Intergenic Variants Upstream of GADD45b Affect Survival of Micropterus salmoides Following LMBV Exposure
Abstract
1. Introduction
2. Results
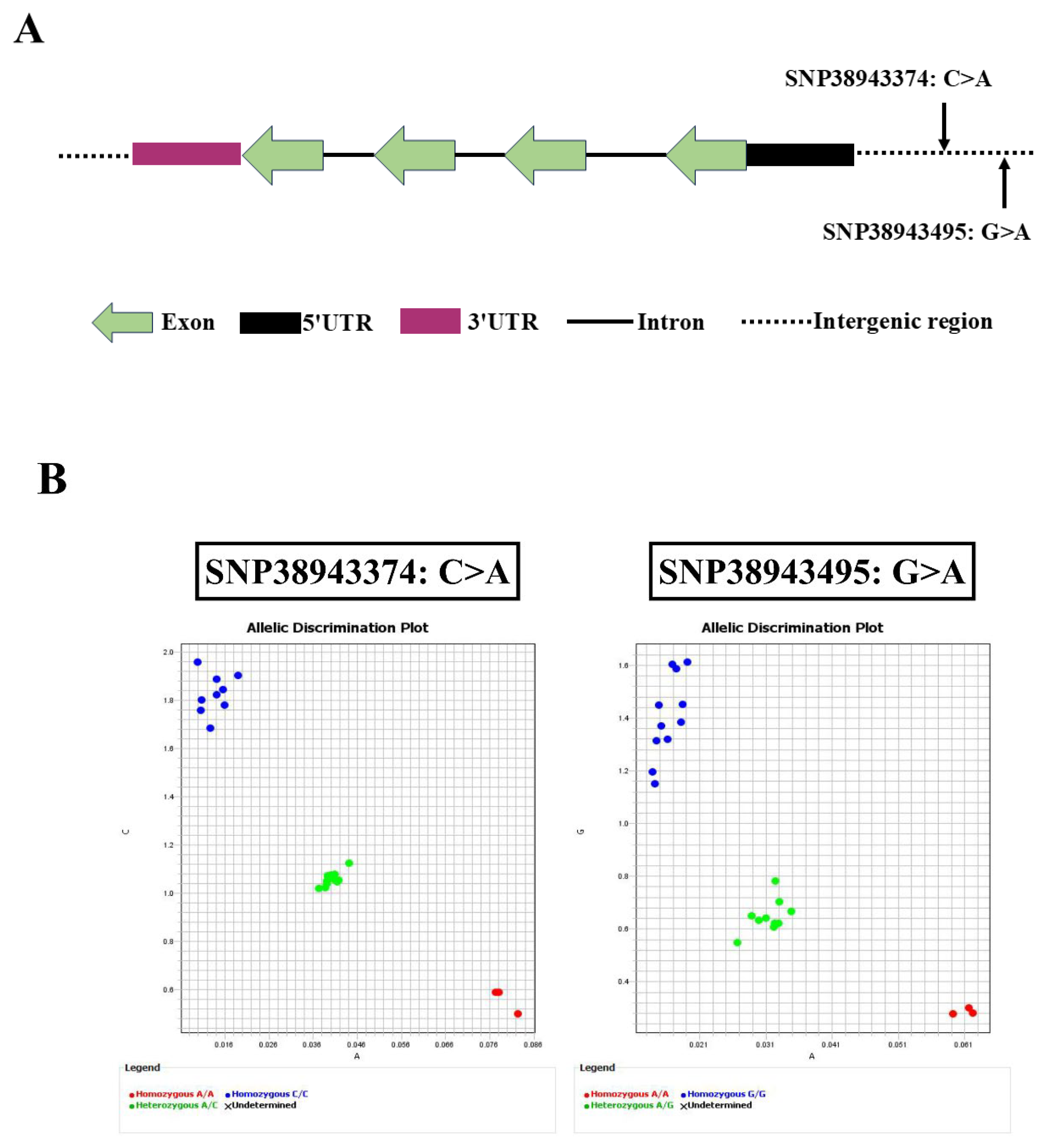
2.1. Polymorphisms in GADD45b Gene and Their KASP-Based Genotyping Profiles
2.2. Association of GADD45b SNPs with the Resistance/Susceptibility to LMBV
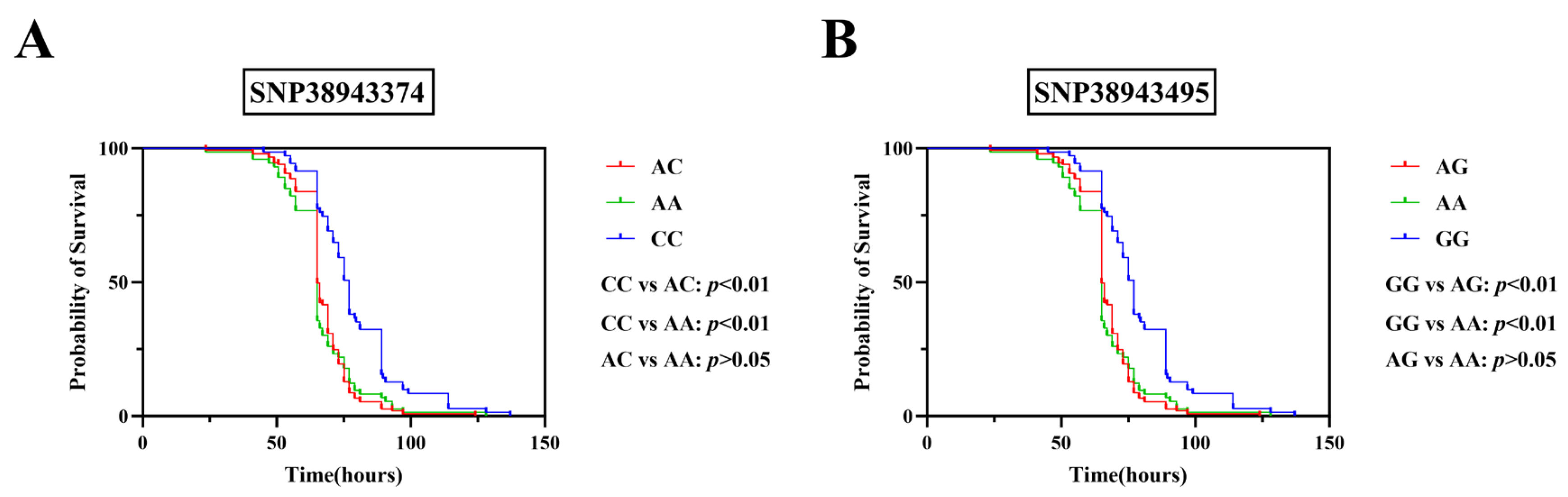
2.3. Survival Time Variation Across Genotypes After LMBV Challenge
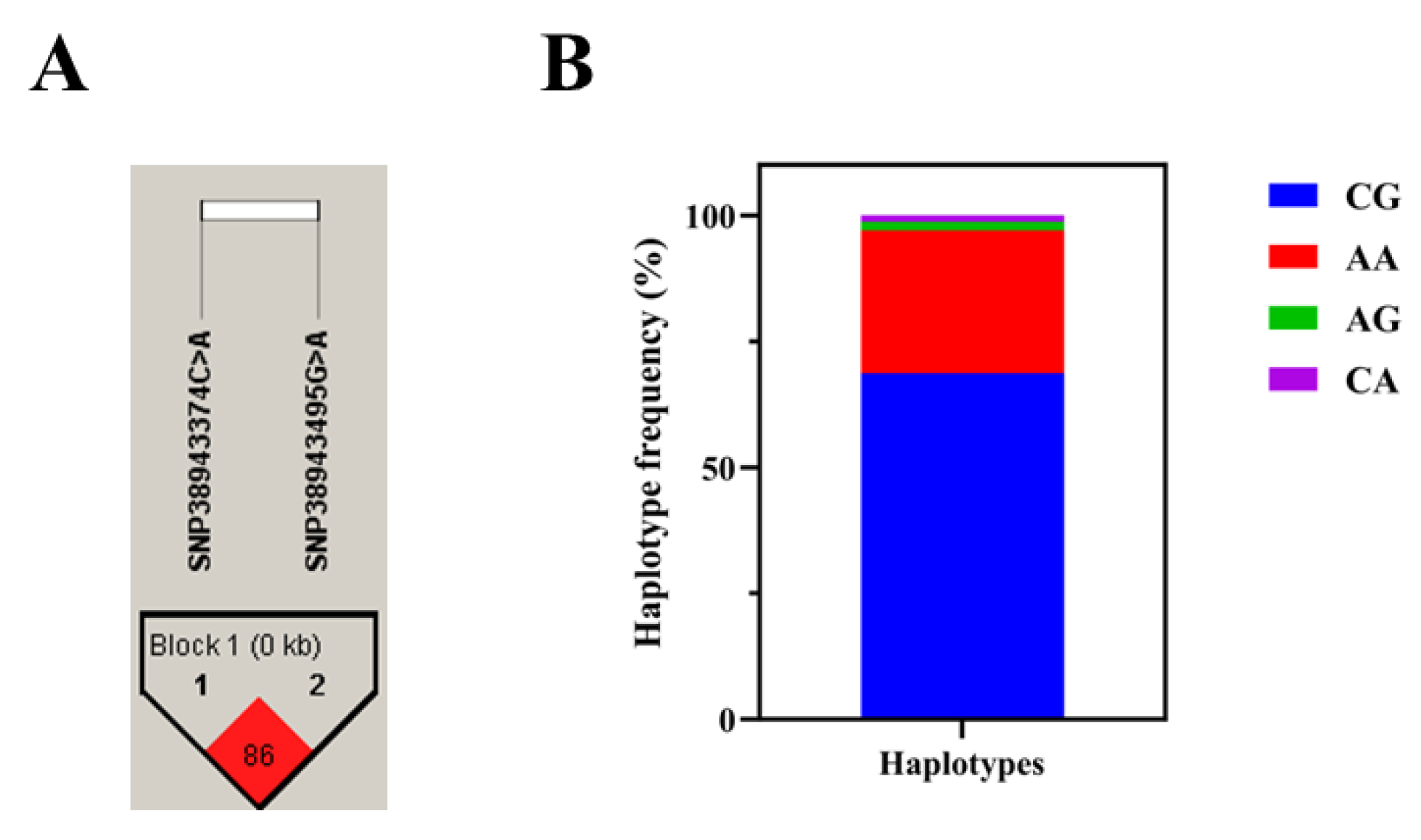
2.4. Linkage Disequilibrium Analysis of GADD45b Gene SNPs
2.5. GADD45b Expression in Different Genotypes After LMBV Infection
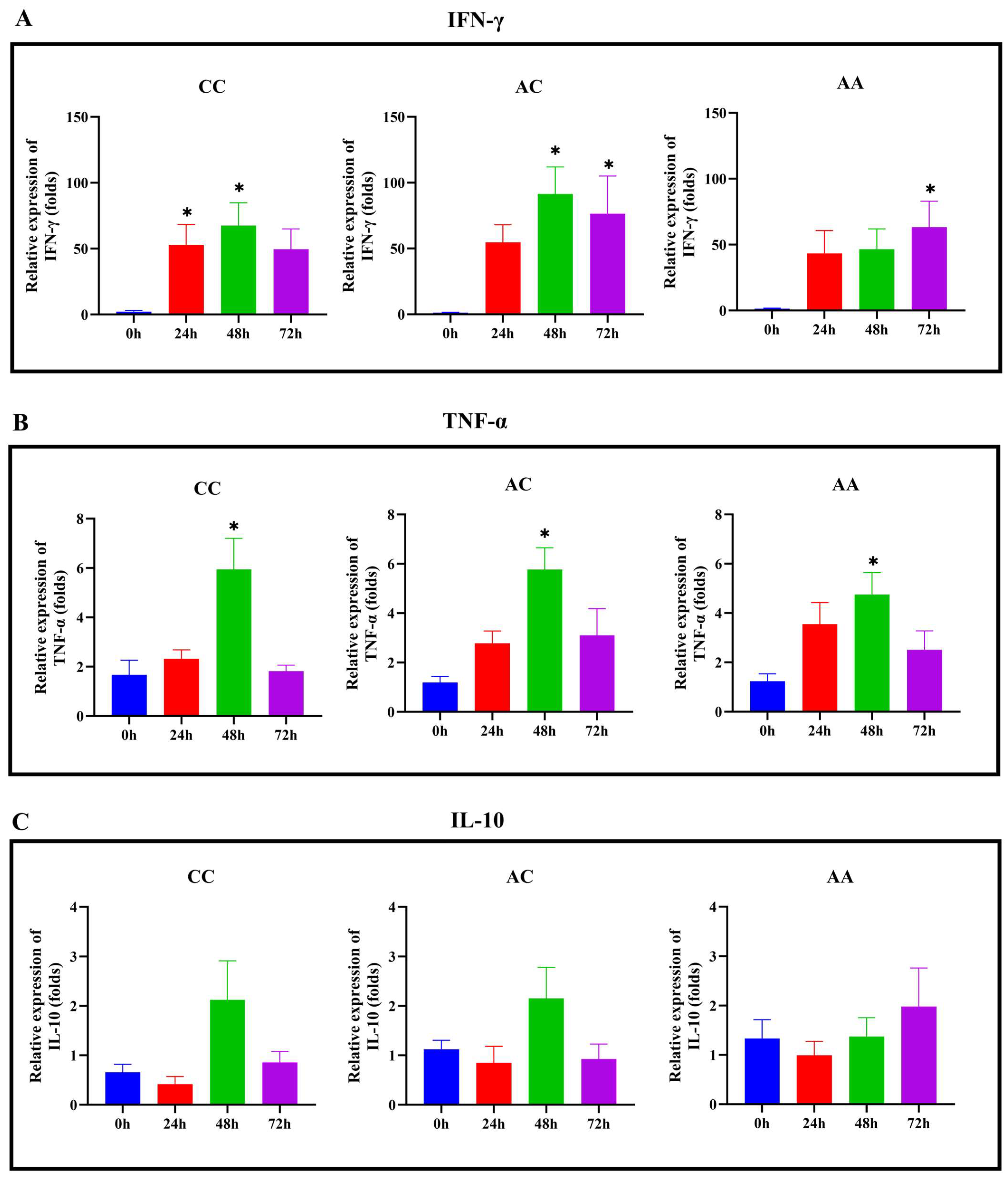
2.6. Immune Gene Expression Profiles Across Genotypes Following LMBV Infection
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Animals and Virus
4.3. LMBV Infection and Sample Collection
4.4. DNA/RNA Extraction and qRT-PCR Analysis
4.5. Genotyping SNPs in the GADD45b Gene with KASP
4.6. Association Analysis Between GADD45b SNPs and Resistance/Susceptibility to LMBV
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rasal, K.D.; Chakrapani, V.; Pandey, A.K.; Rasal, A.R.; Sundaray, J.K.; Ninawe, A.; Jayasankar, P. Status and future perspectives of single nucleotide polymorphisms (SNPs) markers in farmed fishes: Way ahead using next generation sequencing. Gene Rep. 2017, 6, 81–86. [Google Scholar] [CrossRef]
- Jiang, D.L.; Gu, X.H.; Li, B.J.; Zhu, Z.X.; Qin, H.; Meng, Z.; Lin, H.R.; Xia, J.H. Identifying a long QTL cluster across chrLG18 associated with salt tolerance in tilapia using GWAS and QTL-Seq. Mar. Biotechnol. 2019, 21, 250–261. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, T.; Geng, X.; Liu, S.; Chen, A.; Yao, J.; Jiang, C.; Tan, S.; Su, B.; Liu, Z. A Genome-wide association study of heat stress-associated SNPs in catfish. Anim. Genet. 2017, 48, 233–236. [Google Scholar] [CrossRef]
- San, L.-Z.; Liu, B.-S.; Liu, B.; Zhu, K.-C.; Guo, L.; Guo, H.-Y.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Genome-wide association study reveals multiple novel SNPs and putative candidate genes associated with low oxygen tolerance in golden pompano Trachinotus ovatus (Linnaeus 1758). Aquaculture 2021, 544, 737098. [Google Scholar] [CrossRef]
