CD44 as a Central Integrator of Inflammation and Fibrosis: From Molecular Signaling to Environmental Modulation
Abstract
1. Introduction
2. Molecular Characteristics of the CD44 Protein: Structure, Isoforms, Biological Functions
2.1. Biological Functions
2.2. Subcellular Localization and Expression Profile of CD44 Isoforms
2.3. Protein Structure and Isoforms
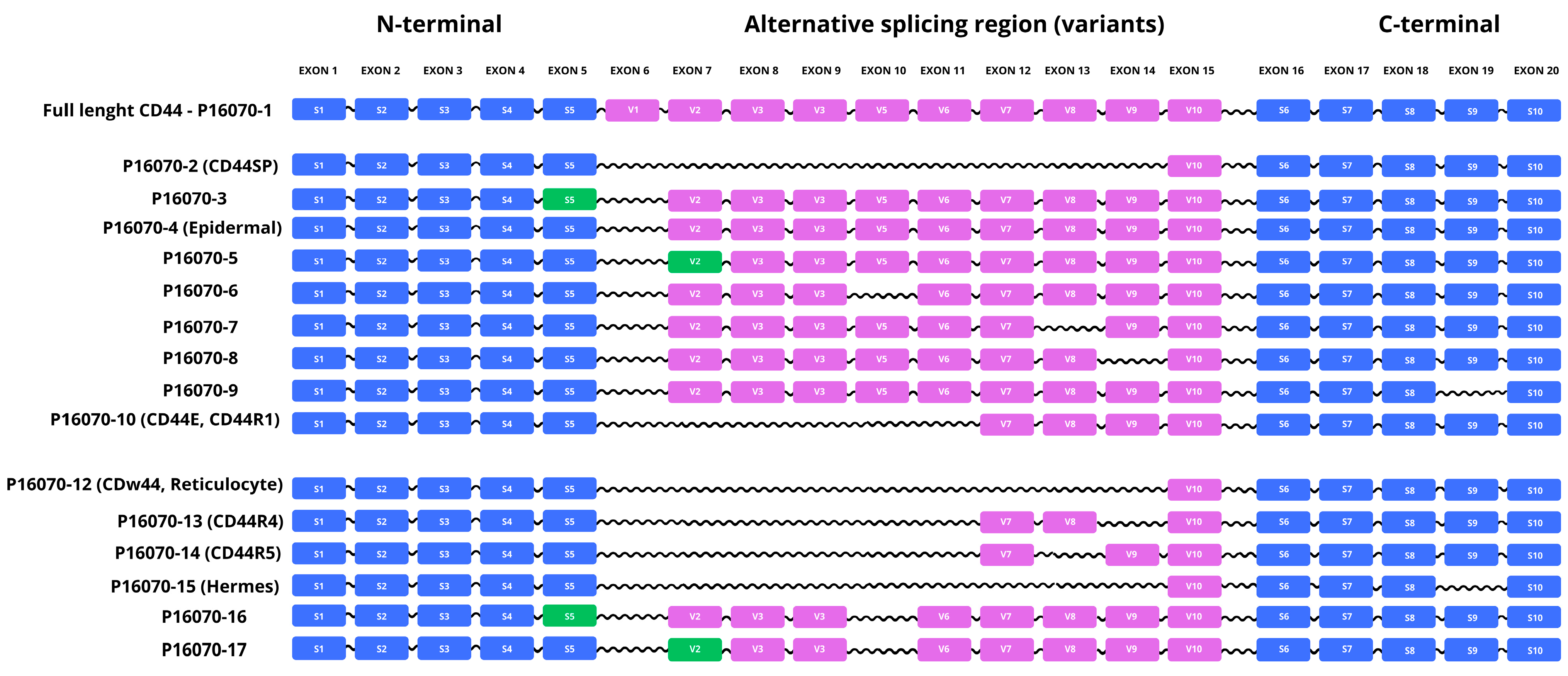
2.3.1. Alternative Splicing of the CD44 Gene and Characterization of Selected Isoforms
| Isoform | Length (aa) | Mass (Da) | pI | % Hydrophilic | % Hydrophobic | Description |
|---|---|---|---|---|---|---|
| P16070-1 | 742 | 81,538 | 4.98 | 57.95% | 42.05% | Canonical isoform. |
| P16070-2 (CD44SP) | 29 | 3327 | 9.37 | 33.33% | 66.67% | Lacks exons 6–14. Differences: 23–29: DLNITCR → GVGRRKS; 30–742: deleted. |
| P16070-3 | 711 | 77,983 | 5.5 | 58.23% | 41.77% | Alternative splice donor/acceptor in exon 5. Differences: 192 G→A; 193–223 deleted. |
| P16070-4 (Epidermal) | 699 | 76,612 | 4.94 | 68.24% | 31.76% | Lacks exon 6. Differences: 223 T→S; 224–266 deleted. |
| P16070-5 | 734 | 80,790 | 4.98 | 67.71% | 32.29% | Alternative splice donor/acceptor in exon 7. Differences: 266–273 deleted. |
| P16070-6 | 699 | 76,705 | 4.95 | 67.38% | 32.62% | Lacks exon 10. Differences: 385 I→T; 386–428 deleted. |
| P16070-7 | 713 | 78,446 | 5.05 | 60.17% | 39.83% | Lacks exon 13. Differences: 506 Q→R; 507–535 deleted. |
| P16070-8 | 674 | 74,388 | 4.88 | 60.83% | 39.17% | Lacks exon 14. Differences: 536 N→R; 537–604 deleted. |
| P16070-9 | 675 | 74,196 | 4.91 | 61.19% | 38.81% | Lacks exon 19. Differences: 675 R→S; 676–742 deleted. |
| P16070-10 (CD44E, CD44R1, Epithelial, Keratinocyte) | 493 | 53,411 | 5.02 | 66.13% | 33.67% | Lacks exons 6–11. Differences: 223 T→N; 224–472 deleted. |
| P16070-11 (CD44R2) | 429 | 46,565 | 5.24 | 56.88% | 43.12% | Lacks 223–535 region. |
| P16070-12 (CDw44, Reticulocyte) | 361 | 39,416 | 5.04 | 50.97% | 49.03% | Lacks exons 6–14. Differences: 223 T→R; 224–604 deleted. |
| P16070-13 (CD44R4) | 425 | 46,261 | 4.86 | 59.06% | 40.94% | Lacks exons 6–11 and 14. Differences: 223 T→N; 224–472 and 537–604 deleted; 536 N→R. |
| P16070-14 (CD44R5) | 396 | 43,169 | 4.99 | 59.60% | 40.40% | Lacks exons 6–11, 13 and 14. Differences: 223 T→N; 224–472, 507–535 and 537–604 deleted; 506 Q→R; 536 N→R. |
