Structural and Dynamic Insights into Acyl Carrier Protein upon Metal Binding and Acylation Revealed by NMR Spectroscopy and MD Simulations
Abstract
1. Introduction
2. Results
2.1. Comparative Sequence Analysis of EcACP with Mesophilic, Thermophilic, and Psychrophilic ACPs
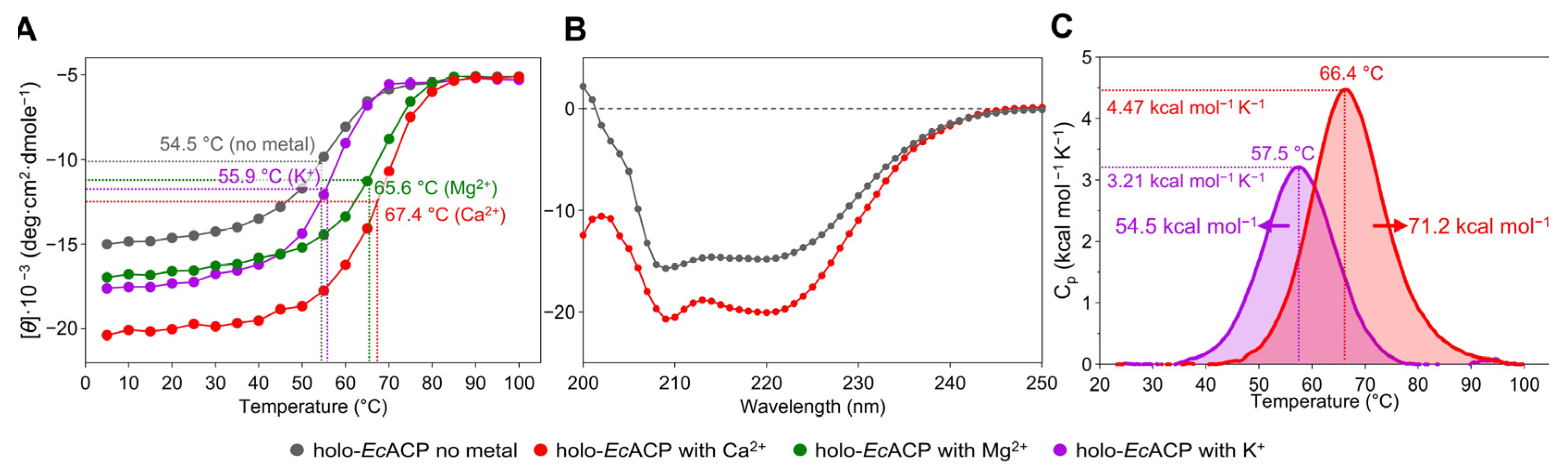
2.2. Thermal Stabilization of EcACP by Different Metal Ions
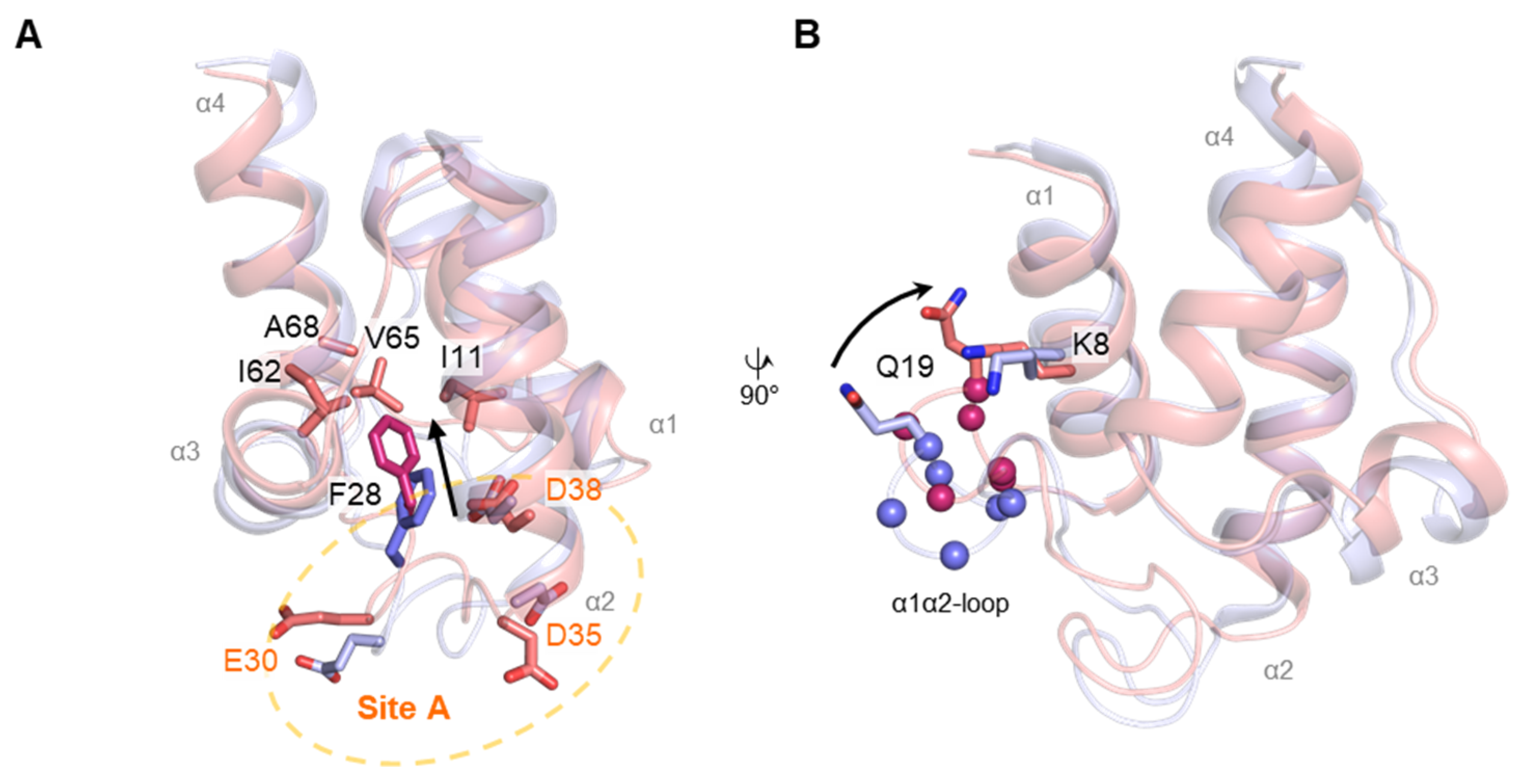
2.3. NMR Structure of EcACP in the Presence of Ca2+
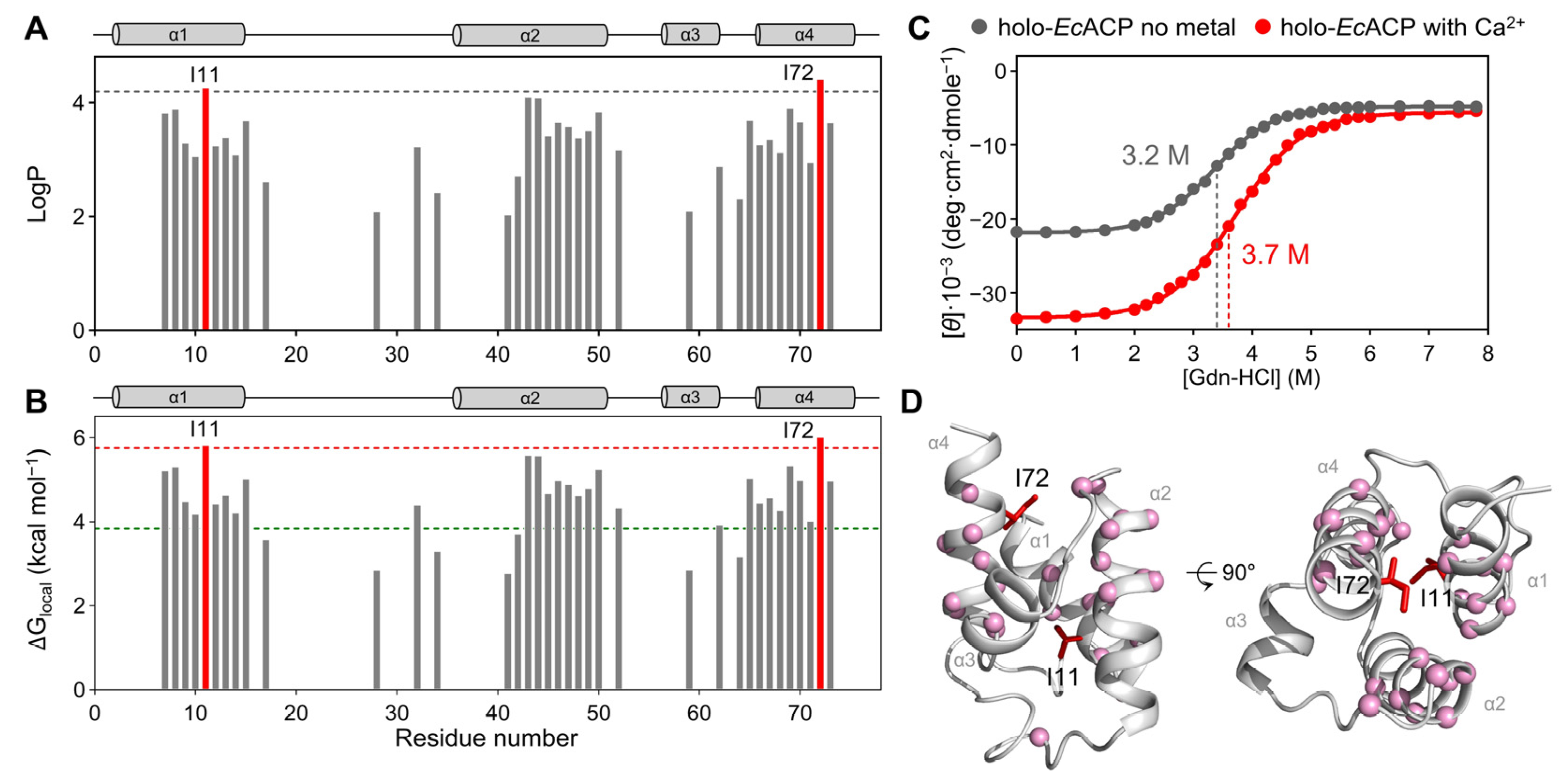
2.4. Structural Stability Analysis Through Hydrogen–Deuterium Exchange and Chemical Denaturation Experiments
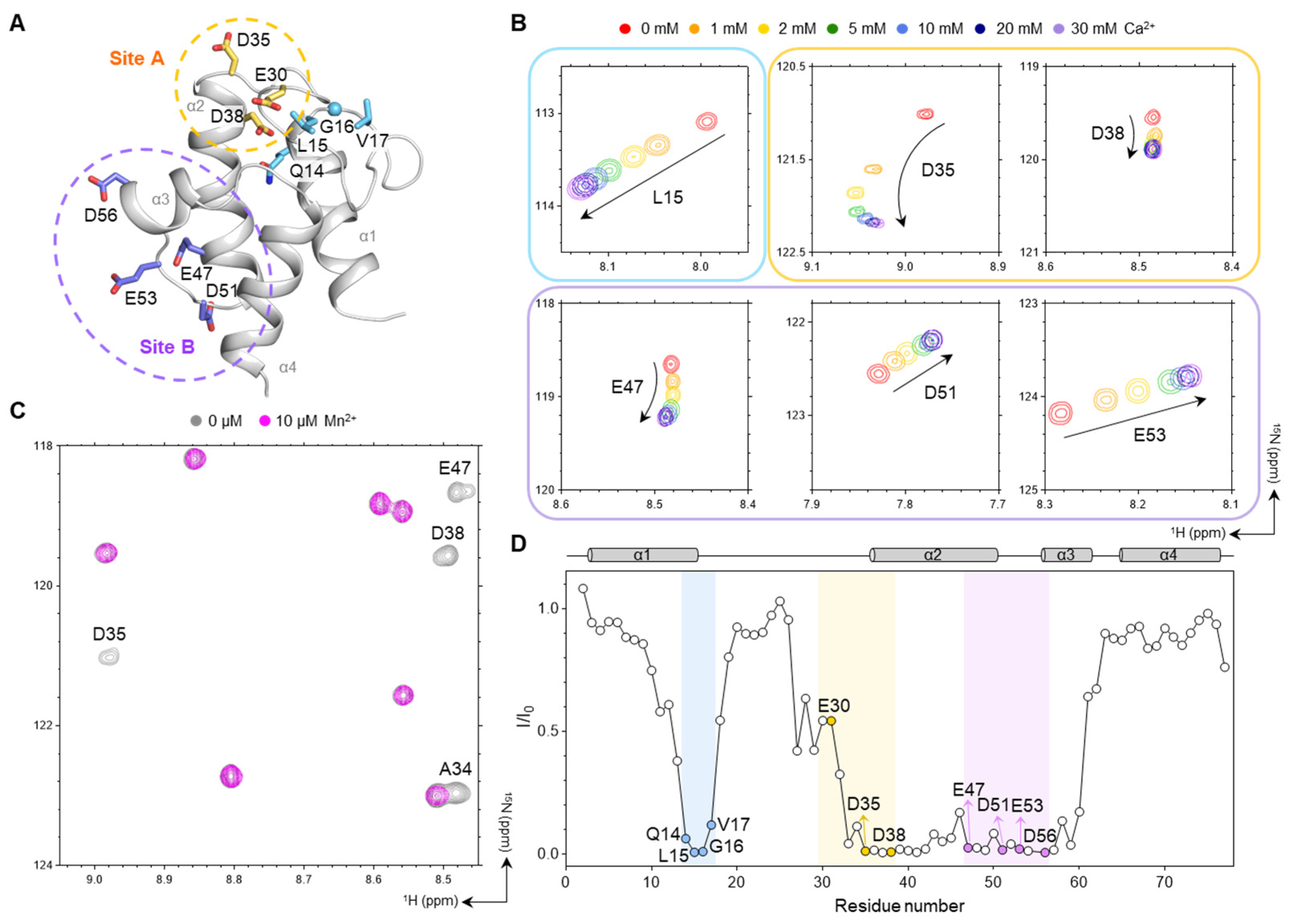
2.5. Analysis of Metal-Binding Sites by Metal Titration HSQC Spectra
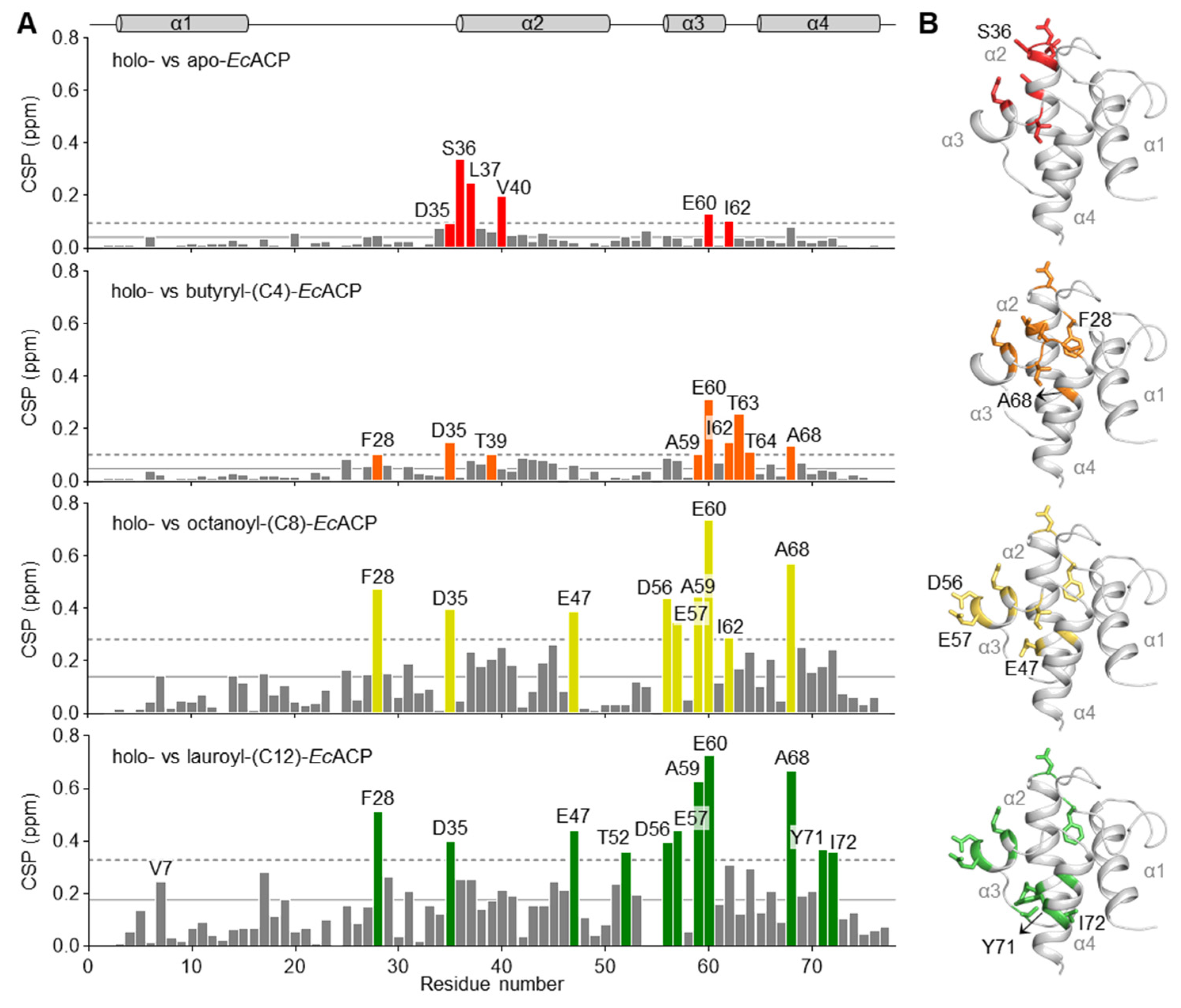
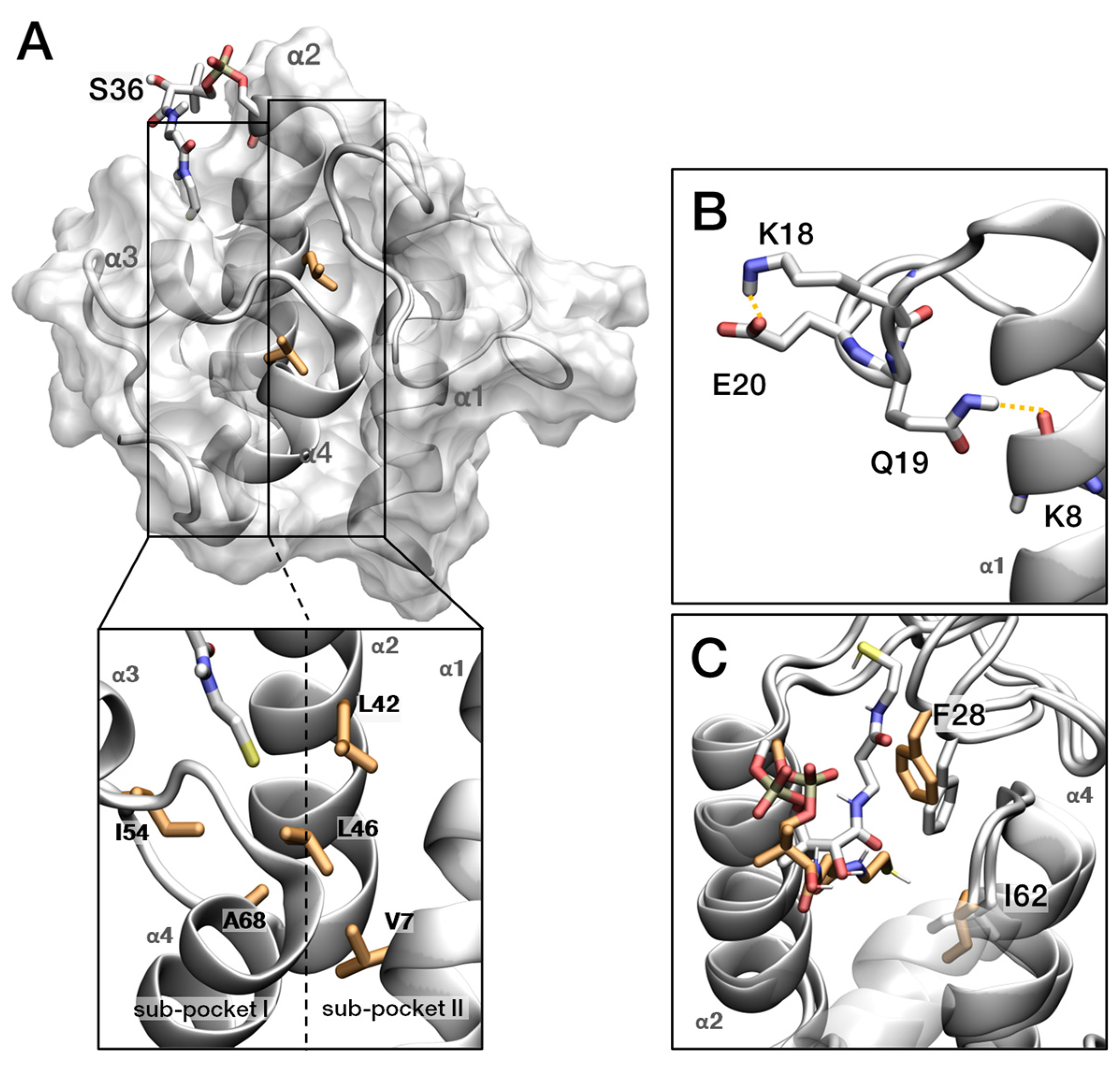
2.6. Chemical Shift Perturbation by Acylation of EcACP
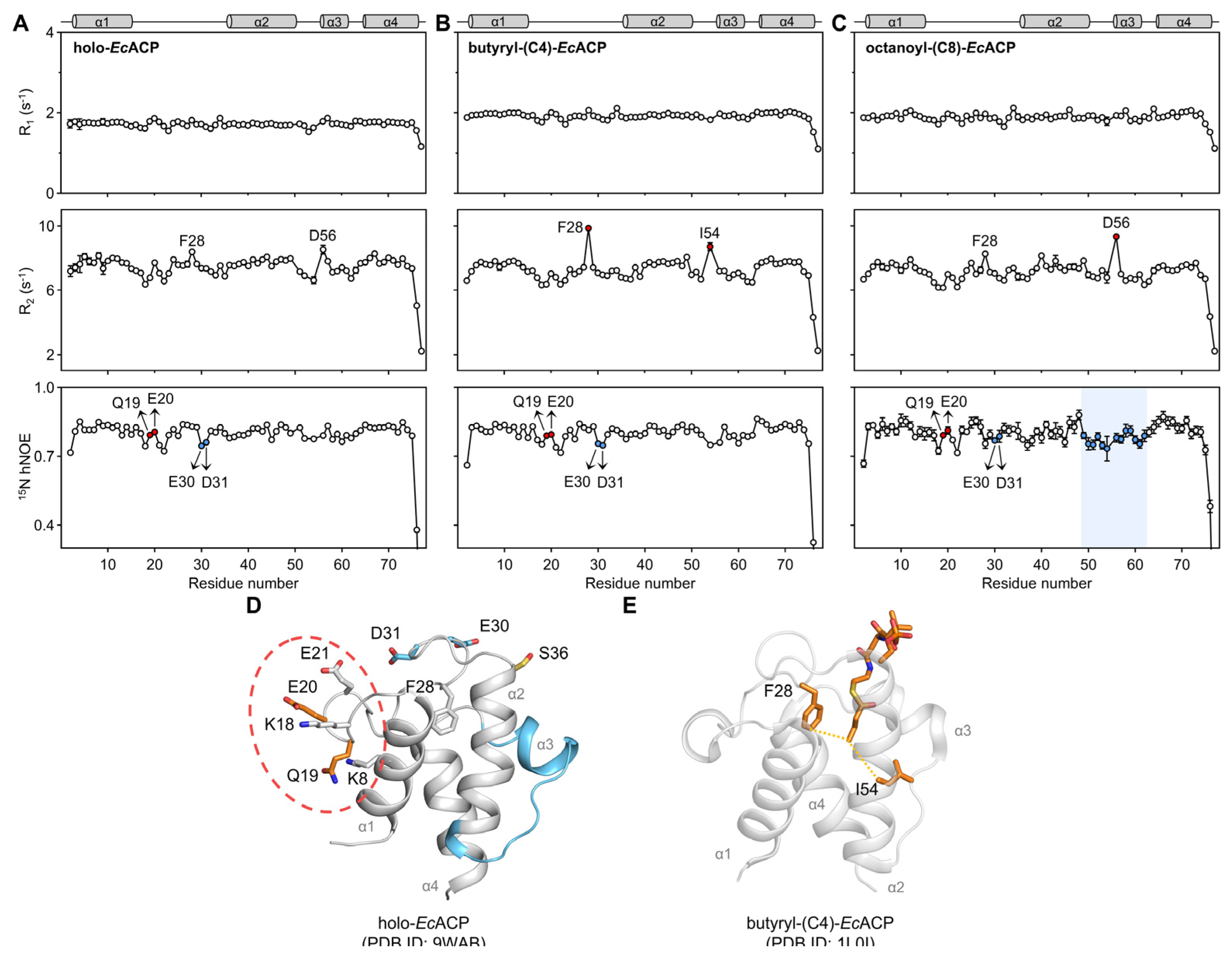
2.7. Backbone Dynamics of Holo-, Butyryl-, and Octanoyl-EcACP
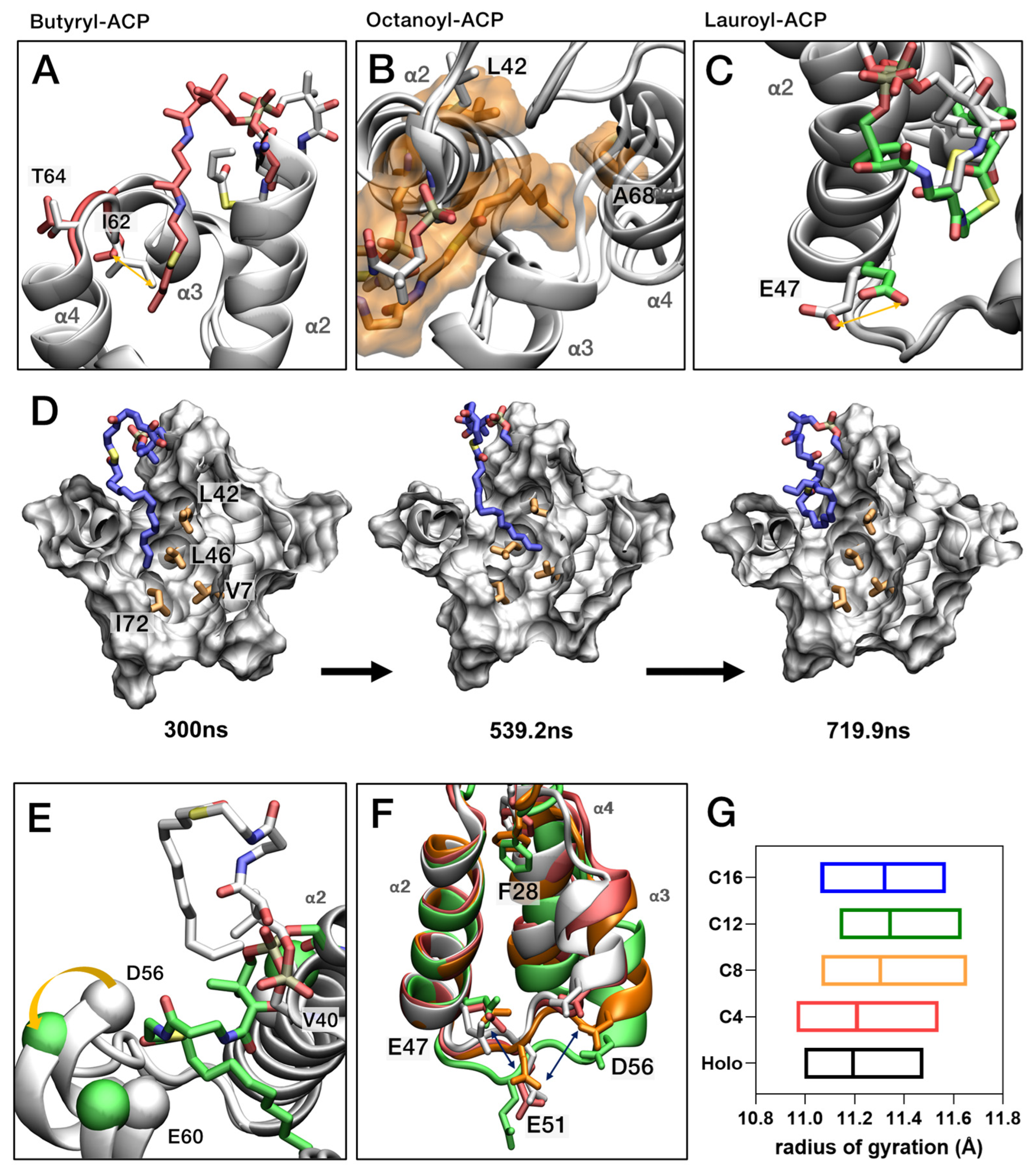
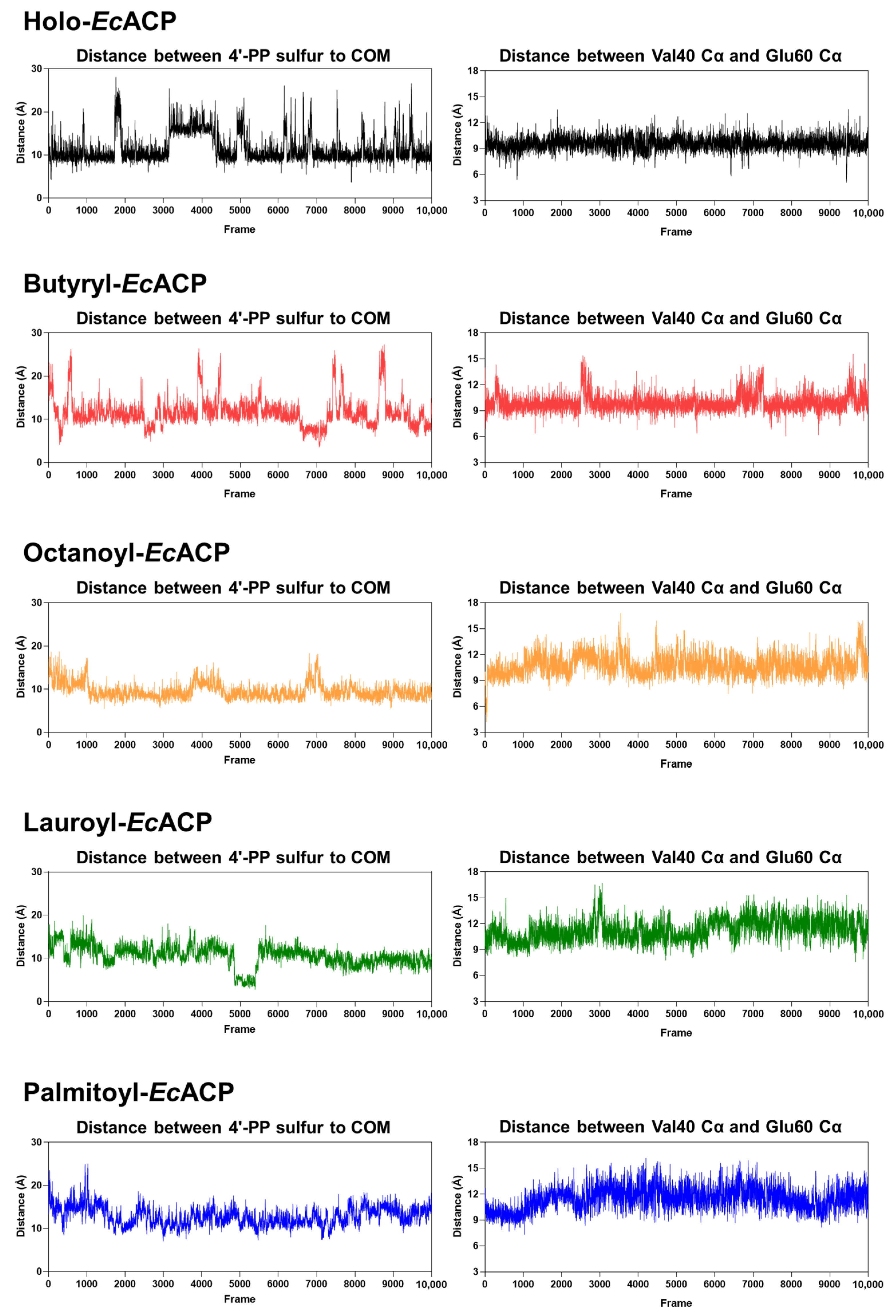
2.8. Molecular Dynamics Simulation of EcACP
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification of EcACP
4.2. Circular Dichroism Experiments
4.3. Differential Scanning Calorimetry
4.4. NMR Experiments and Assignment
4.5. Solution Structure Calculation
4.6. Hydrogen/Deuterium Exchange Experiment
4.7. Chemical Denaturation Experiments
4.8. Metal Titration Experiments
4.9. Chemical Shift Perturbations
4.10. Backbone Dynamics
4.11. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACP | Acyl carrier protein |
| FAS | Fatty acid synthesis |
| MD | Molecular dynamics |
| Ec | Escherichia coli |
| ACPS | Holo-ACP synthase |
| 4′-PP | 4′-phosphopantetheine |
| MDR | Multi-drug-resistant |
| Gdn-HCl | Guanidine hydrochloride |
| RDC | Residual dipolar coupling |
| H/D | Hydrogen–deuterium |
| DSC | Differential scanning calorimetry |
| pI | Isoelectric point |
| Tm | Thermotoga maritima |
| Cp | Colwellia psychrerythraea |
| Ta | Thermus aquaticus |
| Ab | Acinetobacter baumannii |
| Vh | Vibrio harveyi |
| Tm | Melting temperature |
| CD | Circular dichroism |
| Cp | Specific heat capacity |
| ΔHcal | Calorimetric enthalpy |
| RMSD | Root-mean-square deviation |
| LogP | Protection factor |
| kex | Exchange rate |
| kprot | H/D exchange rate |
| krc | Random coil exchange rate |
| ΔGlocal | Local unfolding free energy |