- Hua, J.; Zhong, C.; Chen, W.; Fu, J.; Wang, J.; Wang, Q.; Zhu, G.; Li, Y.; Tao, Y.; Zhang, M.; et al. Single nucleotide polymorphism SNP19140160 A>C is a potential breeding locus for fast-growth largemouth bass (Micropterus salmoides). BMC Genom. 2024, 25, 64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-S.; Ruan, Z.-H.; Lu, Z.-Q.; Li, Y.-F.; Luo, Y.-Y.; Zhang, X.-Q.; Liu, W.-S. Novel SNPs in the 3′UTR region of GHRb gene associated with growth traits in striped catfish (Pangasianodon hypophthalmus), a valuable aquaculture species. Fishes 2022, 7, 230. [Google Scholar] [CrossRef]
- Özcan Gökçek, E.; Işık, R. Associations between genetic variants of the insulin-like growth factor I (IGF-I) gene and growth traits in European sea bass (Dicentrarchus labrax, L.). Fish Physiol. Biochem. 2020, 46, 1131–1138. [Google Scholar] [CrossRef]
- Tran, T.T.H.; Tran, H.S.; Le, B.T.N.; Van Nguyen, S.; Vu, H.-A.; Kim, O.T.P. Novel single nucleotide polymorphisms of insulin-like growth factor-binding protein 7 (IGFBP7) gene significantly associated with growth traits in striped catfish (Pangasianodon hypophthalmus Sauvage, 1878). Mol. Genet. Genom. 2023, 298, 883–893. [Google Scholar] [CrossRef]
- Cáceres, G.; López, M.E.; Cádiz, M.I.; Yoshida, G.M.; Jedlicki, A.; Palma-Véjares, R.; Travisany, D.; Díaz-Domínguez, D.; Maass, A.; Lhorente, J.P.; et al. Fine mapping using whole-genome sequencing confirms anti-müllerian hormone as a major gene for sex determination in farmed nile tilapia (Oreochromis niloticus L.). G3 Genes Genomes Genet. 2019, 9, 3213–3223. [Google Scholar] [CrossRef]
- Gabián, M.; Morán, P.; Fernández, A.I.; Villanueva, B.; Chtioui, A.; Kent, M.P.; Covelo-Soto, L.; Fernández, A.; Saura, M. Identification of genomic regions regulating sex determination in Atlantic salmon using high density SNP data. BMC Genom. 2019, 20, 764. [Google Scholar] [CrossRef]
- Jin, R.-M.; Huang, H.-Z.; Zhou, Y.; Wang, Y.-Y.; Fu, H.-C.; Li, Z.; Fu, X.-Z.; Li, N.-Q. Characterization of mandarin fish (Siniperca chuatsi) IL-6 and IL-6 signal transducer and the association between their SNPs and resistance to ISKNV disease. Fish Shellfish Immunol. 2021, 113, 139–147. [Google Scholar] [CrossRef]