| P16070-15 (Hermes) | 294 | 32,075 | 4.86 | 54.76% | 45.24% | Lacks exons 6–14 and 19. Differences: 223 T→R; 224–604 and 676–742 deleted; 675 R→S. |
| P16070-16 | 668 | 73,150 | 5.03 | 60.63% | 39.37% | Alternative splice donor/acceptor on exon 5; lacks exon 10. Differences: 192 G→A; 193–223 and 386–428 deleted; 385 I→T. |
| P16070-17 | 691 | 75,957 | 4.95 | 67.15% | 32.85% | Alternative splice donor/acceptor on exon 7; lacks exon 10. Differences: 266–273 and 386–428 deleted; 385 I→T. |
| P16070-18 | 340 | 37,278 | 5.15 | 61.47% | 38.53% | Differences: 223 T→R; 224–604 and 605–625 deleted. |
| P16070-19 (CD44RC) | 139 | 15,635 | 7.73 | 61.87% | 38.13% | Soluble isoform; enhanced HA binding. Differences: 78–139 replaced with SLHCSQQSKK...QGVVRNSRPVYDS; 140–742 deleted. |
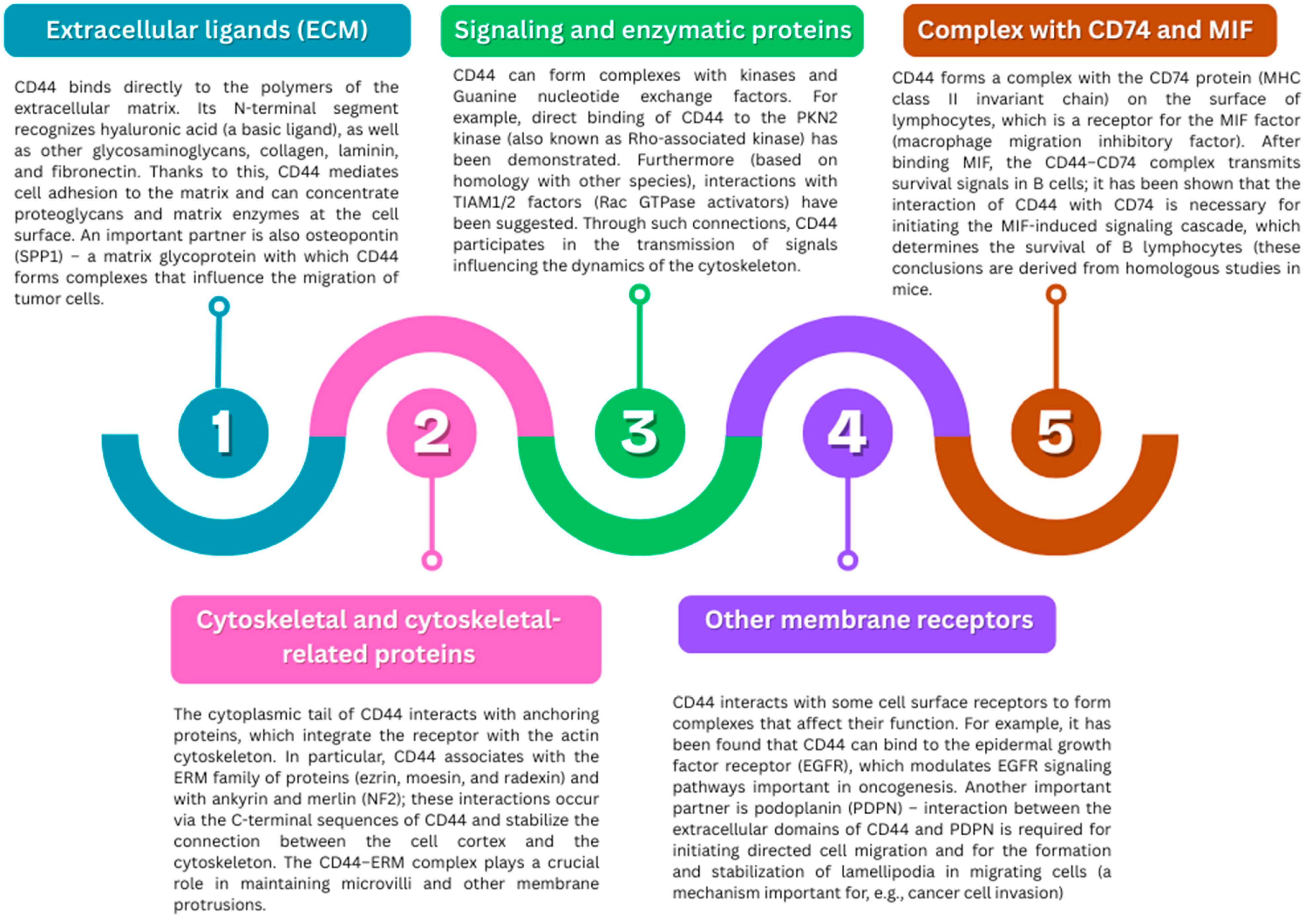
2.3.2. Protein Interactions and Complexes
2.3.3. Post-Translational Modifications (PTMs)
3. The Role of CD44 in Pathologies
| Disease/Cancer Type | Role of CD44 | CD44 Isoforms | Mechanism of Action | Clinical Significance |
|---|---|---|---|---|
| Breast cancer | CSC marker, EMT, therapy resistance | CD44s, CD44^high | STAT3 and PI3K/Akt activation; splicing toward CD44s | Poor prognosis, tumor recurrence |
| Pancreatic cancer | Invasion, metastasis | CD44v6 | MMP-9 localization, interactions with HA | Promotes metastasis, reduces overall survival (OS) |
| Lung cancer | Proliferation, chemoresistance | CD44v | MET/VEGFR2, PI3K/Akt signaling | Accelerated tumor growth |
| Rheumatoid arthritis (RA) | Leukocyte adhesion, fibroblast activation | CD44v (various) | HA interactions, pannus formation | CD44 blockade reduces inflammation |
| Lupus nephritis (LN) | Inflammation, renal fibrosis | CD44s, sCD44 | CD4+, CD19+ recruitment, fibroblast activation | Disease activity biomarker, therapeutic target |
| Crohn’s disease | IL-6 production, Treg deficiency | CD44v7 | Interaction with osteopontin | CD44v7 blockade protects against colitis |
3.1. CD44 as a Regulator of Tissue Fibrosis and Myofibroblast Differentiation
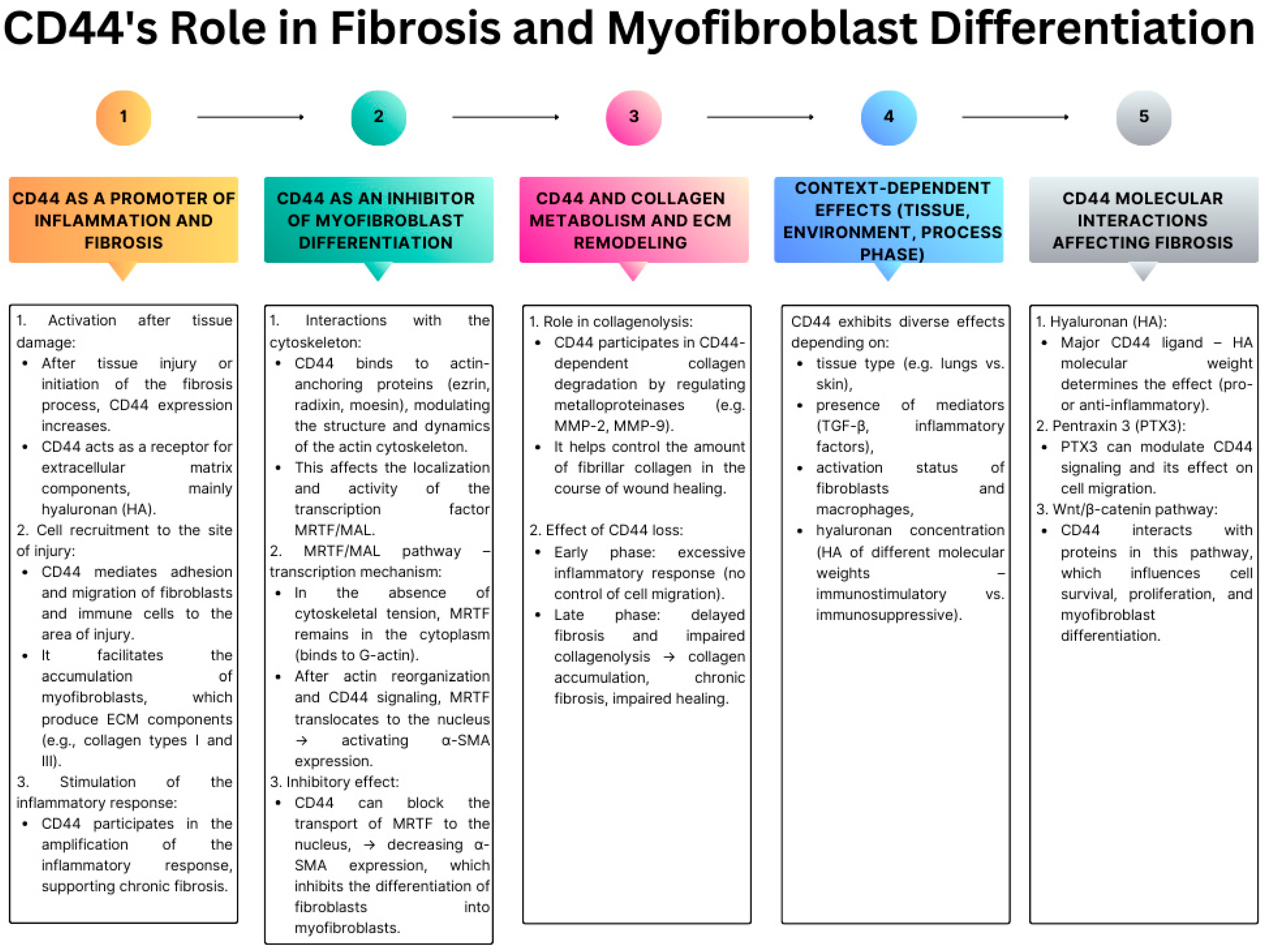
3.2. In Vitro and In Vivo Experimental Evidence for the Role of CD44 in Fibrosis (Lung, Skin, Heart, Liver)
3.3. CD44 and the Invasive Phenotype of Myofibroblasts—Analogies to EMT in Cancer
