Sex-Specific Effects of AQP4 Gene Polymorphisms on Multiple Sclerosis Susceptibility and Response to Multidisciplinary Rehabilitation
Abstract
1. Introduction
2. Results
2.1. AQP4 SNP Genotypic, Allelic, and Haplotypes Distributions in Study Population
2.2. HLA-DRB1*15 Positivity
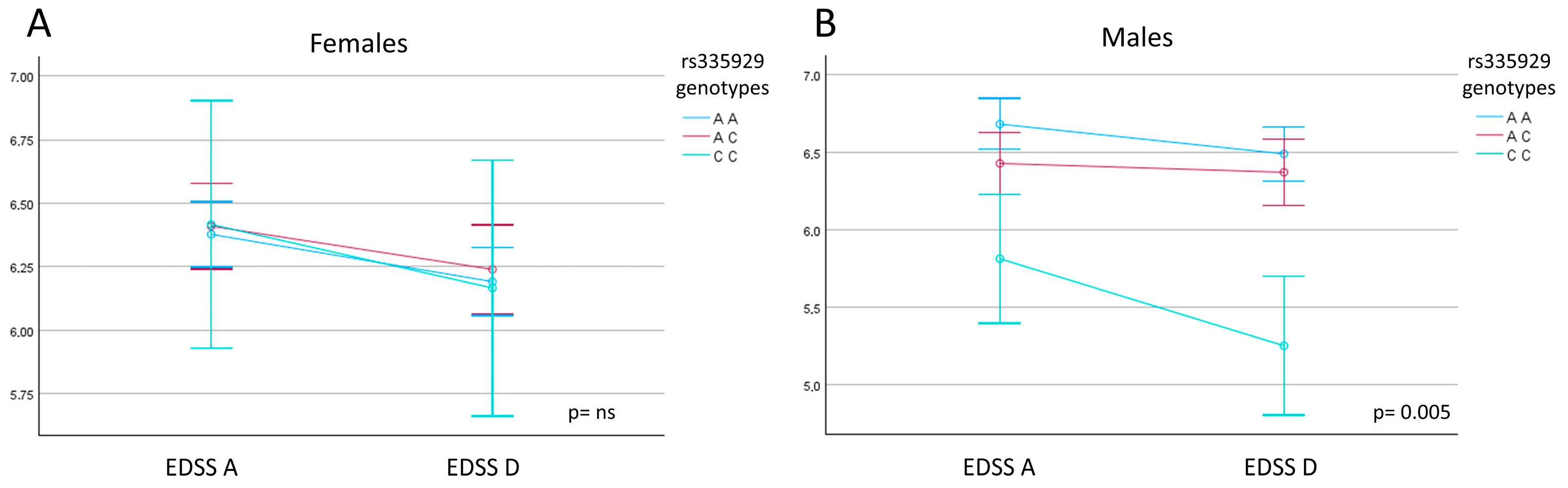
2.3. AQP4 Gene SNPs and Their Impact on Multidisciplinary Rehabilitation Outcomes
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection and DNA Extraction
4.3. AQP4 rs2075575, rs162009, and rs335929 SNP Description and Genotyping
4.4. HLA-DRB1*15 Positivity
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | multiple sclerosis |
| CNS | central nervous system |
| RR | relapsing remitting |
| SP | secondary progressive |
| PP | primary progressive |
| BBB | blood–brain barrier |
| NMO | neuromyelitis optica |
| SNP | single-nucleotide polymorphism |
| EDSS | expanded disability status scale |
| EDSS | A EDSS at admission |
| EDSS | D EDSS at discharge |
| mBI | modified Barthel index |
| mBI A | mBI at admission |
| mBI D | mBI at discharge |
| NRS | Numeric Rating Scale |
| NRS A | NRS at admission |
| NRS D | NRS at discharge |
| IDD | inflammatory demyelinating disease |
| pwMS | people with MS |
| IMR | inpatient multidisciplinary rehabilitation |
| PCR | polymerase chain reaction |
References
- Cañellas, A.R.; Gols, A.R.; Izquierdo, J.R.; Subirana, M.T.; Gairin, X.M. Idiopathic inflammatory-demyelinating diseases of the central nervous system. Neuroradiology 2007, 49, 393–409. [Google Scholar] [CrossRef]
- Faguy, K. Multiple Sclerosis: An Update. Radiol. Technol. 2016, 87, 529–550. [Google Scholar] [PubMed]
- Miller, D.H.; Leary, S.M. Primary-progressive multiple sclerosis. Lancet Neurol. 2007, 6, 903–912. [Google Scholar] [CrossRef]
- Jung, J.S.; Bhat, R.V.; Preston, G.M.; Guggino, W.B.; Baraban, J.M.; Agre, P. Molecular characterization of an aquaporin cDNA from brain: Candidate osmoreceptor and regulator of water balance. Proc. Natl. Acad. Sci. USA 1994, 91, 13052–13056. [Google Scholar] [CrossRef] [PubMed]
- Alghanimy, A.; Work, L.M.; Holmes, W.M. The glymphatic system and multiple sclerosis: An evolving connection. Mult. Scler. Relat. Disord. 2024, 83, 105456. [Google Scholar] [CrossRef]
- Jazaeri, S.Z.; Taghizadeh, G.; Babaei, J.F.; Goudarzi, S.; Saadatmand, P.; Joghataei, M.T.; Khanahmadi, Z. Aquaporin 4 beyond a water channel; participation in motor, sensory, cognitive and psychological performances, a comprehensive review. Physiol. Behav. 2023, 271, 114353. [Google Scholar] [CrossRef] [PubMed]