- Xu, T.; Chen, S.; Zhang, Y. MHC class IIα gene polymorphism and its association with resistance/susceptibility to Vibrio anguillarum in Japanese flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 2010, 34, 1042–1050. [Google Scholar] [CrossRef]
- Xu, Y.; Liang, B.; Zhang, J.; Tang, B.; Su, J. Association study on single nucleotide polymorphisms in the Ctenopharyngodon idella TLR8b gene with resistance/susceptibility to grass carp reovirus. Aquaculture 2025, 599, 742124. [Google Scholar] [CrossRef]
- Ju, S.; Zhu, Y.; Liu, L.; Dai, S.; Li, C.; Chen, E.; He, Y.; Zhang, X.; Lu, B. Gadd45b and Gadd45g are important for anti-tumor immune responses. Eur. J. Immunol. 2009, 39, 3010–3018. [Google Scholar] [CrossRef]
- Tamura, R.E.; de Vasconcellos, J.F.; Sarkar, D.; Fisher, P.B.; Zerbini, L.F. GADD45 Proteins: Central Players in Tumorigenesis. Curr. Mol. Med. 2012, 12, 634–651. [Google Scholar] [CrossRef] [PubMed]
- Balliet, A.G.; Hatton, K.S.; Hoffman, B.; Liebermann, D.A. Comparative analysis of the genetic structure and chromosomal location of the murine MyD118 (GADD45b) gene. DNA Cell Biol. 2001, 20, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Imbeault, M.; Giguère, K.; Ouellet, M.; Tremblay, M.J. Exon level transcriptomic profiling of HIV-1-infected CD4+ T cells reveals virus-induced genes and host environment favorable for viral replication. PLoS Pathog. 2012, 8, e1002861. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Liu, R.; Zhang, H.; Zhang, S.; Hu, X.; Tan, J.; Liang, C.; Qiao, W. GADD45 proteins inhibit HIV-1 replication through specific suppression of HIV-1 transcription. Virology 2016, 493, 1–11. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, X.; Song, C.; Lin, W.; Cheng, Y.; Xie, Z.; Chen, S.; Nie, Y.; Li, A.; Zhang, H.; et al. ALV-J inhibits autophagy through the GADD45β/MEKK4/P38MAPK signaling pathway and mediates apoptosis following autophagy. Cell Death Dis. 2020, 11, 684. [Google Scholar] [CrossRef]
- Deng, K.; Fan, Y.; Liang, Y.; Cai, Y.; Zhang, G.; Deng, M.; Wang, Z.; Lu, J.; Shi, J.; Wang, F.; et al. FTO-mediated demethylation of GADD45B promotes myogenesis through the activation of P38 MAPK pathway. Mol. Ther. Nucleic Acids 2021, 26, 34–48. [Google Scholar] [CrossRef]