4. Impact of Environmental Pollutants on CD44 in Connective Tissue
4.1. Heavy Metals
4.2. Particulate Matter (PM)
4.3. Endocrine-Disrupting Compounds (EDCs)
4.4. Microplastics
5. Limitations and Future Research Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CD44 | Cluster of Differentiation 44 |
| HA | Hyaluronic Acid |
| TGF-β | Transforming Growth Factor Beta |
| EGF | Epidermal Growth Factor |
| VEGF | Vascular Endothelial Growth Factor |
| PI3K | Phosphoinositide 3-Kinase |
| AKT | AKT Serine/Threonine Kinase |
| MAPK | Mitogen-Activated Protein Kinase |
| RhoA | Ras Homolog Family Member A |
| ROCK | Rho-associated Protein Kinase |
| ERM | Ezrin–Radixin–Moesin |
| EMT | Epithelial–Mesenchymal Transition |
| CSC | Cancer Stem Cell |
| ECM | Extracellular Matrix |
| MMP | Matrix Metalloproteinase |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| NF-κB | Nuclear Factor Kappa-light-chain-enhancer of Activated B Cells |
| ALT | Alanine Aminotransferase |
| SSc | Systemic Sclerosis |
| IPF | Idiopathic Pulmonary Fibrosis |
| RZS | Rheumatoid Arthritis (RA) |
| LN | Lupus Nephritis |
| EDCs | Endocrine Disrupting Chemicals |
| BPA | Bisphenol A |
| DEHP | Di(2-ethylhexyl) Phthalate |
| ERRγ | Estrogen-Related Receptor Gamma |
| PM2.5 | Particulate Matter ≤ 2.5 µm |
| 4-MU | 4-Methylumbelliferone |
| OS | Overall Survival |
| KO | Knockout |
| sCD44 | Soluble CD44 |
| DAMP | Damage-Associated Molecular Pattern |
| PCB | Polychlorinated Biphenyls |
References
- Kamrani, P.; Marston, G.; Arbor, T.C.; Jan, A. Anatomy, Connective Tissue. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Di, X.; Gao, X.; Peng, L.; Ai, J.; Jin, X.; Qi, S.; Li, H.; Wang, K.; Luo, D. Cellular Mechanotransduction in Health and Diseases: From Molecular Mechanism to Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 282. [Google Scholar] [CrossRef]
- Boraldi, F.; Lofaro, F.D.; Bonacorsi, S.; Mazzilli, A.; Garcia-Fernandez, M.; Quaglino, D. The Role of Fibroblasts in Skin Homeostasis and Repair. Biomedicines 2024, 12, 1586. [Google Scholar] [CrossRef]
