Reproducibility Crossroads: Impact of Statistical Choices on Proteomics Functional Enrichment
Abstract
1. Introduction
2. Results
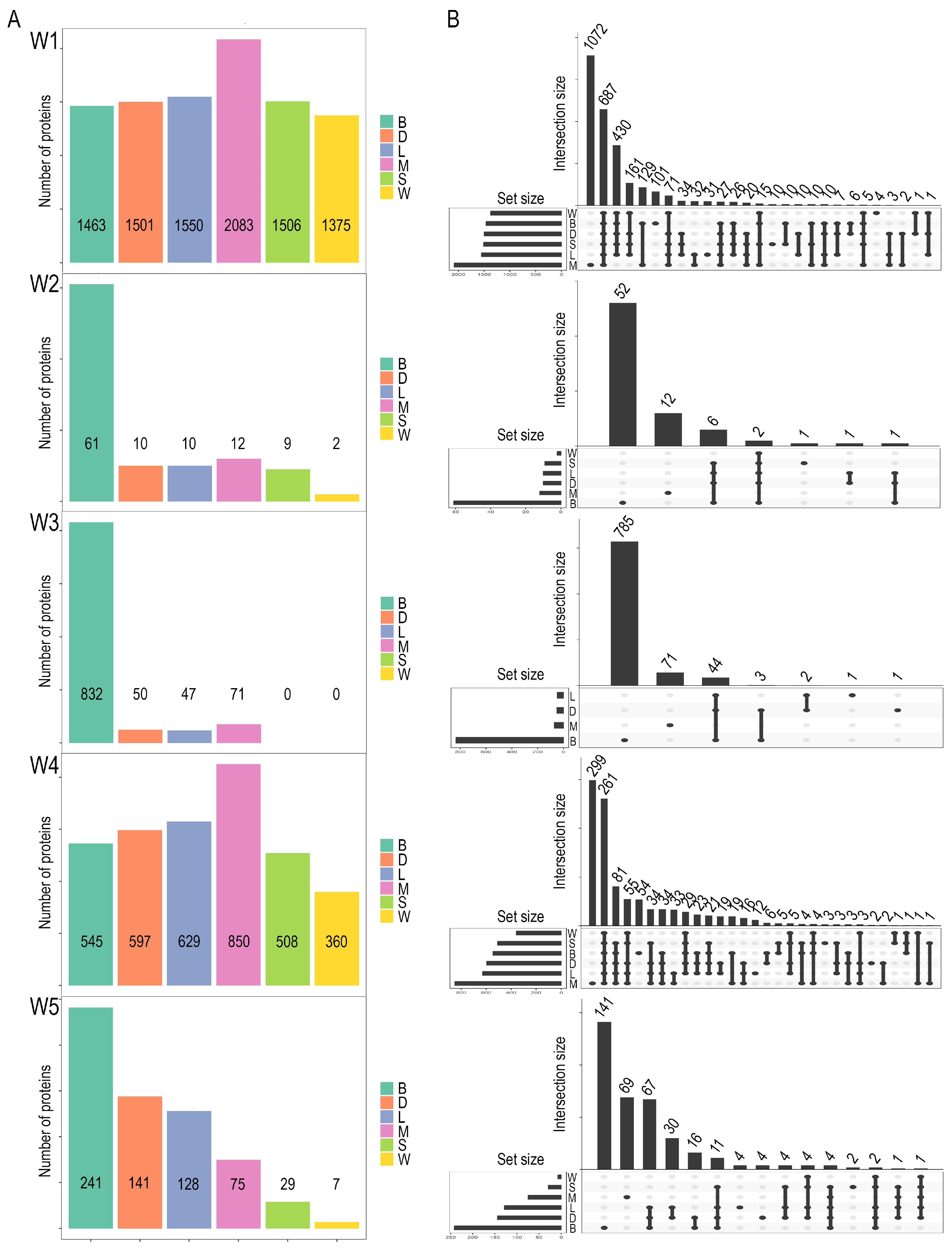
2.1. Characterization of Dataset Variability and Its Influence on Proteomic Outcomes
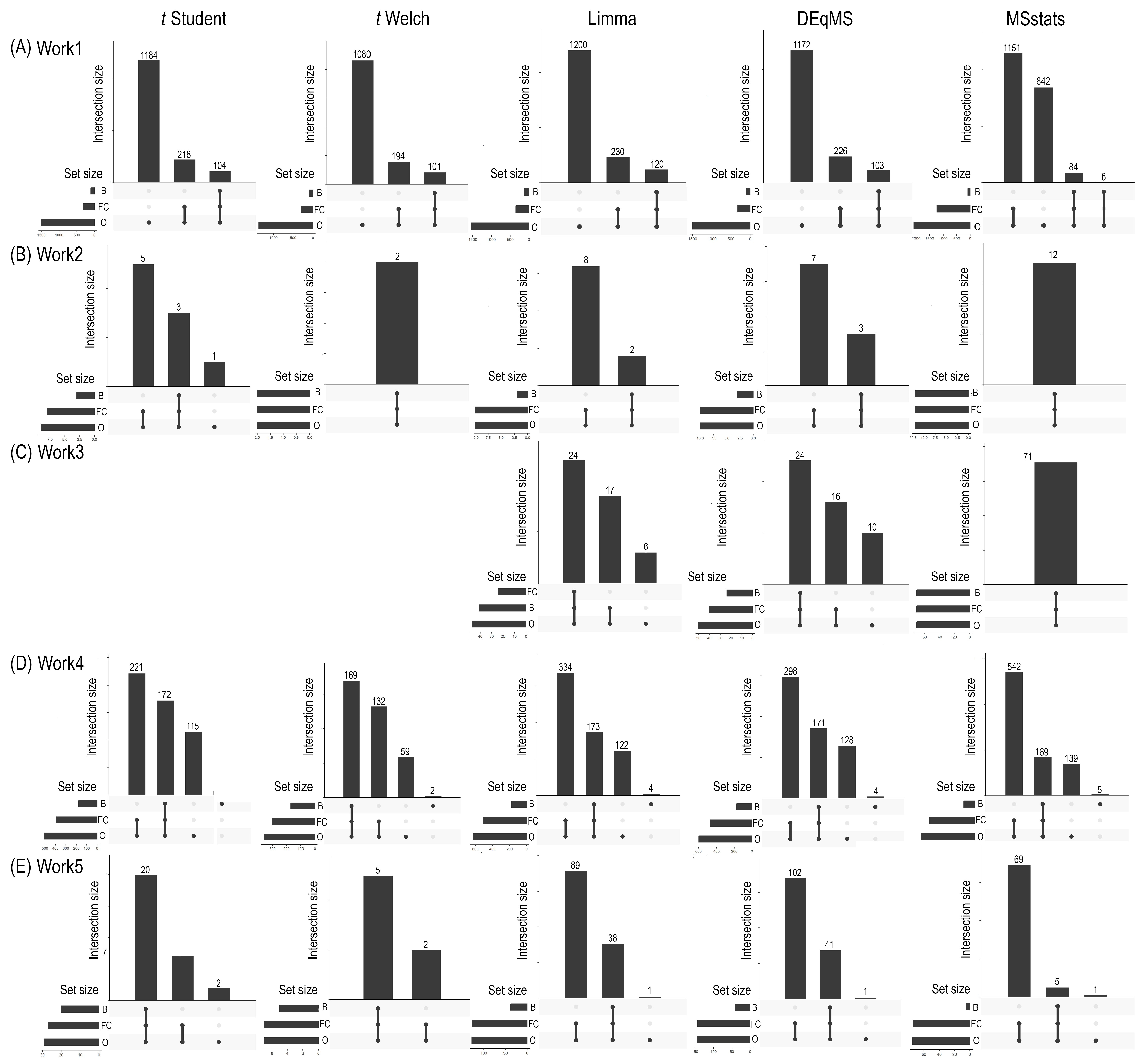
