancer activity. Med. Res. Rev. 2019, 39, 1427–1441. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Antony, S.; Marchand, C.; Pommier, Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 2008, 8, 381–389. [Google Scholar] [CrossRef]
- Meisenberg, C.; Gilbert, D.C.; Chalmers, A.; Haley, V.; Gollins, S.; Ward, S.E.; El-Khamisy, S.F. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan. Mol. Cancer Ther. 2015, 14, 575–585. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, S.; Begum, S.; Sidransky, D.; Westra, W.H.; Brock, M.; Califano, J.A. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer 2007, 55, 303–311. [Google Scholar] [CrossRef]
- Fam, H.K.; Walton, C.; Mitra, S.A.; Chowdhury, M.; Osborne, N.; Choi, K.; Sun, G.; Wong, P.C.; O’Sullivan, M.J.; Turashvili, G.; et al. TDP1 and PARP1 deficiency are cytotoxic to rhabdomyosarcoma cells. Mol. Cancer Res. 2013, 11, 1179–1192. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Das, B.B.; Chatterjee, R.; Abaan, O.D.; Agama, K.; Matuo, R.; Vinson, C.; Meltzer, P.S.; Pommier, Y. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair 2014, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Liu, J.; Baglo, Y.; Rizvi, I.; Anbil, S.; Pigula, M.; Hasan, T. Mechanism-informed Repurposing of Minocycline Overcomes Resistance to Topoisomerase Inhibition for Peritoneal Carcinomatosis. Mol. Cancer Ther. 2018, 17, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Alagoz, M.; Gilbert, D.C.; El-Khamisy, S.; Chalmers, A.J. DNA repair and resistance to topoisomerase I inhibitors: Mechanisms, biomarkers and therapeutic targets. Curr. Med. Chem. 2012, 19, 3874–3885. [Google Scholar] [CrossRef]
- Brettrager, E.J.; van Waardenburg, R.C.A.M. Targeting Tyrosyl-DNA phosphodiesterase I to enhance toxicity of phosphodiester linked DNA-adducts. Cancer Drug Resist. 2019, 2, 1153–1163. [Google Scholar] [CrossRef]
- Leung, E.; Patel, J.; Hollywood, J.A.; Zafar, A.; Tomek, P.; Barker, D.; Pilkington, L.I.; van Rensburg, M.; Langley, R.J.; Helsby, N.A.; et al. Validating TDP1 as an Inhibition Target for the Development of Chemosensitizers for Camptothecin-Based Chemotherapy Drugs. Oncol. Ther. 2021, 9, 541–556. [Google Scholar] [CrossRef]
- Khomenko, T.M.; Zakharenko, A.L.; Kornienko, T.E.; Chepanova, A.A.; Dyrkheeva, N.S.; Artemova, A.O.; Korchagina, D.V.; Achara, C.; Curtis, A.; Reynisson, J.; et al. New 5-Hydroxycoumarin-Based Tyrosyl-DNA Phosphodiesterase I Inhibitors Sensitize Tumor Cell Line to Topotecan. Int. J. Mol. Sci. 2023, 24, 9155. [Google Scholar] [CrossRef]
- Nikolin, V.P.; Popova, N.A.; Kaledin, V.I.; Luzina, O.A.; Zakharenko, A.L.; Salakhutdinov, N.F.; Lavrik, O.I. The influence of an enamine usnic acid derivative (a tyrosyl-DNA phosphodiesterase 1 inhibitor) on the therapeutic effect of topotecan against transplanted tumors in vivo. Clin. Exp. Metastasis 2021, 38, 431–440. [Google Scholar] [CrossRef]
- Koldysheva, E.V.; Men’shchikova, A.P.; Lushnikova, E.L.; Popova, N.A.; Kaledin, V.I.; Nikolin, V.P.; Zakharenko, A.L.; Luzina, O.A.; Salakhutdinov, N.F.; Lavrik, O.I. Antimetastatic Activity of Combined Topotecan and Tyrosyl-DNA Phosphodiesterase-1 Inhibitor on Modeled Lewis Lung Carcinoma. Bull. Exp. Biol. Med. 2019, 166, 661–666. [Google Scholar] [CrossRef]
- Dyrkheeva, N.S.; Zakharenko, A.L.; Malakhova, A.A.; Okorokova, L.S.; Shtokalo, D.N.; Medvedev, S.P.; Tupikin, A.A.; Kabilov, M.R.; Lavrik, O.I. Transcriptomic analysis of HEK293A cells with a CRISPR/Cas9-mediated TDP1 knockout. Biochim. Biophys. Acta Gen. Subj. 2024, 1868, 130616. [Google Scholar] [CrossRef]