- Chaudhry, G.-S.; Akim, A.; Naveed Zafar, M.; Safdar, N.; Sung, Y.Y.; Muhammad, T.S.T. Understanding Hyaluronan Receptor (CD44) Interaction, HA-CD44 Activated Potential Targets in Cancer Therapeutics. Adv. Pharm. Bull. 2021, 11, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Thorne, R.F.; Legg, J.W.; Isacke, C.M. The Role of the CD44 Transmembrane and Cytoplasmic Domains in Co-Ordinating Adhesive and Signalling Events. J. Cell Sci. 2004, 117, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The Biology and Role of CD44 in Cancer Progression: Therapeutic Implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Martegani, M.P.; Del Prete, F.; Gasbarri, A.; Natali, P.G.; Bartolazzi, A. Structural Variability of CD44v Molecules and Reliability of Immunodetection of CD44 Isoforms Using mAbs Specific for CD44 Variant Exon Products. Am. J. Pathol. 1999, 154, 291–300. [Google Scholar] [CrossRef][Green Version]
- Maltseva, D.; Tonevitsky, A. RNA-Binding Proteins Regulating the CD44 Alternative Splicing. Front. Mol. Biosci. 2023, 10, 1326148. [Google Scholar] [CrossRef]
- Gaiteiro, C.; Soares, J.; Relvas-Santos, M.; Peixoto, A.; Ferreira, D.; Brandão, A.; Fernandes, E.; Azevedo, R.; Paulo, P.; Palmeira, C.; et al. Glycoproteogenomics Characterizes the CD44 Splicing Code Driving Bladder Cancer Invasion. Theranostics 2021, 12, 3150–3177. [Google Scholar] [CrossRef]
- Bajorath, J. Molecular Organization, Structural Features, and Ligand Binding Characteristics of CD44, a Highly Variable Cell Surface Glycoprotein with Multiple Functions. Proteins 2000, 39, 103–111. [Google Scholar] [CrossRef]
- Tsuneki, M.; Madri, J.A. CD44 Influences Fibroblast Behaviors Via Modulation of Cell-Cell and Cell-Matrix Interactions, Affecting Survivin and Hippo Pathways. J. Cell. Physiol. 2016, 231, 731–743. [Google Scholar] [CrossRef]
- Jordan, A.R.; Racine, R.R.; Hennig, M.J.P.; Lokeshwar, V.B. The Role of CD44 in Disease Pathophysiology and Targeted Treatment. Front. Immunol. 2015, 6, 182. [Google Scholar] [CrossRef]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a Tumor Biomarker and Therapeutic Target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Herishanu, Y.; Gibellini, F.; Njuguna, N.; Hazan-Halevy, I.; Keyvanfar, K.; Lee, E.; Wilson, W.; Wiestner, A. CD44 Signaling via PI3K/AKT and MAPK/ERK Pathways Protects CLL Cells from Spontaneous and Drug Induced Apoptosis through MCL-1. Leuk. Lymphoma 2011, 52, 1758–1769. [Google Scholar] [CrossRef]