| ΔGglobal | Global unfolding free energy |
| [Gdn-HCl]1/2 | Half-denaturation concentration of Gdn-HCl |
| CSP | Chemical shift perturbation |
| HSQC | Heteronuclear single quantum coherence |
| R1 | Longitudinal spin relaxation |
| R2 | Transverse spin relaxation |
| hNOE | Heteronuclear NOE |
| χ1 | Side-chain torsion angle |
| COM | Center of mass |
| IPTG | Isopropyl β-D-thiogalactopyranoside |
| DTT | Dithiothreitol |
| DSS | 2,2-Dimethyl-2-silapentane-5-sulfonate |
| PDB | Protein Data Bank |
| KBSI | Korea Basic Science Institute |
References
- Chan, D.I.; Vogel, H.J. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 2010, 430, 1–19. [Google Scholar] [CrossRef]
- Shen, B.; Summers, R.G.; Gramajo, H.; Bibb, M.J.; Hutchinson, C.R. Purification and characterization of the acyl carrier protein of the Streptomyces glaucescens tetracenomycin C polyketide synthase. J. Bacteriol. 1992, 174, 3818–3821. [Google Scholar] [CrossRef][Green Version]
- Masoudi, A.; Raetz, C.R.; Zhou, P.; Pemble, C.W.T. Chasing acyl carrier protein through a catalytic cycle of lipid A production. Nature 2014, 505, 422–426. [Google Scholar] [CrossRef]
- Wakil, S.J.; Stoops, J.K.; Joshi, V.C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983, 52, 537–579. [Google Scholar] [CrossRef] [PubMed]
- Jenni, S.; Leibundgut, M.; Boehringer, D.; Frick, C.; Mikolasek, B.; Ban, N. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 2007, 316, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Tsai, S.C. The type I fatty acid and polyketide synthases: A tale of two megasynthases. Nat. Prod. Rep. 2007, 24, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- White, S.W.; Zheng, J.; Zhang, Y.M.; Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Lu, Y.J.; Zhang, Y.M.; Rock, C.O. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol. 2004, 82, 145–155. [Google Scholar] [CrossRef]
- Park, J.; Lee, Y.; Cheon, D.; Kim, Y. Structure and dynamics of human and bacterial acyl carrier proteins and their interactions with fatty acid synthesis proteins. Biochem. Biophys. Res. Commun. 2019, 516, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, J.; Yeon, J.; Jang, A.; Lee, W.C.; Kim, Y. Deciphering the Binding Interactions between Acinetobacter baumannii ACP and β-ketoacyl ACP Synthase III to Improve Antibiotic Targeting Using NMR Spectroscopy. Int. J. Mol. Sci. 2021, 22, 3317. [Google Scholar] [CrossRef]