- Keil, E.; Höcker, R.; Schuster, M.; Essmann, F.; Ueffing, N.; Hoffman, B.; Liebermann, D.A.; Pfeffer, K.; Schulze-Osthoff, K.; Schmitz, I. Phosphorylation of Atg5 by the Gadd45β–MEKK4-P38 pathway inhibits autophagy. Cell Death Differ. 2013, 20, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Sun, H.; Xu, R.; Wang, Y.; Guo, J.; Li, X.; Cheng, Y.; Xu, C.; Tang, C.; Sun, B.; et al. GADD45B promotes glucose-induced renal tubular epithelial-mesenchymal transition and apoptosis via the P38 MAPK and JNK signaling pathways. Front. Physiol. 2020, 11, 1074. [Google Scholar] [CrossRef] [PubMed]
- Salerno, D.M.; Tront, J.S.; Hoffman, B.; Liebermann, D.A. Gadd45a and Gadd45b modulate innate immune functions of granulocytes and macrophages by differential regulation of P38 and JNK Signaling. J. Cell. Physiol. 2012, 227, 3613–3620. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Kohama, Y.; Kuge, A.; Kido, E.; Sakurai, H. GADD45 family proteins suppress JNK signaling by targeting MKK7. Arch. Biochem. Biophys. 2017, 635, 1–7. [Google Scholar] [CrossRef]
- Lu, B.; Ferrandino, A.F.; Flavell, R.A. Gadd45β is important for perpetuating cognate and inflammatory signals in T cells. Nat. Immunol. 2004, 5, 38–44. [Google Scholar] [CrossRef]
- Luo, X.; Wang, R.; Ma, B.; Chang, O.; Fu, X.; Liang, H.; Niu, Y.; Lin, Q.; Liu, L.; Lv, A.; et al. Inactivated LMBV vaccine with nano-aluminum adjuvant provided immune protection in largemouth bass (Micropterus salmoides). Aquaculture 2025, 595, 741634. [Google Scholar] [CrossRef]
- Zhao, R.; Geng, Y.; Qin, Z.; Wang, K.; Ouyang, P.; Chen, D.; Huang, X.; Zuo, Z.; He, C.; Guo, H.; et al. A New ranavirus of the santee-cooper group invades largemouth bass (Micropterus salmoides) culture in southwest China. Aquaculture 2020, 526, 735363. [Google Scholar] [CrossRef]
- Niu, Y.; Fu, X.; Lin, Q.; Liang, H.; Luo, X.; Zuo, S.; Liu, L.; Li, N. In vivo and in vitro, antiviral effects of two mixture of Chinese herbal drug active monomers against MSRV and LMBV in Largemouth bass (Micropterus salmoides). Aquaculture 2023, 577, 739977. [Google Scholar] [CrossRef]