- Lennon, V.A.; Wingerchuk, D.M.; Kryzer, T.J.; Pittock, S.J.; Lucchinetti, C.F.; Fujihara, K.; Nakashima, I.; Weinshenker, B.G. A serum autoantibody marker of neuromyelitis optica: Distinction from multiple sclerosis. Lancet 2004, 364, 2106–2112. [Google Scholar] [CrossRef]
- Lennon, V.A.; Kryzer, T.J.; Pittock, S.J.; Verkman, A.S.; Hinson, S.R. IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J. Exp. Med. 2005, 202, 473–477. [Google Scholar]
- Jarius, S.; Wildemann, B.; Paul, F. Neuromyelitis optica: Clinical features, immunopathogenesis and treatment. Clin. Exp. Immunol. 2014, 176, 149–164. [Google Scholar] [CrossRef]
- Aoki-Yoshino, K.; Uchihara, T.; Duyckaerts, C.; Nakamura, A.; Hauw, J.J.; Wakayama, Y. Enhanced expression of aquaporin 4 in human brain with inflammatory diseases. Acta Neuropathol. 2005, 110, 281–288. [Google Scholar] [CrossRef]
- Rohr, S.O.; Greiner, T.; Joost, S.; Amor, S.; Valk, P.V.; Schmitz, C.; Kipp, M. Aquaporin-4 Expression during Toxic and Autoimmune Demyelination. Cells 2020, 9, 2187. [Google Scholar] [CrossRef]
- Christiansen, P.; Gideon, P.; Thomsen, C.; Stubgaard, M.; Henriksen, O.; Larsson, H.B. Increased water self-diffusion in chronic plaques and in apparently normal white matter in patients with multiple sclerosis. Acta Neurol. Scand. 1993, 87, 195–199. [Google Scholar] [CrossRef]
- Barkhof, F.; van Walderveen, M. Characterization of tissue damage in multiple sclerosis by nuclear magnetic resonance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 1675–1686. [Google Scholar] [CrossRef]
- Lassmann, H.; Reindl, M.; Rauschka, H.; Berger, J.; Aboul-Enein, F.; Berger, T.; Zurbriggen, A.; Lutterotti, A.; Brück, W.; Weber, J.R.; et al. A new paraclinical CSF marker for hypoxia-like tissue damage in multiple sclerosis lesions. Brain 2003, 126, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef] [PubMed]
- Goris, A.; Vandebergh, M.; McCauley, J.L.; Saarela, J.; Cotsapas, C. Genetics of multiple sclerosis: Lessons from polygenicity. Lancet Neurol. 2022, 21, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Zackowski, K.M.; Freeman, J.; Brichetto, G.; Centonze, D.; Dalgas, U.; DeLuca, J.; Ehde, D.; Elgott, S.; Fanning, V.; Feys, P.; et al. Prioritizing progressive MS rehabilitation research: A call from the International Progressive MS Alliance. Mult. Scler. 2021, 27, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Hata, R.; Toku, K.; Yang, L.; Ma, Y.J.; Maeda, N.; Sakanaka, M.; Tanaka, J. Testosterone up-regulates aquaporin-4 expression in cultured astrocytes. J. Neurosci. Res. 2003, 72, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Sayeed, I.; Baronne, L.M.; Hoffman, S.W.; Guennoun, R.; Stein, D.G. Progesterone administration modulates AQP4 expression and edema after traumatic brain injury in male rats. Exp. Neurol. 2006, 198, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Beer, S.; Elmenhorst, D.; Bischof, G.N.; Ramirez, A.; Bauer, A.; Drzezga, A. Alzheimer’s Disease Neuroimaging Initiative. Explainable artificial intelligence identifies an AQP4 polymorphism-based risk score associated with brain amyloid burden. Neurobiol. Aging 2024, 143, 19–29. [Google Scholar] [CrossRef]