- Son, M.; Oh, S.; Oh, Y.; Cheon, D.; Jang, A.; Kim, E.; Kim, N.K.; Kim, Y. Structural and dynamic insights into acyl carrier protein in Cutibacterium acnes reveal mechanisms for fatty acid synthesis and transport. Biochem. Biophys. Res. Commun. 2024, 741, 151090. [Google Scholar] [CrossRef]
- Park, Y.G.; Jung, M.C.; Song, H.; Jeong, K.W.; Bang, E.; Hwang, G.S.; Kim, Y. Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis. J. Biol. Chem. 2016, 291, 1692–1702. [Google Scholar] [CrossRef]
- Lee, Y.; Jang, A.; Jeong, M.C.; Park, N.; Park, J.; Lee, W.C.; Cheong, C.; Kim, Y. Structural Characterization of an ACP from Thermotoga maritima: Insights into Hyperthermal Adaptation. Int. J. Mol. Sci. 2020, 21, 2600. [Google Scholar] [CrossRef]
- Chan, D.I.; Chu, B.C.; Lau, C.K.; Hunter, H.N.; Byers, D.M.; Vogel, H.J. NMR solution structure and biophysical characterization of Vibrio harveyi acyl carrier protein A75H: Effects of divalent metal ions. J. Biol. Chem. 2010, 285, 30558–30566. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Prestegard, J.H. Refinement of the NMR structures for acyl carrier protein with scalar coupling data. Proteins 1990, 8, 377–385. [Google Scholar] [CrossRef]
- Wu, B.N.; Zhang, Y.M.; Rock, C.O.; Zheng, J.J. Structural modification of acyl carrier protein by butyryl group. Protein Sci. 2009, 18, 240–246. [Google Scholar] [CrossRef]
- Roujeinikova, A.; Simon, W.J.; Gilroy, J.; Rice, D.W.; Rafferty, J.B.; Slabas, A.R. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 2007, 365, 135–145. [Google Scholar] [CrossRef]
- Sharma, A.K.; Sharma, S.K.; Surolia, A.; Surolia, N.; Sarma, S.P. Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry 2006, 45, 6904–6916. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jeong, K.W.; Lee, J.U.; Kang, D.I.; Kim, Y. Novel E. coli β-ketoacyl-acyl carrier protein synthase III inhibitors as targeted antibiotics. Bioorg. Med. Chem. 2009, 17, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.W.; Lee, J.; Kang, D.I.; Lee, J.U.; Shin, S.Y.; Kim, Y. Screening of Flavonoids as Candidate Antibiotics against Enterococcus faecalis. J. Nat. Prod. 2009, 72, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeong, K.W.; Shin, S.; Lee, J.U.; Kim, Y. Discovery of novel selective inhibitors of Staphylococcus aureus β-ketoacyl acyl carrier protein