- Yi, W.; Zhang, X.; Zeng, K.; Xie, D.; Song, C.; Tam, K.; Liu, Z.; Zhou, T.; Li, W. Construction of a DNA vaccine and its protective effect on largemouth bass (Micropterus salmoides) challenged with largemouth bass virus (LMBV). Fish Shellfish Immunol. 2020, 106, 103–109. [Google Scholar] [CrossRef]
- Li, P.; Luo, X.; Zuo, S.; Fu, X.; Lin, Q.; Niu, Y.; Liang, H.; Ma, B.; Li, N. Genome-wide association study of resistance to largemouth bass ranavirus (LMBV) in Micropterus salmoides. IJMS 2024, 25, 10036. [Google Scholar] [CrossRef]
- Yue, G.H. Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish. 2014, 15, 376–396. [Google Scholar] [CrossRef]
- Zenger, K.R.; Khatkar, M.S.; Jones, D.B.; Khalilisamani, N.; Jerry, D.R.; Raadsma, H.W. Genomic selection in aquaculture: Application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front. Genet. 2019, 9, 693. [Google Scholar] [CrossRef]
- Sawayama, E.; Kitamura, S.-I.; Nakayama, K.; Ohta, K.; Okamoto, H.; Ozaki, A.; Takagi, M. Development of a novel RSIVD-resistant strain of red sea bream (Pagrus major) by marker-assisted selection combined with DNA-based family selection. Aquaculture 2019, 506, 188–192. [Google Scholar] [CrossRef]
- Fuji, K.; Hasegawa, O.; Honda, K.; Kumasaka, K.; Sakamoto, T.; Okamoto, N. Marker-assisted breeding of a lymphocystis disease-resistant Japanese flounder (Paralichthys olivaceus). Aquaculture 2007, 272, 291–295. [Google Scholar] [CrossRef]
- Lu, S.; Li, Y.Z.; Wang, L.; Liu, Y.; Cheng, X.M.; Zhou, Q.; Yang, Y.M.; Zheng, W.W.; Chen, S.L. Application of genomic selection in the breeding of new variety of Chinese tongue sole (Cynoglossus semilaevis) “Tayou No. 1” . J. Fish. China 2022, 46, 1305–1312. (In Chinese) [Google Scholar] [CrossRef]
- You, X.; Shan, X.; Shi, Q. Research advances in the genomics and applications for molecular breeding of aquaculture animals. Aquaculture 2020, 526, 735357. [Google Scholar] [CrossRef]
- Cao, Z.; Liu, X.; Xiang, Y.; Wang, L.; Sun, Y.; Zhou, Y. Genetic polymorphism of MHC class IIα alleles and its association with resistance/susceptibility to viral nervous necrosis virus (VNNV) in golden pompano (Trachinotus ovatus). Aquaculture 2019, 501, 144–152. [Google Scholar] [CrossRef]