- Ogasawara, M.; Meguro, A.; Sakai, T.; Mizuki, N.; Takahashi, T.; Fujihara, K.; Tsuneoka, H.; Shikishima, K. Genetic analysis of the aquaporin-4 gene for anti-AQP4 antibody-positive neuromyelitis optica in a Japanese population. Jpn. J. Ophthalmol. 2016, 60, 198–205. [Google Scholar] [CrossRef]
- Sun, X.; Tian, Q.; Yang, Z.; Liu, Y.; Li, C.; Hou, B.; Xie, A. Association of AQP4 single nucleotide polymorphisms (rs335929 and rs2075575) with Parkinson’s disease: A case-control study. Neurosci. Lett. 2023, 797, 137062. [Google Scholar] [CrossRef]
- Eidahl, J.M.L.; Stray-Pedersen, A.; Rognum, T.O.; Opdal, S.H. Aquaporin 4 expression in the hippocampus in sudden infant death syndrome and sudden unexplained death in childhood. J. Chem. Neuroanat. 2021, 115, 101962. [Google Scholar] [CrossRef]
- Català-Senent, J.F.; Andreu, Z.; Hidalgo, M.R.; Soler-Sáez, I.; Roig, F.J.; Yanguas-Casás, N.; Neva-Alejo, A.; López-Cerdán, A.; de la Iglesia-Vayá, M.; Stranger, B.E.; et al. A deep transcriptome meta-analysis reveals sex differences in multiple sclerosis. Neurobiol. Dis. 2023, 181, 106113. [Google Scholar] [CrossRef] [PubMed]
- Scharfman, H.E.; Binder, D.K. Aquaporin-4 water channels and synaptic plasticity in the hippocampus. Neurochem. Int. 2013, 63, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Connallon, T.; Czuppon, P.; Olito, C.; Goedert, D.; Kokko, H.; Nava-Bolaños, A.; Nilén, S.; Svensson, E.I.; Zwoinska, M.; Dutoit, L.; et al. Predicting the prevalence of genetic trade-offs among adaptive substitutions. Evolution 2025, 79, 1243–1255. [Google Scholar] [CrossRef]
- van der Knaap, M.S.; Min, R. Multiple sclerosis: An immune attack on astrocyte-mediated ion and water homeostasis. Nat. Rev. Neurol. 2025, 21, 283–289. [Google Scholar] [CrossRef]
- He, X.F.; Liu, D.X.; Zhang, Q.; Liang, F.Y.; Dai, G.Y.; Zeng, J.S.; Pei, Z.; Xu, G.Q.; Lan, Y. Voluntary Exercise Promotes Glymphatic Clearance of Amyloid Beta and Reduces the Activation of Astrocytes and Microglia in Aged Mice. Front. Mol. Neurosci. 2017, 10, 144. [Google Scholar] [CrossRef]