synthase III. Eur. J. Med. Chem. 2012, 47, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeong, M.C.; Jeon, D.; Lee, Y.; Lee, W.C.; Kim, Y. Structure-activity relationship-based screening of antibiotics against Gram-negative Acinetobacter baumannii. Bioorg. Med. Chem. 2017, 25, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Park, J.; Balasubramanian, P.K.; Kim, Y. Elucidation of the crystal structure of FabD from the multidrug-resistant bacterium Acinetobacter baumannii. Biochem. Biophys. Res. Commun. 2018, 505, 208–214. [Google Scholar] [CrossRef]
- Cheon, D.; Lee, W.C.; Lee, Y.; Lee, J.Y.; Kim, Y. Structural basis of branched-chain fatty acid synthesis by Propionibacterium acnes β-ketoacyl acyl Carrier protein synthase. Biochem. Biophys. Res. Commun. 2019, 509, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Jang, M.; Lee, S.; Lee, J.Y.; Lee, W.C.; Bae, S.; Kang, J.; Han, M.; Kim, Y. Identification of inhibitor binding hotspots in Acinetobacter baumannii β-ketoacyl acyl carrier protein synthase III using molecular dynamics simulation. J. Mol. Graph. Model. 2020, 100, 107669. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Chen, A.; Kim, W.E.; Re, R.N.; Davis, T.D.; Noel, J.P.; Burkart, M.D. Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases. Acs Catal. 2021, 11, 6787–6799. [Google Scholar] [CrossRef] [PubMed]
- Misson, L.E.; Mindrebo, J.T.; Davis, T.D.; Patel, A.; McCammon, J.A.; Noel, J.P.; Burkart, M.D. Interfacial plasticity facilitates high reaction rate of E. coli FAS malonyl-CoA:ACP transacylase, FabD. Proc. Natl. Acad. Sci. USA 2020, 117, 24224–24233. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Patel, A.; Kim, W.E.; Davis, T.D.; Chen, A.; Bartholow, T.G.; La Clair, J.J.; McCammon, J.A.; Noel, J.P.; Burkart, M.D. Gating mechanism of elongating beta-ketoacyl-ACP synthases. Nat. Commun. 2020, 11, 1727. [Google Scholar] [CrossRef]
- Cronan, J.E. The chain-flipping mechanism of ACP (acyl carrier protein)-dependent enzymes appears universal. Biochem. J. 2014, 460, 157–163. [Google Scholar] [CrossRef]
- Beld, J.; Cang, H.; Burkart, M.D. Visualizing the Chain-Flipping Mechanism in Fatty-Acid Biosynthesis. Angew. Chem. Int. Ed. 2014, 53, 14456–14461. [Google Scholar] [CrossRef]