2.2. Differential Expression Analysis: The Role of Hypothesis Testing Methods in Protein Identification
2.3. Biological Relevance in Differential Proteomics Analysis
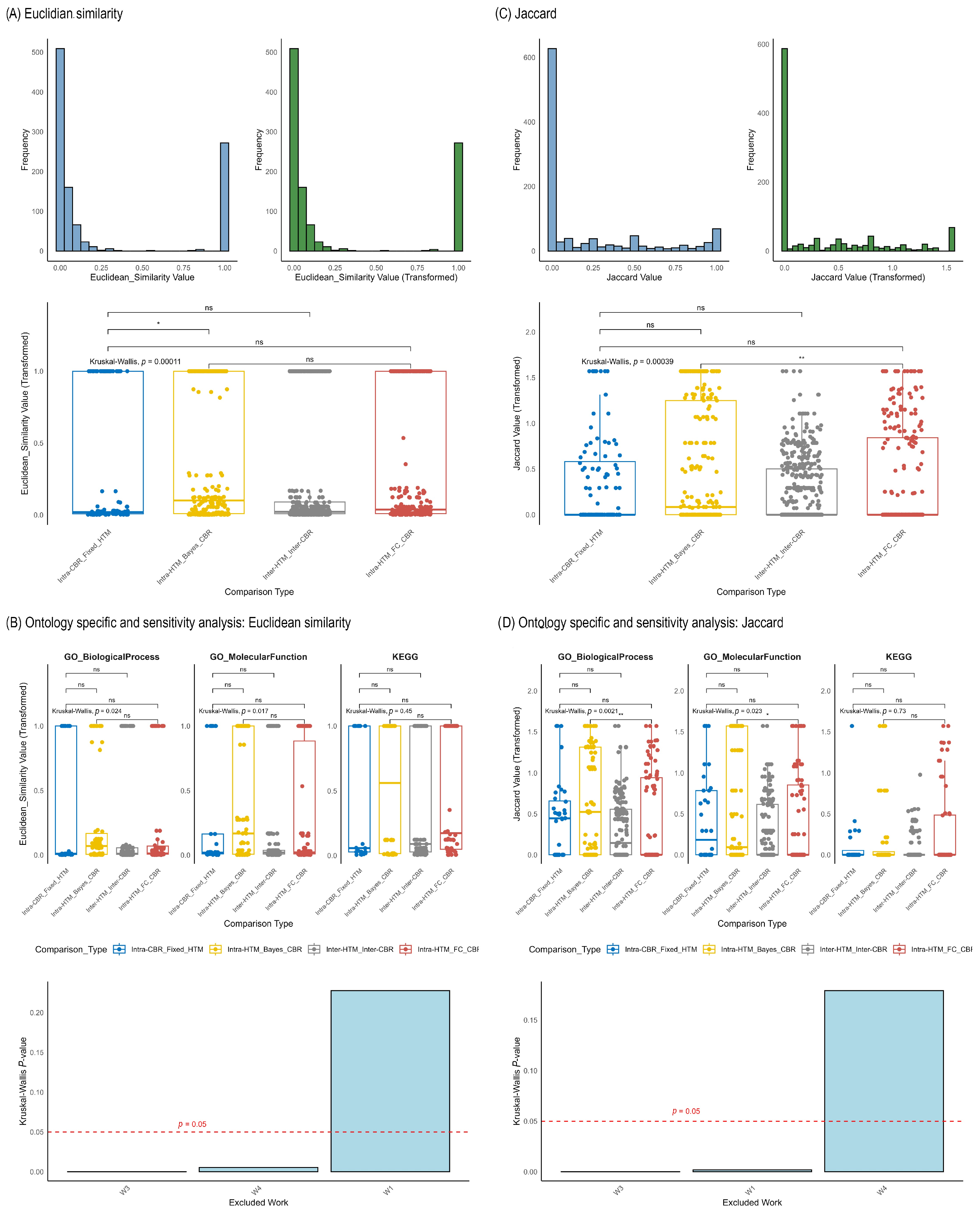
2.4. Functional Enrichment Analysis: Consistency Across Methodological Choices
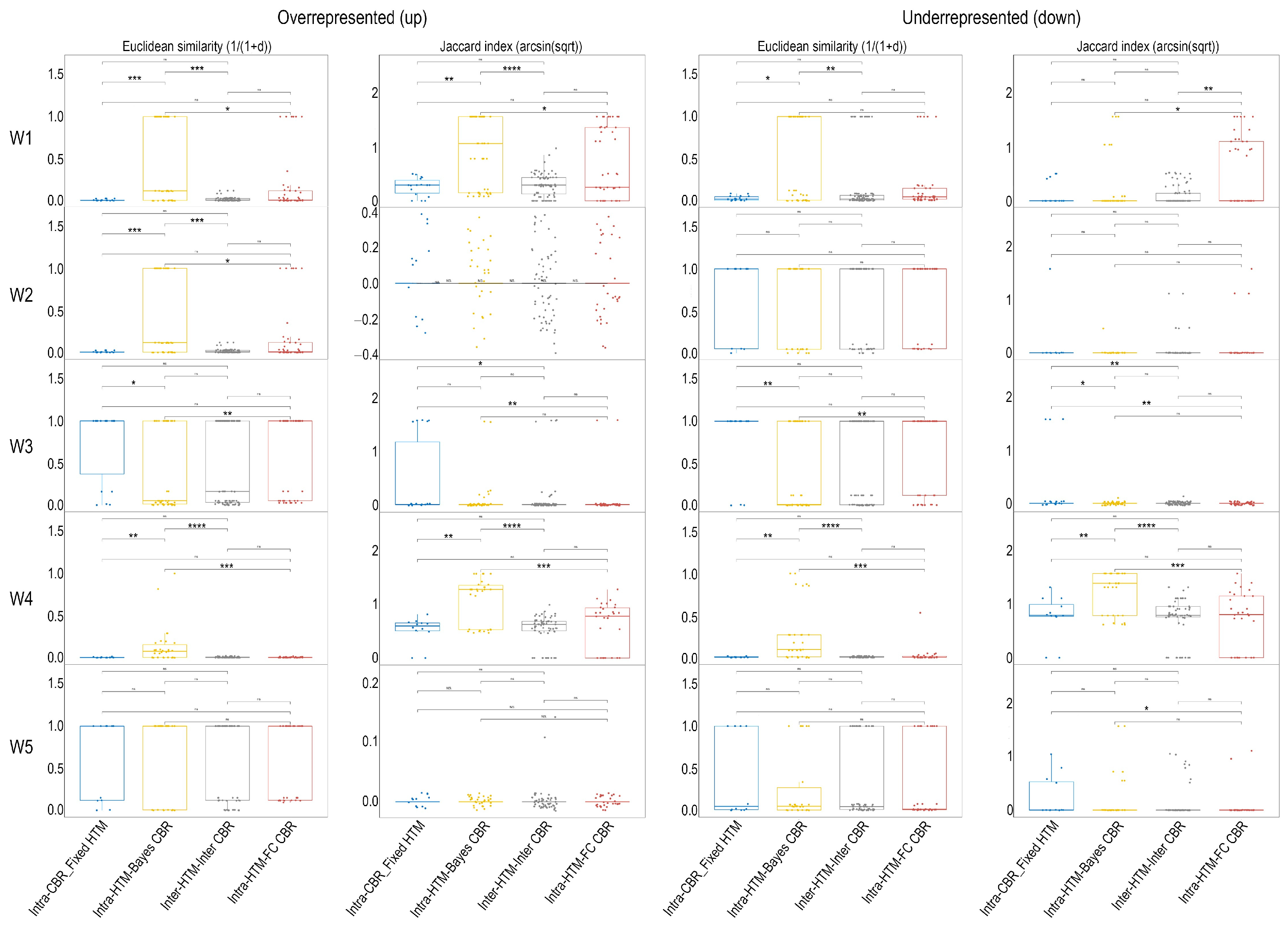
2.5. Meta-Analysis of Functional Enrichment Concordance
3. Discussion
- Does the specific hypothesis testing method (HTM; e.g., t-Student, t-Welch, Limma, DEqMS, MSstats, Bayesian) influence the resulting biological enrichments when the criterion for biological relevance (CBR) is kept constant?
- Does the method used for determining biological relevance (CBR; fold change-based vs. Bayesian posterior probability-based approaches) influence the resulting biological enrichments when the hypothesis testing method (HTM) is kept constant?
- What has a greater influence on the observed biological enrichments: the specific hypothesis testing method (HTM) or the criterion for determining biological relevance (CBR)?