- Zakharenko, A.L.; Luzina, O.A.; Sokolov, D.N.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Patel, J.; Zakharova, O.D.; Chepanova, A.A.; Zafar, A.; et al. Novel Tyrosyl-DNA Phosphodiesterase 1 Inhibitors Enhance the Therapeutic Impact of Topoteñan on in Vivo Tumor Models. Eur. J. Med. Chem. 2019, 161, 581–593. [Google Scholar] [CrossRef]
- Dyrkheeva, N.S.; Malakhova, A.A.; Zakharenko, A.L.; Okorokova, L.S.; Shtokalo, D.N.; Pavlova, S.V.; Medvedev, S.P.; Zakian, S.M.; Nushtaeva, A.A.; Tupikin, A.E.; et al. Transcriptomic Analysis of CRISPR/Cas9-Mediated PARP1-Knockout Cells under the Influence of Topotecan and TDP1 Inhibitor. Int. J. Mol. Sci. 2023, 24, 5148. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef] [PubMed]
- Costes, L.H.; Lippi, Y.; Naylies, C.; Jamin, E.L.; Genthon, C.; Bailly, S.; Oswald, I.P.; Bailly, J.D.; Puel, O. The Solvent Dimethyl Sulfoxide Affects Physiology, Transcriptome and Secondary Metabolism of Aspergillus flavus. J. Fungi 2021, 7, 1055. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Rao, Z.; Han, R.; Jin, L.; Cao, L. Genes involved in DMSO-mediated yield increase of entomopathogenic nematodes. Sci. Rep. 2024, 14, 31670. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shi, J.; Fu, X.; Mao, B.; Wang, W.; Li, W.; Li, G.; Zhou, S. Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget 2018, 9, 37471–37479. [Google Scholar] [CrossRef]
- Kovaleva, I.E.; Garaeva, A.A.; Chumakov, P.M.; Evstafieva, A.G. Intermedin/adrenomedullin 2 is a stress-inducible gene controlled by activating transcription factor 4. Gene 2016, 590, 177–185. [Google Scholar] [CrossRef]
- Shah, S.; Verma, T.; Rashid, M.; Gadewal, N.; Gupta, S. Histone H2A isoforms: Potential implications in epigenome plasticity and diseases in eukaryotes. J. Biosci. 2020, 45, 4. [Google Scholar] [CrossRef]
- Beck, J.S.; Madaj, Z.; Cheema, C.T.; Kara, B.; Bennett, D.A.; Schneider, J.A.; Gordon, M.N.; Ginsberg, S.D.; Mufson, E.J.; Counts, S.E. Co-expression network analysis of frontal cortex during the progression of Alzheimer’s disease. Cereb. Cortex 2022, 32, 5108–5120. [Google Scholar] [CrossRef]
- Szymanowicz, O.; Drużdż, A.; Słowikowski, B.; Pawlak, S.; Potocka, E.; Goutor, U.; Konieczny, M.; Ciastoń, M.; Lewandowska, A.; Jagodziński, P.P.; et al. A Review of the CACNA Gene Family: Its Role in Neurological Disorders. Diseases 2024, 12, 90. [Google Scholar] [CrossRef]
- Pavez Lorie, E.; Stricker, N.; Plitta-Michalak, B.; Chen, I.-P.; Volkmer, B.; Greinert, R.; Jauch, A.; Boukamp, P.; Rapp, A. Characterisation of the novel spontaneously immortalized and invasively growing human skin keratinocyte line HaSKpw. Sci. Rep. 2020, 10, 15196. [Google Scholar] [CrossRef]
- Lin, H.-C.; Yeh, C.-W.; Chen, Y.-F.; Lee, T.-T.; Hsieh, P.-Y.; Rusnac, D.V.; Lin, S.-Y.; Elledge, S.J.; Zheng, N.; Yen, H.-C.S. C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol. Cell 2018, 70, 602–613.e3. [Google Scholar] [CrossRef]
- Chen, S.N.; Peng, X.Y.; Nie, P. The composition and unrevealed immune role of non-RLR DExD/H box RNA helicases in fish. Comp. Immunol. Rep. 2024, 7, 200172. [Google Scholar] [CrossRef]
- Gisina, A.; Yarygin, K.; Lupatov, A. The Impact of Glycosylation on the Functional Activity of CD133 and the Accuracy of Its Immunodetection. Biology 2024, 13, 449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, X.C.; Huang, Y.Y.; Ge, Y.P.; Sun, M.; Chen, W.L.; Liu, W.B.; Li, X.F. Carbonyl cyanide 3-chlorophenylhydrazone induced the imbalance of mitochondrial homeostasis in the liver of Megalobrama amblycephala: A dynamic study. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 244, 109003. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. DNA single-strand break repair and human genetic disease. Trends Cell Biol. 2022, 32, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Moor, N.A.; Vasil’eva, I.A.; Anarbaev, R.O.; Antson, A.A.; Lavrik, O.I. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015, 43, 6009–6022. [Google Scholar] [CrossRef]