- Arya, R.; Sharma, B.; Dhembla, C.; Pal, R.K.; Patel, A.K.; Sundd, M.; Ghosh, B.; Makde, R.D.; Kundu, S. A conformational switch from a closed apo- to an open holo-form equips the acyl carrier protein for acyl chain accommodation. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 163–174. [Google Scholar] [CrossRef]
- Sztain, T.; Bartholow, T.G.; Lee, D.J.; Casalino, L.; Mitchell, A.; Young, M.A.; Wang, J.; McCammon, J.A.; Burkart, M.D. Decoding allosteric regulation by the acyl carrier protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2025597118. [Google Scholar] [CrossRef]
- Colizzi, F.; Masetti, M.; Recanatini, M.; Cavalli, A. Atomic-Level Characterization of the Chain-Flipping Mechanism in Fatty-Acids Biosynthesis. J. Phys. Chem. Lett. 2016, 7, 2899–2904. [Google Scholar] [CrossRef]
- Colizzi, F.; Recanatini, M.; Cavalli, A. Mechanical Features of Plasmodium falciparum Acyl Carrier Protein in the Delivery of Substrates. J. Chem. Inf. Model. 2008, 48, 2289–2293. [Google Scholar] [CrossRef]
- Frederick, A.F.; Kay, L.E.; Prestegard, J.H. Location of divalent ion sites in acyl carrier protein using relaxation perturbed 2D NMR. FEBS Lett. 1988, 238, 43–48. [Google Scholar] [CrossRef]
- Gong, H.; Murphy, A.; McMaster, C.R.; Byers, D.M. Neutralization of acidic residues in helix II stabilizes the folded conformation of acyl carrier protein and variably alters its function with different enzymes. J. Biol. Chem. 2007, 282, 4494–4503. [Google Scholar] [CrossRef] [PubMed]
- Horvath, L.A.; Sturtevant, J.M.; Prestegard, J.H. Kinetics and thermodynamics of thermal denaturation in acyl carrier protein. Protein Sci. 1994, 3, 103–108. [Google Scholar] [CrossRef]
- Oh, S.; Lee, C.; Son, M.; Yeon, J.; Kim, Y. Dynamics of acyl carrier protein in de novo fatty acid synthesis by Enterococcus faecalis based on NMR spectroscopy and molecular dynamics simulation. J. Anal. Sci. Technol. 2024, 15, 31. [Google Scholar] [CrossRef]
- Beld, J.; Sonnenschein, E.C.; Vickery, C.R.; Noel, J.P.; Burkart, M.D. The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 2014, 31, 61–108. [Google Scholar] [CrossRef] [PubMed]
- Stenstroem, O.; Champion, C.; Lehner, M.; Bouvignies, G.; Riniker, S.; Ferrage, F. How does it really move? Recent progress in the investigation of protein nanosecond dynamics by NMR and simulation. Curr. Opin. Struct. Biol. 2022, 77, 102459. [Google Scholar] [CrossRef] [PubMed]