3.1. Methodological Implications of Hypothesis Testing in Proteomics
3.2. The Criterion for Biological Relevance as the Key Driver
3.3. Reproducibility, Transparency, and Community Standards
3.4. Limitations and Future Perspectives
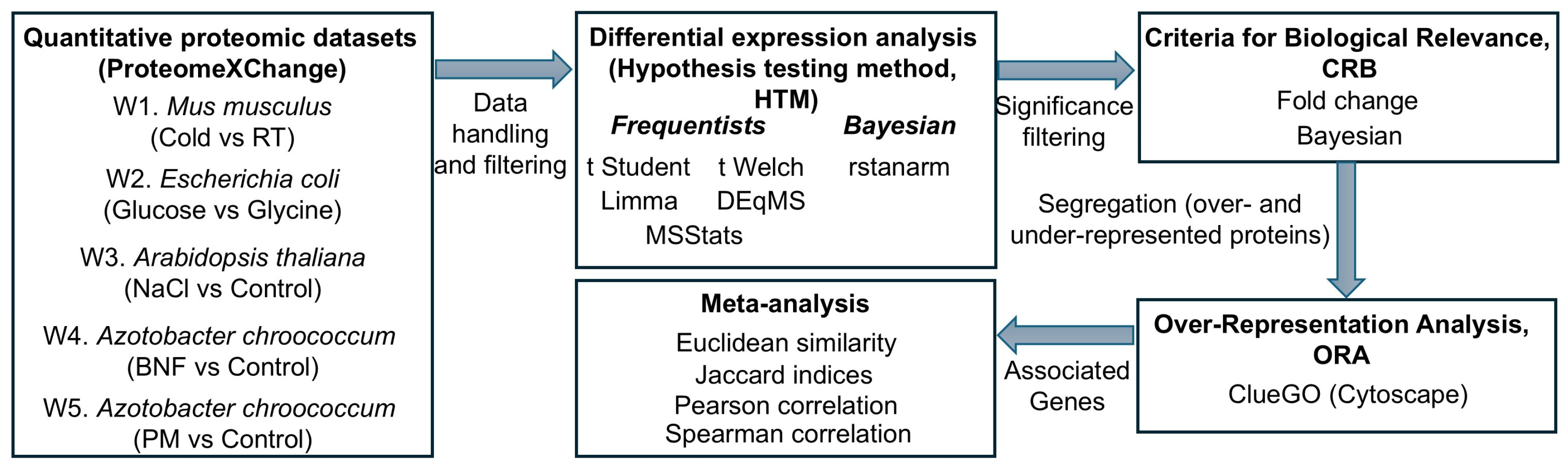
4. Materials and Methods
4.1. Dataset Selection
- -
- Work 1 (ProteomeXchange: PXD051640) originated from a study on brown adipose tissue and liver in a cold-exposed cardiometabolic mouse model [30]. The protein database used for identification was Mus musculus (C57BL/6J) (UP000000589).
- -
- Work 2 (ProteomeXchange: PXD041209) investigated the Escherichia coli protein acetylome under three growth conditions [31]. Protein identification relied on the Escherichia coli K12 (UP000000625) protein database.
- -
- Work 3 (ProteomeXchange: PXD019139) explored quantitative proteome and PTMome responses in Arabidopsis thaliana roots to osmotic and salinity stress [32]. The corresponding protein database was Arabidopsis thaliana (UP000006548).
- -
- Works 4 and 5 (ProteomeXchange: PXD034112) were derived from a comprehensive study on biological nitrogen fixation and phosphorus mobilization in Azotobacter chroococcum NCIMB 8003 [33]. For dataset 4, raw files from control and biological nitrogen fixation conditions were used, while for dataset 5, raw files corresponding to control conditions were compared to phosphorous mobilization conditions. The protein database for these works was UP000068210”.
4.2. Differential Abundance Analysis
- -
- Student’s and Welch’s t-tests: Performed on the base-2 logarithm of protein intensity data to compare means between two conditions. Both Student’s t-test (assuming equal variances) and Welch’s t-test (not assuming equal variances) were applied using pairwise complete observations; that is, proteins were retained if they had at least two valid (non-missing) values per group.
- -
- Limma: The Limma R package [17] was used to fit a linear model to log2-transformed protein intensity data. This method employs empirical Bayes moderation of variances and tolerates missing values, provided sufficient replicate data are available. In this way, statistical power in enhanced and variance stabilized, which is particularly critical in experiments with low biological replicates.
- -
- DEqMS: The DEqMS R package [18] was employed, extending the Limma framework by incorporating peptide count information to refine variance estimation in differential protein abundance analysis. It leverages the observation that proteins identified with more peptide-spectrum matches (PSMs) yield more reliable intensity measurements, leading to improved statistical power. As with Limma, DEqMS accepts missing values natively during model fitting and uses empirical variance estimation without requiring imputation.
- -
- MSstats: The MSstats R package [13] is specifically designed for quantitative mass spectrometry data. Uniquely among these methods, MSstats requires the original proteinGroups.txt file (not the filtered version) along with the evidence.txt file as input. Data was pre-processed using the dataProcess function in MSstats, and group comparisons were performed using linear mixed-effects models. This approach accounts for various sources of variability (e.g., biological/technical replicates, batch effects) by explicitly modeling them as random effects, thus providing robust variance estimates and increased statistical power. Missing values were handled using MSstats’ default model-based approach, which treats them as censored (i.e., non-random) and does not perform global imputation.