- Das, B.B.; Huang, S.Y.; Murai, J.; Rehman, I.; Amé, J.C.; Sengupta, S.; Das, S.K.; Majumdar, P.; Zhang, H.; Biard, D.; et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014, 42, 4435–4449. [Google Scholar] [CrossRef]
- Jubin, T.; Kadam, A.; Jariwala, M.; Bhatt, S.; Sutariya, S.; Gani, A.R.; Gautam, S.; Begum, R. The PARP family: Insights into functional aspects of poly (ADP-ribose) polymerase-1 in cell growth and survival. Cell Prolif. 2016, 49, 421–437. [Google Scholar] [CrossRef]
- Kwong, S.P.; Wang, C. Review: Usnic acid-induced hepatotoxicity and cell death. Environ. Toxicol. Pharmacol. 2020, 80, 103493. [Google Scholar] [CrossRef]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Wu, W.; Gou, H.; Dong, J.; Yang, X.; Zhao, Y.; Peng, H.; Chen, D.; Geng, R.; Chen, L.; Liu, J. Usnic Acid Inhibits Proliferation and Migration through ATM Mediated DNA Damage Response in RKO Colorectal Cancer Cell. Curr. Pharm. Biotechnol. 2021, 22, 1129–1138. [Google Scholar] [CrossRef]
- Vasarri, M.; Ponti, L.; Degl’Innocenti, D.; Bergonzi, M.C. Usnic Acid-Loaded Polymeric Micelles: An Optimal Migrastatic-Acting Formulation in Human SH-SY5Y Neuroblastoma Cells. Pharmaceuticals 2022, 15, 1207. [Google Scholar] [CrossRef]
- Gimła, M.; Herman-Antosiewicz, A. Multifaceted Properties of Usnic Acid in Disrupting Cancer Hallmarks. Biomedicines 2024, 12, 2199. [Google Scholar] [CrossRef]
- Yang, Y.; Nguyen, T.T.; Jeong, M.H.; Crişan, F.; Yu, Y.H.; Ha, H.H.; Choi, K.H.; Jeong, H.G.; Jeong, T.C.; Lee, K.Y.; et al. Inhibitory Activity of (+)-Usnic Acid against Non-Small Cell Lung Cancer Cell Motility. PLoS ONE 2016, 11, e0146575. [Google Scholar] [CrossRef]
- Petrová, K.; Bačkorová, M.; Demčišáková, Z.; Petrovová, E.; Goga, M.; Vilková, M.; Frenák, R.; Bačkor, M.; Mojžiš, J.; Kello, M. Usnic Acid Isolated from Usnea antarctica (Du Rietz) Reduced In Vitro Angiogenesis in VEGF- and bFGF-Stimulated HUVECs and Ex Ovo in Quail Chorioallantoic Membrane (CAM) Assay. Life 2022, 12, 1444. [Google Scholar] [CrossRef] [PubMed]
- Joseph, A.; Lee, T.; Moland, C.L.; Branham, W.S.; Fuscoe, J.C.; Leakey, J.E.; Allaben, W.T.; Lewis, S.M.; Ali, A.A.; Desai, V.G. Effect of (+)-usnic acid on mitochondrial functions as measured by mitochondria-specific oligonucleotide microarray in liver of B6C3F1 mice. Mitochondrion 2009, 9, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Lu, C.; Huang, H.; Zhang, W.; Song, S.; Liu, B. (+)-Usnic Acid Induces ROS-dependent Apoptosis via Inhibition of Mitochondria Respiratory Chain Complexes and Nrf2 Expression in Lung Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 876. [Google Scholar] [CrossRef]
- Min, D.J.; Zhao, Y.; Monks, A.; Palmisano, A.; Hose, C.; Teicher, B.A.; Doroshow, J.H.; Simon, R.M. Identification of pharmacodynamic biomarkers and common molecular mechanisms of response to genotoxic agents in cancer cell lines. Cancer Chemother. Pharmacol. 2019, 84, 771–780. [Google Scholar] [CrossRef]
- Chepanova, A.A.; Zakharenko, A.L.; Dyrkheeva, N.S.; Chernyshova, I.A.; Zakharova, O.D.; Ilina, E.S.; Luzina, O.A.; Salakhutdinov, N.F.; Lavrik, O.I. Influence of Tyrosyl-DNA Phosphodiesterase 1 Inhibitor on the Proapoptotic and Genotoxic Effects of Anticancer Agent Topotecan. Dokl. Biochem. Biophys. 2023, 508, 25–30. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 September 2025).