- Herishanu, Y.; Gibellini, F.; Njuguna, N.; Keyvanfar, K.; Wiestner, A. CD44 Signaling Via PI3K/AKT and MAPK/ERK Pathways Protects CLL Cells from Spontaneous and Drug Induced Apoptosis. Blood 2008, 112, 541. [Google Scholar] [CrossRef]
- Robbins, E.W.; Travanty, E.A.; Yang, K.; Iczkowski, K.A. MAP Kinase Pathways and Calcitonin Influence CD44 Alternate Isoform Expression in Prostate Cancer Cells. BMC Cancer 2008, 8, 260. [Google Scholar] [CrossRef]
- Bai, R.-J.; Liu, D.; Li, Y.-S.; Tian, J.; Yu, D.-J.; Li, H.-Z.; Zhang, F.-J. OPN Inhibits Autophagy through CD44, Integrin and the MAPK Pathway in Osteoarthritic Chondrocytes. Front. Endocrinol. 2022, 13, 919366. [Google Scholar] [CrossRef] [PubMed]
- Ohata, H.; Ishiguro, T.; Aihara, Y.; Sato, A.; Sakai, H.; Sekine, S.; Taniguchi, H.; Akasu, T.; Fujita, S.; Nakagama, H.; et al. Induction of the Stem-like Cell Regulator CD44 by Rho Kinase Inhibition Contributes to the Maintenance of Colon Cancer-Initiating Cells. Cancer Res. 2012, 72, 5101–5110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, H.; Ge, X.; Chen, Q.; Yuan, D.; Chen, Q.; Leng, W.; Chen, L.; Tang, Q.; Bi, F. CD44 Acts through RhoA to Regulate YAP Signaling. Cell. Signal. 2014, 26, 2504–2513. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Dai, Y.; Yang, T.; Zhang, Y.; Li, M.; Xu, X. Decreased Erythrocyte CD44 and CD58 Expression Link E-Waste Pb Toxicity to Changes in Erythrocyte Immunity in Preschool Children. Sci. Total Environ. 2019, 664, 690–697. [Google Scholar] [CrossRef]
- Avivar-Valderas, A. Inhibition of PI3Kβ and mTOR influence the immune response and the defense mechanism against pathogens. Int. J. Infect. 2023, 7, 46–49. [Google Scholar]
- Toniato, E. IL-37 is an inhibitory cytokine that could be useful for treating infections. Int. J. Infect. 2024, 8, 1–2. [Google Scholar]
- Chou, Y.-E.; Hsieh, M.-J.; Hsin, C.-H.; Chiang, W.-L.; Lai, Y.-C.; Lee, Y.-H.; Huang, S.-C.; Yang, S.-F.; Lin, C.-W. CD44 Gene Polymorphisms and Environmental Factors on Oral Cancer Susceptibility in Taiwan. PLoS ONE 2014, 9, e93692. [Google Scholar] [CrossRef]
- Cirillo, N. The Hyaluronan/CD44 Axis: A Double-Edged Sword in Cancer. Int. J. Mol. Sci. 2023, 24, 15812. [Google Scholar] [CrossRef]
- UniProt. Available online: https://www.uniprot.org/uniprotkb/P16070/entry (accessed on 10 July 2025).