- Zhu, A.; Lin, Y.; Hu, X.; Lin, Z.; Lin, Y.; Xie, Q.; Ni, S.; Cheng, H.; Lu, Q.; Lai, S.; et al. Treadmill exercise decreases cerebral edema in rats with local cerebral infarction by modulating AQP4 polar expression through the caveolin-1/TRPV4 signaling pathway. Brain Res. Bull. 2022, 188, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Groppo, E.; Signori, A.; Sormani, M.P.; Grosso, C.; Mantia, L.; Cattaneo, D.; Rovaris, M. Predictors of hospital-based multidisciplinary rehabilitation effects in persons with multiple sclerosis: A large-scale, single-centre study. Mult. Scler. J. Exp. Transl. Clin. 2019, 5, 2055217319843673. [Google Scholar] [CrossRef] [PubMed]
- Alsubiheen, A.M.; Choi, W.; Yu, W.; Lee, H. The Effect of Task-Oriented Activities Training on Upper-Limb Function, Daily Activities, and Quality of Life in Chronic Stroke Patients: A Randomized Controlled Trial. Int. J. Environ. Res. Public Health 2022, 19, 14125. [Google Scholar] [CrossRef]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef]
- de Bakker, P.I.; McVean, G.; Sabeti, P.C.; Miretti, M.M.; Green, T.; Marchini, J.; Ke, X.; Monsuur, A.J.; Whittaker, P.; Delgado, M.; et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006, 38, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, Z.; Chen, J.; Song, Z.; Zhou, Z.; Shi, Y. SHEsisPlus, a toolset for genetic studies on polyploid species. Sci. Rep. 2016, 6, 24095. [Google Scholar] [CrossRef] [PubMed]

| rs2075575 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Males | Females | |||||||||||
| pwMS | HC | p Value | OR (95% CI) | pwMS | HC | p Value | OR (95% CI) | |||||
| Genotype | N | (%) | N | (%) | N | (%) | N | (%) | ||||
| AA | 26 | (27.4) | 51 | (26.0) | 0.803 | 1.07 (0.61–1.86) | 29 | (20.4) | 65 | (24.5) | 0.353 | 0.79 (0.48–1.29) |
| GA | 52 | (54.7) | 88 | (44.9) | 0.118 | 1.48 (0.91–2.44) | 73 | (51.4) | 138 | (52.1) | 0.898 | 0.97 (0.65–1.47) |
| GG | 17 | (17.9) | 57 | (29.1) | 0.078 | 0.53 (0.28–0.97) | 40 | (28.2) | 62 | (23.4) | 0.293 | 1.28 (0.80–2.04) |
| Total | 95 | 196 | 0.106 | 142 | 265 | 0.467 | ||||||
| Allele | ||||||||||||
| A | 104 | (54.7) | 190 | (48.5) | 131 | (46.1) | 268 | (50.6) | ||||
| G | 86 | (45.3) | 202 | (51.5) | 153 | (53.9) | 262 | (49.4) | ||||
| Total | 190 | 392 | 0.158 | 1.29 (0.91–1.82) | 284 | 530 | 0.228 | 0.84 (0.63–1.12) | ||||
| rs162009 | ||||||||||||
| Males | Females | |||||||||||
| pwMS | HC | p value | OR (95% CI) | pwMS | HC | p value | OR (95% CI) | |||||
| Genotype | N | (%) | N | (%) | N | (%) | N | (%) | ||||
| AA | 10 | (10.5) | 21 | (10.7) | 0.976 | 0.98 (0.43–2.16) | 14 | (9.9) | 26 | (9.8) | 0.977 | 1.01 (0.50–1.98) |
| GA | 45 | (47.4) | 90 | (45.9) | 0.817 | 1.06 (0.65–1.74) | 64 | (45.1) | 113 | (42.6) | 0.639 | 1.10 (0.73–1.67) |
| GG | 40 | (42.1) | 85 | (43.4) | 0.841 | 0.95 (0.58–1.56) | 64 | (45.1) | 126 | (47.5) | 0.635 | 0.91 (0.60–1.34) |
| Total | 95 | 196 | 0.973 | 142 | 265 | 0.884 | ||||||
| Allele | ||||||||||||