- Kümmerer, F.; Orioli, S.; Harding-Larsen, D.; Hoffmann, F.; Gavrilov, Y.; Teilum, K.; Lindorff-Larsen, K. Fitting Side-Chain NMR Relaxation Data Using Molecular Simulations. J. Chem. Theory Comput. 2021, 17, 5262–5275. [Google Scholar] [CrossRef] [PubMed]
- Champion, C.; Lehner, M.; Smith, A.A.; Ferrage, F.; Bolik-Coulon, N.; Riniker, S. Unraveling motion in proteins by combining NMR relaxometry and molecular dynamics simulations: A case study on ubiquitin. J. Chem. Phys. 2024, 160, 104105. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Barnwal, R.P.; Kaur, M.; Heckert, A.; Gartia, J.; Varani, G. Comparative structure, dynamics and evolution of acyl-carrier proteins from Borrelia burgdorferi, Brucella melitensis and Rickettsia prowazekii. Biochem. J. 2020, 477, 491–508. [Google Scholar] [CrossRef]
- Paul, S.; Ishida, H.; Nguyen, L.T.; Liu, Z.; Vogel, H.J. Structural and dynamic characterization of a freestanding acyl carrier protein involved in the biosynthesis of cyclic lipopeptide antibiotics. Protein Sci. 2017, 26, 946–959. [Google Scholar] [CrossRef]
- Rivas, M.; Courouble, V.C.; Baker, M.C.; Cookmeyer, D.L.; Fiore, K.E.; Frost, A.J.; Godbe, K.N.; Jordan, M.R.; Krasnow, E.N.; Mollo, A.; et al. The Effect of Divalent Cations on the Thermostability of Type II Polyketide Synthase Acyl Carrier Proteins. AIChE J. 2018, 64, 4308–4318. [Google Scholar] [CrossRef]
- Chan, D.I.; Stockner, T.; Tieleman, D.P.; Vogel, H.J. Molecular Dynamics Simulations of the Apo-, Holo-, and Acyl-forms of Escherichia coli Acyl Carrier Protein. J. Biol. Chem. 2008, 283, 33620–33629. [Google Scholar] [CrossRef]
- Andrec, M.; Hill, R.B.; Prestegard, J.H. Amide exchange rates in Escherichia coli acyl carrier protein: Correlation with protein structure and dynamics. Protein Sci. 1995, 4, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.Z.; Liu, Z.Y.; Qin, H.W.; Zhao, H.; Zhang, W.Q.; Xiao, C.L.; Wang, X.; Yang, X.M.; Wang, F.J. Ultraviolet Photodissociation Mass Spectrometry Captures the Acyl Chain Length-Dependent Conformation Dynamics of Acyl Carrier Protein. J. Am. Chem. Soc. 2025, 147, 16760–16765. [Google Scholar] [CrossRef]
- Mayo, K.H.; Prestegard, J.H. Acyl Carrier Protein from Escherichia Coli—Structural Characterization of Short-Chain Acylated Acyl Carrier Proteins by NMR. Biochemistry 1985, 24, 7834–7838. [Google Scholar] [CrossRef]