- -
- Bayesian Analysis: Differential protein abundance was also assessed using a Bayesian framework, implemented with the rstanarm R package [25]. This package provides an interface to Stan for Hamiltonian Monte Carlo (HMC) sampling. For each protein, a Bayesian linear regression model was fitted to the log2-transformed intensity data, utilizing the experimental condition as predictor. Only proteins with at least two valid values per group were considered. Missing values were implicitly handled through marginalization over the posterior, without requiring imputation. Model fitting employed four Markov Chain Monte Carlo (MCMC) chains with 4000 iterations (including 2000 warm-up iterations) and an adapt_delta of 0.99. Convergence of the chains was rigorously monitored using Rhat values (ideally ≤1.01) and effective sample size (ESS, ideally ≥200). This probabilistic approach yields full posterior distributions for the model parameters, which enables direct statements about effect sizes and their associated uncertainties. Weakly informative priors were incorporated to regularize parameter estimates and enhance model stability, particularly beneficial for proteins with limited measurements [35].
4.3. Biological Relevance Filtering and Overlap Analysis
4.4. Segregation and Functional Enrichment Analysis
4.5. Similarity and Correlation Analysis of Functional Enrichment Outcomes
- -
- Jaccard Index (J(A,B) = ∣A∪B∣∣A∩B∣): This metric was used to quantify the overlap between the sets of enriched terms (defined as terms with % Associated Genes > 0) derived from different method combinations. For statistical analysis, Jaccard Index values were transformed using the arcsin square root transformation (arcsin(x)) to stabilize variance.
- -
- Pearson Correlation Coefficient: This metric assessed the linear relationship between the quantitative profiles (vectors of % Associated Genes for all terms within a given ontology) generated by different method combinations.
- -
- Spearman Correlation Coefficient: This non-parametric metric evaluated the monotonic relationship between the quantitative profiles of % Associated Genes, making it robust to non-linear associations and outliers.
- -
- Euclidean Similarity: Derived from Euclidean distance, this metric quantified the closeness of the quantitative profiles of % Associated Genes between different method combinations. Similarity was calculated as 1/(1 + d), where d is the Euclidean distance, resulting in values ranging from 0 (maximal dissimilarity) to 1 (maximal similarity).
- -
- Intra-HTM_FC_CBR: Comparisons between different HTMs where the CBR was consistently fold change-based (e.g., deqms_FC vs. Limma_FC). This category assesses the variability introduced solely by the choice of HTM when a fixed FC relevance criterion is applied.
- -
- Intra-HTM_Bayes_CBR: Comparisons between different HTMs where the CBR was consistently Bayesian posterior probability-based (e.g., deqms_Bayes vs. Limma_Bayes). This category assesses the variability introduced solely by the choice of HTM when a Bayesian relevance criterion is applied.
- -
- Intra-CBR_Fixed_HTM: Comparisons between the two different CBRs (fold change-based vs. Bayesian posterior probability-based), where the HTM was kept constant (e.g., tstudent_FC vs. tstudent_Bayes). This category directly evaluates the influence of the biological relevance criterion itself, controlling for the HTM.
- -
- Inter-HTM_Inter-CBR: Comparisons between combinations where both the HTM and the CBR differed (e.g., tstudent_FC vs. Limma_Bayes). This category represents the cumulative variability from changing both methodological aspects.
4.6. Meta-Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schubert, O.; Röst, H.; Collins, B.; Rosenberger, G.; Aebersold, R. Quantitative proteomics: Challenges and opportunities in basic and applied research. Nat. Protoc. 2017, 12, 1289–1294. [Google Scholar] [CrossRef]
- Bantscheff, M.; Lemeer, S.; Savitski, M.M.; Kuster, B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–965. [Google Scholar] [CrossRef] [PubMed]
- Old, W.M.; Meyer-Arendt, K.; Aveline-Wolf, L.; Pierce, K.G.; Mendoza, A.; Sevinsky, J.R.; Resing, K.A.; Ahn, N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteom. 2005, 4, 1487–1502. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wang, H.; Kong, W.; Li, J.; Bin Goh, W.W. Optimizing differential expression analysis for proteomics data via high-performing rules and ensemble inference. Nat. Commun. 2024, 15, 3922. [Google Scholar] [CrossRef] [PubMed]
- Woodland, B.; Farrell, L.A.; Brockbals, L.; Rezcallah, M.; Brennan, A.; Sunnucks, E.J.; Gould, S.T.; Stanczak, A.M.; O’Rourke, M.B.; Padula, M.P. Sample Preparation for Multi-Omics Analysis: Considerations and Guidance for Identifying the Ideal Workflow. Proteomics 2025, 25, e13983. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28, 710–721. [Google Scholar] [CrossRef]