- Lee, H.B.; Schwab, T.L.; Koleilat, A.; Ata, H.; Daby, C.L.; Cervera, R.L.; McNulty, M.S.; Bostwick, H.S.; Clark, K.J. Allele-specific quantitative PCR for accurate, rapid, and cost-effective genotyping. Hum. Gene Ther. 2016, 27, 425–435. [Google Scholar] [CrossRef]
- Vossen, R.H.A.M. Genotyping DNA variants with high-resolution melting analysis. In Genotyping: Methods and Protocols; White, S.J., Cantsilieris, S., Eds.; Springer: New York, NY, USA, 2017; pp. 17–28. ISBN 978-1-4939-6442-0. [Google Scholar]
- Yu, S.; Li, X.; Liu, X.; Wang, Y.; Yu, F.; Xue, Y.; Mao, Z.; Wang, C.; Li, W. Characteristic and influencing factors of taqman genotyping calling error. Clin. Lab. Anal. 2018, 32, e22613. [Google Scholar] [CrossRef]
- Majeed, U.; Darwish, E.; Rehman, S.U.; Zhang, X. Kompetitive allele specific PCR (KASP): A singleplex genotyping platform and its application. JAS 2018, 11, 11. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Lin, Y.L.; Qin, H.; Xiong, Y.Y.; Jiang, D.L.; Lin, H.R.; Yu, Z.L.; Xia, J.H. Identifying a Genome-Wide QTL interval controlling for ammonia-nitrogen tolerance on chrLG1 of nile tilapia. Aquaculture 2021, 543, 736946. [Google Scholar] [CrossRef]
- Bartonicek, N.; Clark, M.B.; Quek, X.C.; Torpy, J.R.; Pritchard, A.L.; Maag, J.L.V.; Gloss, B.S.; Crawford, J.; Taft, R.J.; Hayward, N.K.; et al. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 2017, 18, 241. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Rao, S.; Du, T.; Hu, H.; Liu, Z.; Shen, Y.; Xu, Q. Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene 2017, 601, 21–26. [Google Scholar] [CrossRef]
- Baloch, A.A.; Abdelsalam, E.E.E.; Piačková, V. Cytokines studied in carp (Cyprinus carpio L.) in response to important diseases. Fishes 2022, 7, 3. [Google Scholar] [CrossRef]
- Ding, G.; Zheng, C.; Wang, B.; Zhang, L.; Deng, D.; Li, Q.; Guo, H.; Zhang, S.; Xu, Q. Transcriptome sequencing reveals the antiviral innate immunity by IFN-γ in Chinese sturgeon macrophages. Front. Immunol. 2022, 13, 854689. [Google Scholar] [CrossRef]