- Roujeinikova, A.; Baldock, C.; Simon, W.J.; Gilroy, J.; Baker, P.J.; Stuitje, A.R.; Rice, D.W.; Slabas, A.R.; Rafferty, J.B. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure 2002, 10, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E., Jr. Molecular properties of short chain acyl thioesters of acyl carrier protein. J. Biol. Chem. 1982, 257, 5013–5017. [Google Scholar] [CrossRef]
- Zornetzer, G.A.; Tanem, J.; Fox, B.G.; Markley, J.L. The length of the bound fatty acid influences the dynamics of the acyl carrier protein and the stability of the thioester bond. Biochemistry 2010, 49, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Finzel, K.; Burkart, M.D. Versatility of acyl-acyl carrier protein synthetases. Chem. Biol. 2014, 21, 1293–1299. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Chou, J.J.; Gaemers, S.; Howder, B.; Louis, J.M.; Bax, A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR 2001, 21, 377–382. [Google Scholar] [CrossRef]
- Sass, H.J.; Musco, G.; Stahl, S.J.; Wingfield, P.T.; Grzesiek, S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR 2000, 18, 303–309. [Google Scholar] [CrossRef]
- Cordier, F.; Dingley, A.J.; Grzesiek, S. A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR 1999, 13, 175–180. [Google Scholar] [CrossRef]
- Lee, W.; Stark, J.L.; Markley, J.L. PONDEROSA-C/S: Client-server based software package for automated protein 3D structure determination. J. Biomol. NMR 2014, 60, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Cornilescu, G.; Dashti, H.; Eghbalnia, H.R.; Tonelli, M.; Westler, W.M.; Butcher, S.E.; Henzler-Wildman, K.A.; Markley, J.L. Integrative NMR for biomolecular research. J. Biomol. NMR 2016, 64, 307–332. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins 2007, 66, 778–795. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, version 1.8; Schrodinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Bai, Y.; Milne, J.S.; Mayne, L.; Englander, S.W. Primary structure effects on peptide group hydrogen exchange. Proteins 1993, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.M.; Bolen, D.W. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry 1992, 31, 4901–4907. [Google Scholar] [CrossRef]