- Dowell, J.A.; Wright, L.J.; Armstrong, E.A.; Denu, J.M. Benchmarking quantitative performance in label-free proteomics. ACS Omega 2021, 6, 2494–2504. [Google Scholar] [CrossRef]
- Ruggles, K.V.; Krug, K.; Wang, X.; Clauser, K.R.; Wang, J.; Payne, S.H.; Fenyö, D.; Zhang, B.; Mani, D.R. Methods, tools and current perspectives in proteogenomics. Mol. Cell. Proteom. 2017, 16, 959–981. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I. Protein identification by tandem mass spectrometry and sequence database searching. In Methods in Molecular Biology; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2007; Volume 367, pp. 87–119. [Google Scholar] [CrossRef]
- Shteynberg, D.; Nesvizhskii, A.I.; Moritz, R.L.; Deutsch, E.W. Combining results of multiple search engines in proteomics. Mol. Cell. Proteom. 2013, 12, 2383–2393. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteom. 2010, 73, 2092–2123. [Google Scholar] [CrossRef]
- Välikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Choi, M.; Chang, C.Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef]
- Choi, H.; Fermin, D.; Nesvizhskii, A.I. Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteom. 2008, 7, 2373–2385. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Fornasiero, I.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2523. [Google Scholar] [CrossRef]
- Langley, S.R.; Mayr, M. Comparative analysis of statistical methods used for detecting differential expression in label-free mass spectrometry proteomics. J. Proteom. 2015, 129, 83–92. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Orre, L.M.; Zhou Tran, Y.; Mermelekas, G.; Johansson, H.J.; Malyutina, A.; Anders, S.; Lehtiö, J. DEqMS: A method for accurate variance estimation in differential protein expression analysis. Mol. Cell. Proteom. 2020, 19, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Jow, H.; Boys, R.J.; Wilkinson, D.J. Bayesian identification of protein differential expression in multi-group isobaric labelled mass spectrometry data. Stat. Appl. Genet. Mol. Biol. 2014, 13, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Pavelka, N.; Pelizzola, M.; Vizzardelli, C.; Cap-Noya, B.; Giudice, G.; Stupka, E. A power law global error model for the identification of differentially expressed genes in microarray data. BMC Bioinform. 2004, 5, 203. [Google Scholar] [CrossRef]
- Lee, K.H.; Assassi, S.; Mohan, C.; Pedroza, C. Addressing statistical challenges in the analysis of proteomics data with extremely small sample size: A simulation study. BMC Genom. 2024, 25, 1086. [Google Scholar] [CrossRef]
- Crook, O.M.; Chung, C.; Deane, C.M. Challenges and opportunities for Bayesian statistics in proteomics. J. Proteome Res. 2022, 21, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Student. The probable error of a mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Welch, B.L. The generalization of “Student’s” problem when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, B.; Gabry, J.; Ali, I.; Brilleman, S. rstanarm: Bayesian Applied Regression Modeling via Stan (Version 2.21.3) [R package]. 2022. Available online: https://mc-stan.org/rstanarm/ (accessed on 10 November 2024).
- Bürkner, P.-C. brms: An R Package for Bayesian Multilevel Models Using Stan. J. Stat. Softw. 2017, 80, 1–28. [Google Scholar] [CrossRef]
- Crook, O.M.; Mulvey, C.M.; Kirk, P.D.W.; Lilley, K.S.; Gatto, L. A Bayesian mixture modelling approach for spatial proteomics. PLoS Comput. Biol. 2018, 14, e1006516. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Roden, J.C.; King, B.W.; Trout, D.; Mortazavi, A.; Wold, B.J.; Hart, C.E. Mining gene expression data by interpreting principal components. BMC Bioinform. 2006, 7, 194. [Google Scholar] [CrossRef]
- Amor, M.; Diaz, M.; Bianco, V.; Svecla, M.; Schwarz, B.; Rainer, S.; Pirchheim, A.; Schooltink, L.; Mukherjee, S.; Grabner, G.F.; et al. Identification of regulatory networks and crosstalk factors in brown adipose tissue and liver of a cold-exposed cardiometabolic mouse model. Cardiovasc. Diabetol. 2024, 23, 298. [Google Scholar] [CrossRef]
- Lozano-Terol, G.; Chiozzi, R.Z.; Gallego-Jara, J.; Sola-Martínez, R.A.; Vivancos, A.M.; Ortega, Á.; Heck, A.J.R.; Díaz, M.C.; de Diego Puente, T. Relative impact of three growth conditions on the Escherichia coli protein acetylome. iScience 2024, 27, 109017. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Mehta, D.; Tan, M.; Uhrig, R.G. Quantitative Proteome and PTMome Analysis of Arabidopsis thaliana Root Responses to Persistent Osmotic and Salinity Stress. Plant Cell Physiol. 2021, 62, 1012–1029. [Google Scholar] [CrossRef]
- Biełło, K.A.; Lucena, C.; López-Tenllado, F.J.; Hidalgo-Carrillo, J.; Rodríguez-Caballero, G.; Cabello, P.; Sáez, L.P.; Luque-Almagro, V.; Roldán, M.D.; Moreno-Vivián, C.; et al. Holistic view of biological nitrogen fixation and phosphorus mobilization in Azotobacter chroococcum NCIMB 8003. Front. Microbiol. 2023, 14, 1129721. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2024. Available online: https://www.R-project.org/ (accessed on 10 November 2024).