- Kalaichelvan, A.; Nadarajapillai, K.; Sellaththurai, S.R.; Arachchi, U.P.E.; Kim, M.-J.; Jung, S.; Lee, J. CRISPR/Cas9-induced knockout of tumor necrosis factor-alpha-type I augments viral infection in zebrafish. Fish. Shellfish Immunol. 2025, 157, 110092. [Google Scholar] [CrossRef]
- Yang, S.; Ma, Y.; Lou, X.; Zhou, Z.; Zhang, H.; Yi, S.; Cheng, Y.; Qian, S.; Huang, M.; Fei, H. The role of TNF-α in the phagocytosis of largemouth bass (Micropterus salmoides) leukocytes. Fish. Shellfish Immunol. 2023, 132, 108488. [Google Scholar] [CrossRef]
- De Smaele, E.; Zazzeroni, F.; Papa, S.; Nguyen, D.U.; Jin, R.; Jones, J.; Cong, R.; Franzoso, G. Induction of Gadd45β by NF-κB downregulates pro-apoptotic JNK signaling. Nature 2001, 414, 308–313. [Google Scholar] [CrossRef]
- Grayfer, L.; Hodgkinson, J.W.; Hitchen, S.J.; Belosevic, M. Characterization and functional analysis of goldfish (Carassius auratus L.) interleukin-10. Mol. Immunol. 2011, 48, 563–571. [Google Scholar] [CrossRef]
- Huo, H.J.; Chen, S.N.; Li, L.; Nie, P. Functional characterization of IL-10 and its receptor subunits in a perciform fish, the mandarin fish, Siniperca chuatsi. Dev. Comp. Immunol. 2019, 97, 64–75. [Google Scholar] [CrossRef]
- Fu, X.; Li, N.; Lai, Y.; Luo, X.; Wang, Y.; Shi, C.; Huang, Z.; Wu, S.; Su, J. A novel fish cell line derived from the brain of Chinese perch Siniperca chuatsi: Development and characterization. J. Fish Biol. 2015, 86, 32–45. [Google Scholar] [CrossRef]
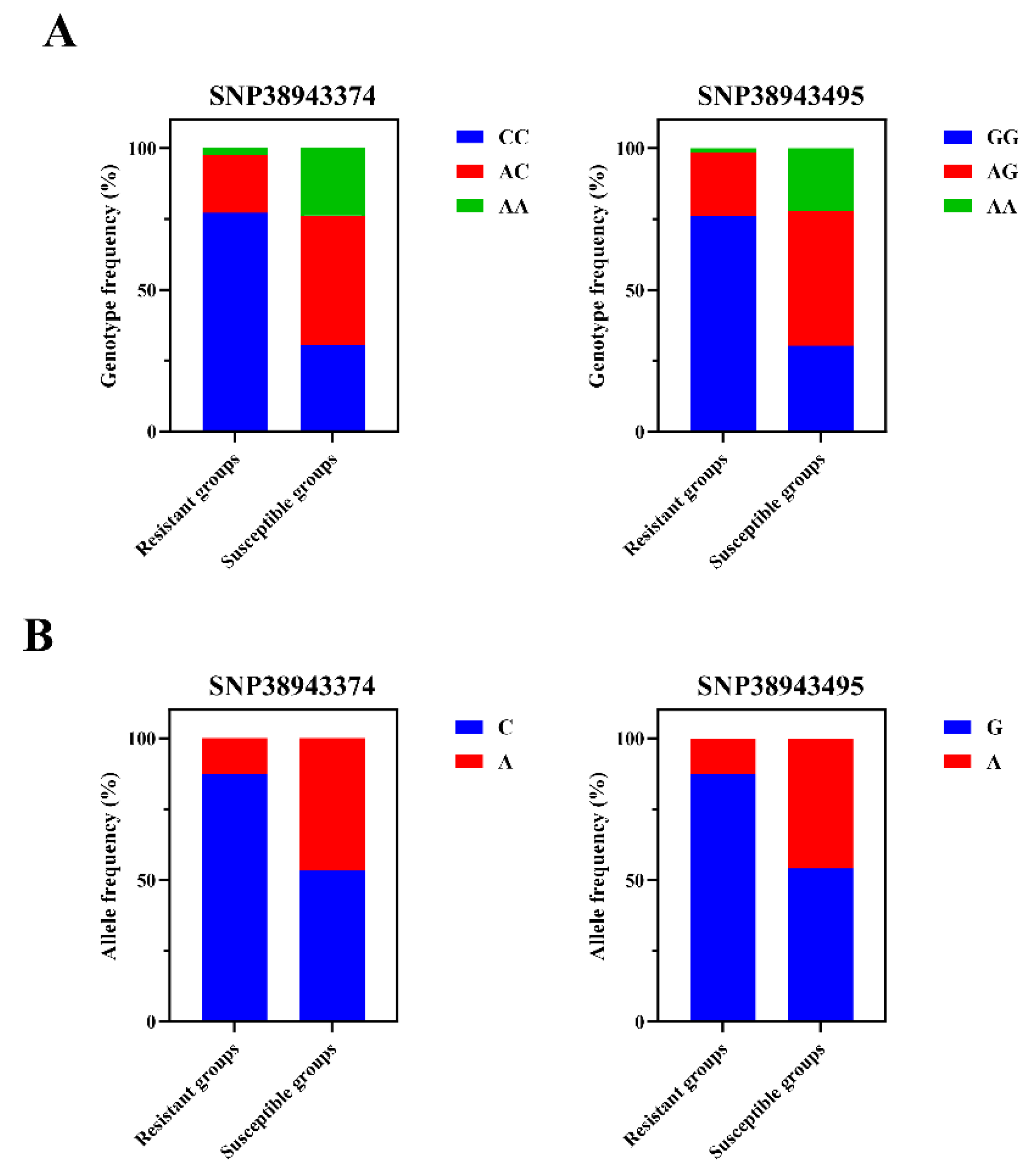

| Locus | Genotype | Resistant Groups No. (%) | Susceptible Groups No. (%) | χ2 (p) |
|---|---|---|---|---|
| SNP38943374 C>A | CC | 91 (0.771) | 37 (0.303) | 56.655 (p < 0.01) |
| AC | 24 (0.203) | 56 (0.459) | ||
| AA | 3 (0.025) | 29 (0.238) | ||
| SNP38943495 G>A | GG | 90 (0.763) | 37 (0.303) | 55.809 (p < 0.01) |
| AG | 26 (0.220) | 58 (0.475) | ||
| AA | 2 (0.017) | 27 (0.221) |
| Locus | Allele | Resistant Groups No. (%) | Susceptible Groups No. (%) | χ2 (p) |
|---|---|---|---|---|
| SNP38943374 C>A | C | 206 (0.873) | 130 (0.533) | 66.075 (p < 0.01) |
| A | 30 (0.127) | 114 (0.467) | ||
| SNP38943495 G>A | G | 206 (0.873) | 132 (0.541) | 63.438 (p < 0.01) |
| A | 30 (0.127) | 112 (0.459) |
| Primer | Sequence (5′-3′) |
|---|---|