| A | 65 | (34.2) | 132 | (33.7) | 92 | (32.4) | 165 | (31.1) | ||||
| G | 125 | (65.8) | 260 | (66.3) | 192 | (67.6) | 365 | (68.9) | ||||
| Total | 190 | 392 | 0.896 | 1.02 (0.71–1.48) | 284 | 530 | 0.711 | 1.06 (0.78–1.44) | ||||
| rs335929 | ||||||||||||
| Males | Females | |||||||||||
| pwMS | HC | p value | OR (95% CI) | pwMS | HC | p value | OR (95% CI) | |||||
| Genotype | N | (%) | N | (%) | N | (%) | N | (%) | ||||
| AA | 52 | (54.7) | 121 | (61.7) | 0.258 | 0.75 (0.46–1.24) | 86 | (60.6) | 163 | (61.5) | 0.851 | 0.96 (0.63–1.46) |
| CA | 35 | (36.8) | 66 | (33.7) | 0.595 | 1.15 (0.68–1.92) | 50 | (35.2) | 92 | (34.7) | 0.919 | 1.02 (0.66–1.57) |
| CC | 8 | (8.4) | 9 | (4.6) | 0.211 | 1.91 (0.68–5.24) | 6 | (4.2) | 10 | (3.8) | 0.813 | 1.13 (0.37–3.17) |
| Total | 95 | 196 | 0.314 | 142 | 265 | 0.967 | ||||||
| Allele | ||||||||||||
| A | 139 | (73.2) | 308 | (78.6) | 222 | (78.2) | 418 | (78.9) | ||||
| C | 51 | (26.8) | 84 | (21.4) | 62 | (21.8) | 112 | (21.1) | ||||
| Total | 190 | 392 | 0.151 | 0.74 (0.50–1.12) | 284 | 530 | 0.814 | 0.96 (0.68–1.37) | ||||
| rs2075575 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Males | Females | |||||||||||
| PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | |
| Genotype | N (%) | N (%) | N (%) | p Value, OR (95% CI) | p Value, OR (95% CI) | p Value, OR (95% CI) | N (%) | N (%) | N (%) | p Value, OR (95% CI) | p Value, OR (95% CI) | p Value, OR (95% CI) |
| AA | 5 (29.4) | 21 (26.9) | 51 (26.0) | 0.823 | 0.746 | 0.872 | 3 (17.6) | 26 (20.8) | 65 (24.5) | 0.805 | 0.556 | 0.422 |
| GA | 7 (41.2) | 45 (57.7) | 88 (44.9) | 0.232 | 0.780 | 0.058 | 10 (58.8) | 63 (50.4) | 138 (52.1) | 0.530 | 0.603 | 0.759 |
| GG | 5 (29.4) | 12 (15.4) | 57 (29.1) | 0.292 | 0.954 | 0.033 *, 0.44 (0.22–0.87) | 4 (23.5) | 36 (28.8) | 62 (23.4) | 0.682 | 0.957 | 0.255 |
| Total | 17 | 78 | 196 | 0.323 | 0.943 | 0.048 | 17 | 125 | 265 | 0.808 | 0.798 | 0.463 |
| Allele | ||||||||||||
| A | 17 (50.0) | 87 (55.8) | 190 (48.5) | 16 (47.1) | 115 (46.0) | 268 (50.6) | ||||||
| G | 17 (50.0) | 69 (44.2) | 202 (51.5) | 18 (52.9) | 135 (54.0) | 262 (49.4) | ||||||
| Total | 34 | 156 | 392 | 0.546 | 0.865 | 0.125 | 34 | 250 | 530 | 0.907 | 0.697 | 0.235 |
| rs162009 | ||||||||||||
| Males | Females | |||||||||||
| PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | |
| Genotype | N (%) | N (%) | N (%) | p value, OR (95% CI) | p value, OR (95% CI) | p value, OR (95% CI) | N (%) | N (%) | N (%) | p value, OR (95% CI) | p value, OR (95% CI) | p value, OR (95% CI) |
| AA | 2 (11.8) | 8 (10.3) | 21 (1.7) | 0.822 | 0.844 | 0.932 | 1 (5.9) | 13 (10.4) | 26 (9.8) | 0.630 | 0.672 | 0.846 |