- Gelman, A.; Jakulin, A.; Pittau, M.G.; Su, Y.S. A weakly informative default prior distribution for logistic and other regression models. Ann. Appl. Stat. 2008, 2, 1360–1383. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2923–2924. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
| Experimental Design | Most Commonly Used Test | Other Possible Test |
|---|---|---|
| Simple Comparison (A vs. B) | Student’s t-test [16] | Limma (moderated t-test) [17], DEqMS [18], Bayesian models [19] |
| Multiple Conditions | One-way ANOVA [16] | Limma [16], DEqMS [18], Bayesian models [19] |
| Time Series Experiments | ANOVA/Linear Regression [16] | Linear mixed-effects models (MSstats) [19], Limma [17], DEqMS [18], Bayesian [19] |
| Multifactorial (e.g., treatment × time) | Factorial ANOVA [16] | Mixed-effects models (MSstats) [13], Limma [17], DEqMS [18], Bayesian [19] |
| Controlled Reference Mixtures | ANOVA/t-test [16] | Limma [17], DEqMS [18], Bayesian [19] |
| Spectral Count Data | QSpec [14] | QSpec [14], hierarchical Bayesian count models [19] |
| Extended Time Series (>4 points) | Regression/Clustering [16] | Linear mixed-effects models (MSstats) [13], Bayesian time series [19] |
| Low Replication Designs | t-test/PLGEM-STN [20] | PLGEM-STN [20], Limma [17], DEqMS [18], Bayesian [19] |
| Work | Median CV (Cond1) (%) | Median CV (Cond2) (%) | SD del|Log2FC (Cond1 vs. Cond2)| | Levene (p Value) |
|---|---|---|---|---|
| 1 | 13.17 | 12.78 | 4.28 | 0.3694 |
| 2 | 26.79 | 25.50 | 3.00 | 0.9991 |
| 3 | 10.16 | 8.34 | 2.75 | 0.0221 |
| 4 | 18.62 | 31.95 | 4.72 | 0.0000 |
| 5 | 36.42 | 31.99 | 4.21 | 0.1310 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biełło, K.A.; Die, J.V.; Amil, F.; Fuentes-Almagro, C.; Pérez-Rodríguez, J.; Olaya-Abril, A. Reproducibility Crossroads: Impact of Statistical Choices on Proteomics Functional Enrichment. Int. J. Mol. Sci. 2025, 26, 9232. https://doi.org/10.3390/ijms26189232
Biełło KA, Die JV, Amil F, Fuentes-Almagro C, Pérez-Rodríguez J, Olaya-Abril A. Reproducibility Crossroads: Impact of Statistical Choices on Proteomics Functional Enrichment. International Journal of Molecular Sciences. 2025; 26(18):9232. https://doi.org/10.3390/ijms26189232
Chicago/Turabian StyleBiełło, Karolina A., José V. Die, Francisco Amil, Carlos Fuentes-Almagro, Javier Pérez-Rodríguez, and Alfonso Olaya-Abril. 2025. "Reproducibility Crossroads: Impact of Statistical Choices on Proteomics Functional Enrichment" International Journal of Molecular Sciences 26, no. 18: 9232. https://doi.org/10.3390/ijms26189232
APA StyleBiełło, K. A., Die, J. V., Amil, F., Fuentes-Almagro, C., Pérez-Rodríguez, J., & Olaya-Abril, A. (2025). Reproducibility Crossroads: Impact of Statistical Choices on Proteomics Functional Enrichment. International Journal of Molecular Sciences, 26(18), 9232. https://doi.org/10.3390/ijms26189232