- Xu, Q. The Indian Blood Group System. Immunohematology 2011, 27, 89–93. [Google Scholar] [CrossRef]
- Joshi, S.R.; Sheladiya, A.; Mendapara-Dobariya, K.V. INRA, a New High-Frequency Antigen in the INDIAN (IN023) Blood Group System. Asian J. Transfus. Sci. 2017, 11, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Poole, J.; Tilley, L.; Warke, N.; Spring, F.A.; Overbeeke, M.A.M.; van der Mark-Zoet, J.A.C.M.; Ahrens, N.; Armstrong, D.; Williams, M.; Daniels, G. Two Missense Mutations in the CD44 Gene Encode Two New Antigens of the Indian Blood Group System. Transfusion 2007, 47, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Telen, M.J.; Udani, M.; Washington, M.K.; Levesque, M.C.; Lloyd, E.; Rao, N. A Blood Group-Related Polymorphism of CD44 Abolishes a Hyaluronan-Binding Consensus Sequence without Preventing Hyaluronan Binding. J. Biol. Chem. 1996, 271, 7147–7153. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, D.S.; Wagner, L.; Feingold, E.A.; Shenmen, C.M.; Grouse, L.H.; Schuler, G.; Klein, S.L.; Old, S.; Rasooly, R.; Good, P.; et al. The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC). Genome Res. 2004, 14, 2121–2127. [Google Scholar] [CrossRef]
- Günthert, U. CD44: A Multitude of Isoforms with Diverse Functions. In Adhesion in Leukocyte Homing and Differentiation; Dunon, D., Mackay, C.R., Imhof, B.A., Eds.; Springer: Berlin/Heidelberg, Germany, 1993; pp. 47–63. ISBN 978-3-642-78253-4. [Google Scholar]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef]
- Klement, J.D.; Paschall, A.V.; Redd, P.S.; Ibrahim, M.L.; Lu, C.; Yang, D.; Celis, E.; Abrams, S.I.; Ozato, K.; Liu, K. An Osteopontin/CD44 Immune Checkpoint Controls CD8+ T Cell Activation and Tumor Immune Evasion. J. Clin. Investig. 2018, 128, 5549–5560. [Google Scholar] [CrossRef]
- Shirasaki, T.; Honda, M.; Yamashita, T.; Nio, K.; Shimakami, T.; Shimizu, R.; Nakasyo, S.; Murai, K.; Shirasaki, N.; Okada, H.; et al. The Osteopontin-CD44 Axis in Hepatic Cancer Stem Cells Regulates IFN Signaling and HCV Replication. Sci. Rep. 2018, 8, 13143. [Google Scholar] [CrossRef]
- Ou, J.; Deng, J.; Wei, X.; Xie, G.; Zhou, R.; Yu, L.; Liang, H. Fibronectin Extra Domain A (EDA) Sustains CD133(+)/CD44(+) Subpopulation of Colorectal Cancer Cells. Stem Cell Res. 2013, 11, 820–833. [Google Scholar] [CrossRef]
- Ozer, E.; Canda, T.; Kurtodlu, B. The Role of Angiogenesis, Laminin and CD44 Expression in Metastatic Behavior of Early-Stage Low-Grade Invasive Breast Carcinomas. Cancer Lett. 1997, 121, 119–123. [Google Scholar] [CrossRef]
- Song, T.; Yang, Y.; Wang, Y.; Ni, Y.; Yang, Y.; Zhang, L. Bulk and Single-Cell RNA Sequencing Reveal the Contribution of Laminin Γ2 -CD44 to the Immune Resistance in Lymphocyte-Infiltrated Squamous Lung Cancer Subtype. Heliyon 2024, 10, e31299. [Google Scholar] [CrossRef] [PubMed]
- Rousselle, P.; Beck, K. Laminins and Matrix Metalloproteinases Connection: A Subtle Relationship That Can Go Wrong in a Tumor Context, Particularly If CD44 Gets Involved. In The Extracellular Matrix and the Tumor Microenvironment; Kovalszky, I., Franchi, M., Alaniz, L.D., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 219–246. ISBN 978-3-030-99708-3. [Google Scholar]
- Govindaraju, P.; Todd, L.; Shetye, S.; Monslow, J.; Puré, E. CD44-Dependent Inflammation, Fibrogenesis, and Collagenolysis Regulates Extracellular Matrix Remodeling and Tensile Strength during Cutaneous Wound Healing. Matrix Biol. 2019, 75–76, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Onyszczuk, M.; Rynkiewicz, M.; Kiczmer, P.; Drozdzowska, B. Prognostic and Clinicopathological Significance of CD44, MMP-2, and MMP-9 Expression in Clear Cell Renal Cell Carcinoma. Med. Res. J. 2025, 10, 9–18. [Google Scholar] [CrossRef]