- Pace, C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986, 131, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Schellman, J.A. The thermodynamic stability of proteins. Annu. Rev. Biophys. Biophys. Chem. 1987, 16, 115–137. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Laurents, D.V.; Fernandez, A.M.; Elias-Arnanz, M.; Ruiz-Sanz, J.; Mateo, P.L.; Rico, M.; Filimonov, V.V. Thermodynamic analysis of the structural stability of phage 434 Cro protein. Biochemistry 1999, 38, 15536–15547. [Google Scholar] [CrossRef][Green Version]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L., 3rd; Lee, H.S.; Im, W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., 3rd; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Eastman, P.; Galvelis, R.; Pelaez, R.P.; Abreu, C.R.A.; Farr, S.E.; Gallicchio, E.; Gorenko, A.; Henry, M.M.; Hu, F.; Huang, J.; et al. OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials. arXiv 2023, arXiv:2310.03121. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
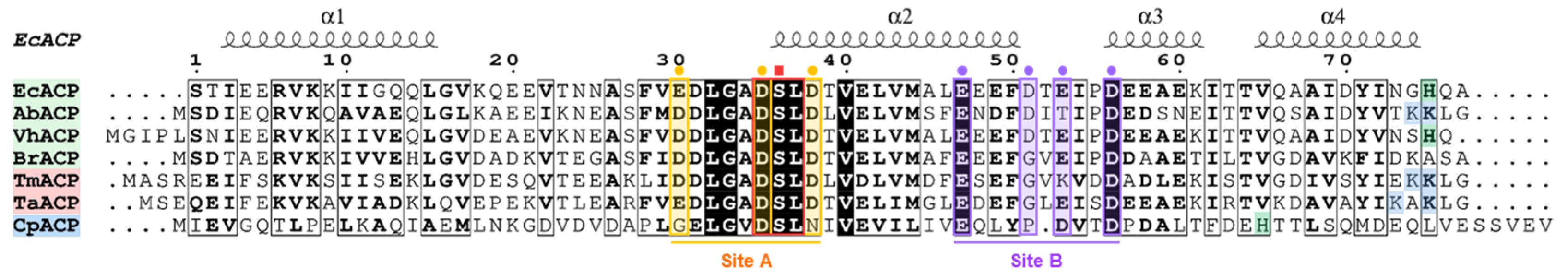

| EcACP | AbACP a | VhACP b | BrACP c | TmACP d | TaACP e | CpACP f | |
|---|---|---|---|---|---|---|---|
| Acidic | 19 | 19 | 22 | 20 | 21 | 21 | 18 |
| Basic | 5 | 6 | 5 | 6 | 9 | 10 | 2 |
| pI g | 3.98 | 3.92 | 3.79 | 3.97 | 4.13 | 4.29 | 3.70 |
| Tm (°C) h | 67.4 | 68.0 | 66.4 | 60.8 | 100.4 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.Y.; Jang, S.; Cho, H.; Jeong, M.-C.; Oh, Y.; Kim, Y. Structural and Dynamic Insights into Acyl Carrier Protein upon Metal Binding and Acylation Revealed by NMR Spectroscopy and MD Simulations. Int. J. Mol. Sci. 2025, 26, 9005. https://doi.org/10.3390/ijms26189005
Lee CY, Jang S, Cho H, Jeong M-C, Oh Y, Kim Y. Structural and Dynamic Insights into Acyl Carrier Protein upon Metal Binding and Acylation Revealed by NMR Spectroscopy and MD Simulations. International Journal of Molecular Sciences. 2025; 26(18):9005. https://doi.org/10.3390/ijms26189005
Chicago/Turabian StyleLee, Chae Yeong, Sungchan Jang, Hyunjoon Cho, Min-Cheol Jeong, Yoojin Oh, and Yangmee Kim. 2025. "Structural and Dynamic Insights into Acyl Carrier Protein upon Metal Binding and Acylation Revealed by NMR Spectroscopy and MD Simulations" International Journal of Molecular Sciences 26, no. 18: 9005. https://doi.org/10.3390/ijms26189005
APA StyleLee, C. Y., Jang, S., Cho, H., Jeong, M.-C., Oh, Y., & Kim, Y. (2025). Structural and Dynamic Insights into Acyl Carrier Protein upon Metal Binding and Acylation Revealed by NMR Spectroscopy and MD Simulations. International Journal of Molecular Sciences, 26(18), 9005. https://doi.org/10.3390/ijms26189005