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Picard Toolkit, Broad Institute, GitHub Repository. Available online: https://broadinstitute.github.io/picard/ (accessed on 1 September 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Packaage, Version 1.18.0. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 1 September 2025).
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling, and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2019. [Google Scholar] [CrossRef]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]

| Gene | C6 | G6 | F7 | OL9-119 WT | OL9-119 C6 | OL9-119 G6 | OL9-119 F7 |
|---|---|---|---|---|---|---|---|
| VEGFA | 0.913 | 1.182 | 1.017 | 1.575 | 0.783 | 0.789 | 0.477 |
| RPSA | −0.377 | −0.356 | −0.588 | −0.29 | - | - | - |
| MTSS1 | 1.219 | 1.316 | 1.915 | 1.184 | - | - | - |
| MMP2 | - | −1.232 | −1.065 | −0.642 | - | - | - |
| MMP9 | - | 1.832 | 1.957 | 2.272 | - | - | - |
| NME4 | −0.733 | - | −0.507 | −0.517 | - | - | - |
| TGFB1 | −0.708 | −0.599 | −1.178 | - | - | - | - |
| MDM2 | −0.824 | −0.454 | −0.596 | - | - | - | - |
| HGF | 4389 | 3188 | 3675 | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakharenko, A.L.; Dyrkheeva, N.S.; Markov, A.V.; Kleshchev, M.A.; Ryabchikova, E.I.; Malakhova, A.A.; Orishchenko, K.E.; Okorokova, L.S.; Shtokalo, D.N.; Medvedev, S.P.; et al. Transcriptomic Analysis of TDP1-Knockout HEK293A Cells Treated with the TDP1 Inhibitor (Usnic Acid Derivative). Int. J. Mol. Sci. 2025, 26, 9291. https://doi.org/10.3390/ijms26199291
Zakharenko AL, Dyrkheeva NS, Markov AV, Kleshchev MA, Ryabchikova EI, Malakhova AA, Orishchenko KE, Okorokova LS, Shtokalo DN, Medvedev SP, et al. Transcriptomic Analysis of TDP1-Knockout HEK293A Cells Treated with the TDP1 Inhibitor (Usnic Acid Derivative). International Journal of Molecular Sciences. 2025; 26(19):9291. https://doi.org/10.3390/ijms26199291
Chicago/Turabian StyleZakharenko, Alexandra L., Nadezhda S. Dyrkheeva, Andrey V. Markov, Maxim A. Kleshchev, Elena I. Ryabchikova, Anastasia A. Malakhova, Konstantin E. Orishchenko, Larisa S. Okorokova, Dmitriy N. Shtokalo, Sergey P. Medvedev, and et al. 2025. "Transcriptomic Analysis of TDP1-Knockout HEK293A Cells Treated with the TDP1 Inhibitor (Usnic Acid Derivative)" International Journal of Molecular Sciences 26, no. 19: 9291. https://doi.org/10.3390/ijms26199291
APA StyleZakharenko, A. L., Dyrkheeva, N. S., Markov, A. V., Kleshchev, M. A., Ryabchikova, E. I., Malakhova, A. A., Orishchenko, K. E., Okorokova, L. S., Shtokalo, D. N., Medvedev, S. P., Zakian, S. M., Tupikin, A. A., Kabilov, M. R., Luzina, O. A., Deyev, S. M., & Lavrik, O. I. (2025). Transcriptomic Analysis of TDP1-Knockout HEK293A Cells Treated with the TDP1 Inhibitor (Usnic Acid Derivative). International Journal of Molecular Sciences, 26(19), 9291. https://doi.org/10.3390/ijms26199291