- Chrabańska, M.; Rynkiewicz, M.; Kiczmer, P.; Drozdzowska, B. Immunohistochemical Expression of CD44, MMP-2, MMP-9, and Ki-67 as the Prognostic Markers in Non-Clear Cell Renal Cell Carcinomas-A Prospective Cohort Study. J. Clin. Med. 2022, 11, 5196. [Google Scholar] [CrossRef]
- Baaten, B.J.; Li, C.-R.; Bradley, L.M. Multifaceted Regulation of T Cells by CD44. Commun. Integr. Biol. 2010, 3, 508–512. [Google Scholar] [CrossRef]
- Gutjahr, J.C.; Greil, R.; Hartmann, T.N. The Role of CD44 in the Pathophysiology of Chronic Lymphocytic Leukemia. Front. Immunol. 2015, 6, 177. [Google Scholar] [CrossRef]
- Martin, T.A.; Harrison, G.; Mansel, R.E.; Jiang, W.G. The Role of the CD44/Ezrin Complex in Cancer Metastasis. Crit. Rev. Oncol. Hematol. 2003, 46, 165–186. [Google Scholar] [CrossRef]
- AbuSamra, D.B.; Al-Kilani, A.; Hamdan, S.M.; Sakashita, K.; Gadhoum, S.Z.; Merzaban, J.S. Quantitative Characterization of E-Selectin Interaction with Native CD44 and P-Selectin Glycoprotein Ligand-1 (PSGL-1) Using a Real Time Immunoprecipitation-Based Binding Assay. J. Biol. Chem. 2015, 290, 21213–21230. [Google Scholar] [CrossRef] [PubMed]
- Sackstein, R. The Bone Marrow Is Akin to Skin: HCELL and the Biology of Hematopoietic Stem Cell Homing. J. Investig. Dermatol. 2004, 122, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Sackstein, R. The Biology of CD44 and HCELL in Hematopoiesis: The “Step 2-Bypass Pathway” and Other Emerging Perspectives. Curr. Opin. Hematol. 2011, 18, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.P.; Sackstein, R. CD44 and HCELL: Preventing Hematogenous Metastasis at Step 1. FEBS Lett. 2011, 585, 3148–3158. [Google Scholar] [CrossRef]
- Cao, H.; Heazlewood, S.Y.; Williams, B.; Cardozo, D.; Nigro, J.; Oteiza, A.; Nilsson, S.K. The Role of CD44 in Fetal and Adult Hematopoietic Stem Cell Regulation. Haematologica 2016, 101, 26–37. [Google Scholar] [CrossRef]
- Zöller, M. CD44, Hyaluronan, the Hematopoietic Stem Cell, and Leukemia-Initiating Cells. Front. Immunol. 2015, 6, 235. [Google Scholar] [CrossRef]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a Mere Stem Cell Marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef]
- Lee-Sayer, S.S.M.; Dougan, M.N.; Cooper, J.; Sanderson, L.; Dosanjh, M.; Maxwell, C.A.; Johnson, P. CD44-Mediated Hyaluronan Binding Marks Proliferating Hematopoietic Progenitor Cells and Promotes Bone Marrow Engraftment. PLoS ONE 2018, 13, e0196011. [Google Scholar] [CrossRef]
- Pokharel, D.; Padula, M.P.; Lu, J.F.; Jaiswal, R.; Djordjevic, S.P.; Bebawy, M. The Role of CD44 and ERM Proteins in Expression and Functionality of P-Glycoprotein in Breast Cancer Cells. Molecules 2016, 21, 290. [Google Scholar] [CrossRef]
- Neisch, A.L.; Fehon, R.G. Ezrin, Radixin and Moesin: Key Regulators of Membrane-Cortex Interactions and Signaling. Curr. Opin. Cell Biol. 2011, 23, 377–382. [Google Scholar] [CrossRef]
- Ilangumaran, S.; Briol, A.; Hoessli, D.C. CD44 Selectively Associates with Active Src Family Protein Tyrosine Kinases Lck and Fyn in Glycosphingolipid-Rich Plasma Membrane Domains of Human Peripheral Blood Lymphocytes. Blood 1998, 91, 3901–3908. [Google Scholar] [CrossRef]
- Tsai, T.; Wu, S.; Lai, Y.; Wang, H.; Hou, P.; Huang, Y.; Chen, H.H.; Su, W. CD44-Hyaluronan Mediating Endocytosis of Iron-Platinum Alloy Nanoparticles Induces Ferroptotic Cell Death in Mesenchymal-State Lung Cancer Cells with Tyrosine Kinase Inhibitor Resistance. Acta Biomater. 2024, 186, 396–410. [Google Scholar] [CrossRef]