| GA | 11 (64.7) | 34 (43.6) | 90 (45.9) | 0.126 | 0.149 | 0.730 | 7 (41.2) | 57 (45.6) | 113 (42.6) | 0.744 | 0.917 | 0.584 |
| GG | 4 (23.5) | 36 (46.2) | 85 (43.4) | 0.093 | 0.117 | 0.804 | 9 (52.9) | 55 (44.0) | 126 (47.5) | 0.500 | 0.675 | 0.515 |
| Total | 17 | 78 | 196 | 0.219 | 0.265 | 0.916 | 17 | 125 | 265 | 0.726 | 0.834 | 0.806 |
| Allele | ||||||||||||
| A | 15 (44.1) | 50 (32.1) | 132 (33.7) | 9 (26.5) | 83 (33.2) | 165 (31.1) | ||||||
| G | 19 (55.9) | 106 (67.9) | 260 (66.3) | 25 (73.5) | 167 (66.8) | 365 (68.9) | ||||||
| Total | 34 | 156 | 392 | 0.190 | 0.229 | 0.721 | 34 | 250 | 530 | 0.445 | 0.586 | 0.563 |
| rs335929 | ||||||||||||
| Males | Females | |||||||||||
| PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | PP-MS | RR-MS (RR-MS + SP-MS) | HCs | PP-MS vs. RR-MS | PP-MS vs. HCs | RR-MS vs. HCs | |
| Genotype | N (%) | N (%) | N (%) | p value, OR (95% CI) | p value, OR (95% CI) | p value, OR (95% CI) | N (%) | N (%) | N (%) | p value, OR (95% CI) | p value, OR (95% CI) | p value, OR (95% CI) |
| AA | 7 (41.2) | 45 (57.7) | 121 (61.7) | 0.232 | 0.110 | 0.539 | 12 (70.6) | 74 (59.2) | 163 (61.5) | 0.386 | 0.475 | 0.663 |
| AC | 8 (47.1) | 27 (34.6) | 66 (33.7) | 0.352 | 0.283 | 0.878 | 5 (29.4) | 45 (36.0) | 92 (34.7) | 0.618 | 0.681 | 0.803 |
| CC | 2 (11.8) | 6 (7.7) | 9 (4.6) | 0.588 | 0.262 | 0.327 | 0 (0.0) | 6 (4.8) | 10 (3.8) | 0.456 | 0.531 | 0.631 |
| Total | 17 | 78 | 196 | 0.459 | 0.177 | 0.564 | 17 | 125 | 265 | 0.517 | 0.611 | 0.847 |
| Allele | ||||||||||||
| A | 22 (64.7) | 117 (75.0) | 308 (78.6) | 29 (85.3) | 193 (77.2) | 418 (78.9) | ||||||
| C | 12 (35.3) | 39 (25.0) | 84 (21.4) | 5 (14.7) | 57 (22.8) | 112 (21.1) | ||||||
| Total | 34 | 156 | 392 | 0.233 | 0.078 | 0.368 | 34 | 250 | 530 | 0.293 | 0.386 | 0.596 |
| Haplotype | pwMS (M + F) (freq) | HCs (M + F) (freq) | Chi2 | Pearson’s p | OR (95% CI) |
|---|---|---|---|---|---|
| GGA | 118 (0.25) | 222 (0.24) | 0.113 | 0.74 | 1.05 (0.81–1.35) |
| GAA | 51 (0.11) | 106 (0.11) | 0.170 | 0.68 | 0.93 (0.65–1.32) |
| AGA | 192 (0.41) | 389 (0.42) | 0.365 | 0.55 | 0.93 (0.74–1.17) |
| AAC | 39 (0.08) | 49 (0.05) | 4.498 | 0.03 | 1.60 (1.03–2.47) |
| GAC | 67 (0.14) | 133 (0.14) | 0.021 | 0.88 | 0.98 (0.71–1.34) |
| Global | 4.64 | 0.33 | |||
| Haplotype | pwMS (F) (freq) | HCs (F) (freq) | Chi2 | Pearson’s p | OR (95% CI) |
| GGA | 74 (0.26) | 126 (0.24) | 0.52 | 0.47 | 1.13 (0.81–1.57) |
| GAA | 34 (0.12) | 58 (0.11) | 0.20 | 0.66 | 1.11 (0.71–1.74) |
| AGA | 114 (0.40) | 228 (0.43) | 0.63 | 0.43 | 0.89 (0.66–1.19) |
| AAC | 16 (0.06) | 30 (0.06) | 2.5 × 10−4 | 0.99 | 1.00 (0.53–1.86) |
| GAC | 42 (0.15) | 71 (0.13) | 0.30 | 0.58 | 1.12 (0.74–1.69) |
| Global | 1.12 | 0.89 | |||
| Haplotype | pwMS (M) (freq) | HCs (M) (freq) | Chi2 | Pearson’s p | OR (95% CI) |