| SNP38943374-F1 | GAAGGTGACCAAGTTCATGCTGATGATGAGGTGCCAGGGTGGAGGC |
| SNP38943374-F2 | GAAGGTCGGAGTCAACGGATTGATGATGAGGTGCCAGGGTGGAGGA |
| SNP38943374-R | GAAGGCTGTAACTGCTGAGCGGAAGTGTT |
| SNP38943495-F1 | GAAGGTGACCAAGTTCATGCTACATTAAAATATCACATGACATAAG |
| SNP38943495-F2 | GAAGGTCGGAGTCAACGGATTACATTAAAATATCACATGACATAAA |
| SNP38943495-R | TTTCAGCTCTGGTTTGGTTGTAAGAGTAT |
| GADD45b-RT-F | CTTTCTGCTGCGACAACGAC |
| GADD45b-RT-R | GAGGGTTCGTGACCAGGATG |
| IFN-γ-RT-F | TGCAGGCTCTCAAACACATC |
| IFN-γ-RT-R | TGTTTTCGGTCAGTGTGCTC |
| TNF-α-RT-F | ATCTGCTGTGAATGCCGTGA |
| TNF-α-RT-R | CGTCAGCCTGGATAGACGAC |
| IL-10-RT-F | CTAGACCAGAGCGTCGAGGA |
| IL-10-RT-R | CCAAGGCTGTTGGCAGAATC |
| 18s-RT-F | TGACGGAAGGGCACCACCAG |
| 18s-RT-R | GCACCACCACCCACAGAATCG |
| SNP | Chromosome | Locus | Ref | Alt | Region |
|---|---|---|---|---|---|
| SNP38943374 C>A | NW_024044237.1 | 38943374 | C | A | Intergenic |
| SNP38943495 G>A | 38943495 | G | A | Intergenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Luo, X.; Li, W.; Fu, X.; Lin, Q.; Niu, Y.; Liang, H.; Ma, B.; Xiao, W.; Li, N. Intergenic Variants Upstream of GADD45b Affect Survival of Micropterus salmoides Following LMBV Exposure. Int. J. Mol. Sci. 2025, 26, 9281. https://doi.org/10.3390/ijms26199281
Li P, Luo X, Li W, Fu X, Lin Q, Niu Y, Liang H, Ma B, Xiao W, Li N. Intergenic Variants Upstream of GADD45b Affect Survival of Micropterus salmoides Following LMBV Exposure. International Journal of Molecular Sciences. 2025; 26(19):9281. https://doi.org/10.3390/ijms26199281
Chicago/Turabian StyleLi, Pinhong, Xia Luo, Wenxian Li, Xiaozhe Fu, Qiang Lin, Yinjie Niu, Hongru Liang, Baofu Ma, Wenwen Xiao, and Ningqiu Li. 2025. "Intergenic Variants Upstream of GADD45b Affect Survival of Micropterus salmoides Following LMBV Exposure" International Journal of Molecular Sciences 26, no. 19: 9281. https://doi.org/10.3390/ijms26199281
APA StyleLi, P., Luo, X., Li, W., Fu, X., Lin, Q., Niu, Y., Liang, H., Ma, B., Xiao, W., & Li, N. (2025). Intergenic Variants Upstream of GADD45b Affect Survival of Micropterus salmoides Following LMBV Exposure. International Journal of Molecular Sciences, 26(19), 9281. https://doi.org/10.3390/ijms26199281