| GGA | 44 (0.23) | 92 (0.23) | 0.006 | 0.93 | 0.98 (0.65–1.48) |
| GAA | 17 (0.09) | 48 (0.12) | 1.40 | 0.24 | 0.70 (0.39–1.26) |
| AGA | 78 (0.41) | 165 (0.42) | 0.06 | 0.81 | 0.96 (0.67–1.36) |
| AAC | 23 (0.12) | 19 (0.05) | 10.07 | 0.001 | 2.70 (1.43–5.10) |
| GAC | 25 (0.13) | 62 (0.16) | 0.71 | 0.40 | 0.81 (0.49–1.33) |
| Global | 11.23 | 0.02 |
| Population Characteristics | |||
|---|---|---|---|
| pwMS | HCs | p | |
| N | 237 | 461 | |
| Males/females, (%/%) | 95/142, (40.1/59.9) | 196/265, (42.5/57.5) | 0.54 |
| Age at enrollment (years), mean ± SD | 50.84 ± 12.13 | 69.95 ± 11.77 | <0.001 |
| MS type: RR-MS, PP-MS, SP-MS n (%) | 92 (38.8), 34 (14.3), 111 (46.8) | n.a. | |
| Age at onset (years), mean ± SD | 29.23 ± 11.42 | n.a. | |
| Disease duration (years), median, IQR | 20.0, 14.0 | n.a. | |
| Duration of admission, (days) median, IQR | 35.0, 13.0 | n.a. | |
| N of interventions, mean ± SD | 3.63 ± 1 | n.a | |
| EDSS I, median, IQR | 6.5, 1.5 | n.a. | |
| EDSS D, median, IQR | 6.5, 1.0 | n.a. | |
| BI I, median, IQR | 65.0, 27.25 | n.a. | |
| BI D, median, IQR | 75.0, 26.25 | n.a. | |
| VNS I, median, IQR | 5.0, 4.0 | n.a. | |
| VNS D, median, IQR | 3.0, 4.0 | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agliardi, C.; Guerini, F.R.; Zanzottera, M.; Bolognesi, E.; Caputo, D.; Groppo, E.; Rovaris, M.; Clerici, M. Sex-Specific Effects of AQP4 Gene Polymorphisms on Multiple Sclerosis Susceptibility and Response to Multidisciplinary Rehabilitation. Int. J. Mol. Sci. 2025, 26, 8915. https://doi.org/10.3390/ijms26188915
Agliardi C, Guerini FR, Zanzottera M, Bolognesi E, Caputo D, Groppo E, Rovaris M, Clerici M. Sex-Specific Effects of AQP4 Gene Polymorphisms on Multiple Sclerosis Susceptibility and Response to Multidisciplinary Rehabilitation. International Journal of Molecular Sciences. 2025; 26(18):8915. https://doi.org/10.3390/ijms26188915
Chicago/Turabian StyleAgliardi, Cristina, Franca Rosa Guerini, Milena Zanzottera, Elisabetta Bolognesi, Domenico Caputo, Elisabetta Groppo, Marco Rovaris, and Mario Clerici. 2025. "Sex-Specific Effects of AQP4 Gene Polymorphisms on Multiple Sclerosis Susceptibility and Response to Multidisciplinary Rehabilitation" International Journal of Molecular Sciences 26, no. 18: 8915. https://doi.org/10.3390/ijms26188915
APA StyleAgliardi, C., Guerini, F. R., Zanzottera, M., Bolognesi, E., Caputo, D., Groppo, E., Rovaris, M., & Clerici, M. (2025). Sex-Specific Effects of AQP4 Gene Polymorphisms on Multiple Sclerosis Susceptibility and Response to Multidisciplinary Rehabilitation. International Journal of Molecular Sciences, 26(18), 8915. https://doi.org/10.3390/ijms26188915