- Wang, S.J.; Bourguignon, L.Y.W. Hyaluronan-CD44 Promotes Phospholipase C-Mediated Ca2+ Signaling and Cisplatin Resistance in Head and Neck Cancer. Arch. Otolaryngol. Head Neck Surg. 2006, 132, 19–24. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Gilad, E.; Brightman, A.; Diedrich, F.; Singleton, P. Hyaluronan-CD44 Interaction with Leukemia-Associated RhoGEF and Epidermal Growth Factor Receptor Promotes Rho/Ras Co-Activation, Phospholipase C Epsilon-Ca2+ Signaling, and Cytoskeleton Modification in Head and Neck Squamous Cell Carcinoma Cells. J. Biol. Chem. 2006, 281, 14026–14040. [Google Scholar] [CrossRef]
- Wong, N.K.Y.; Lai, J.C.Y.; Maeshima, N.; Johnson, P. CD44-Mediated Elongated T Cell Spreading Requires Pyk2 Activation by Src Family Kinases, Extracellular Calcium, Phospholipase C and Phosphatidylinositol-3 Kinase. Cell. Signal. 2011, 23, 812–819. [Google Scholar] [CrossRef]
- Chellaiah, M.A.; Biswas, R.S.; Rittling, S.R.; Denhardt, D.T.; Hruska, K.A. Rho-Dependent Rho Kinase Activation Increases CD44 Surface Expression and Bone Resorption in Osteoclasts. J. Biol. Chem. 2003, 278, 29086–29097. [Google Scholar] [CrossRef]
- Lee, M.N.; Song, J.H.; Oh, S.-H.; Tham, N.T.; Kim, J.-W.; Yang, J.-W.; Kim, E.-S.; Koh, J.-T. The Primary Cilium Directs Osteopontin-Induced Migration of Mesenchymal Stem Cells by Regulating CD44 Signaling and Cdc42 Activation. Stem Cell Res. 2020, 45, 101799. [Google Scholar] [CrossRef] [PubMed]
- Murai, T.; Miyazaki, Y.; Nishinakamura, H.; Sugahara, K.N.; Miyauchi, T.; Sako, Y.; Yanagida, T.; Miyasaka, M. Engagement of CD44 Promotes Rac Activation and CD44 Cleavage during Tumor Cell Migration. J. Biol. Chem. 2004, 279, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Ge, H.; Güngör, C.; Gong, X.; Chen, Y. Cytoskeletal and Cytoskeleton-Associated Proteins: Key Regulators of Cancer Stem Cell Properties. Pharmaceuticals 2022, 15, 1369. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting Integrin Pathways: Mechanisms and Advances in Therapy. Signal Transduct. Target. Ther. 2023, 8, 1. [Google Scholar] [CrossRef]
- Trapasso, S.; Allegra, E. Role of CD44 as a Marker of Cancer Stem Cells in Head and Neck Cancer. Biologics 2012, 6, 379–383. [Google Scholar] [CrossRef][Green Version]
- Poltavets, V.; Kochetkova, M.; Pitson, S.M.; Samuel, M.S. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front. Oncol. 2018, 8, 431. [Google Scholar] [CrossRef]
- Nonnast, E.; Mira, E.; Mañes, S. The Role of Laminins in Cancer Pathobiology: A Comprehensive Review. J. Transl. Med. 2025, 23, 83. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Singleton, P.A.; Diedrich, F. Hyaluronan-CD44 Interaction with Rac1-Dependent Protein Kinase N-γ Promotes Phospholipase Cγ1 Activation, Ca2+ Signaling, and Cortactin-Cytoskeleton Function Leading to Keratinocyte Adhesion and Differentiation. J. Biol. Chem. 2004, 279, 29654–29669. [Google Scholar] [CrossRef]
- Vikesaa, J.; Hansen, T.V.; Jønson, L.; Borup, R.; Wewer, U.M.; Christiansen, J.; Nielsen, F.C. RNA-binding IMPs Promote Cell Adhesion and Invadopodia Formation. EMBO J. 2006, 25, 1456–1468. [Google Scholar] [CrossRef]
- Casalino-Matsuda, S.M.; Monzon, M.E.; Day, A.J.; Forteza, R.M. Hyaluronan Fragments/CD44 Mediate Oxidative Stress–Induced MUC5B Up-Regulation in Airway Epithelium. Am. J. Respir. Cell Mol. Biol. 2009, 40, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Crosby, H.A.; Lalor, P.F.; Ross, E.; Newsome, P.N.; Adams, D.H. Adhesion of Human Haematopoietic (CD34+) Stem Cells to Human Liver Compartments Is Integrin and CD44 Dependent and Modulated by CXCR3 and CXCR4. J. Hepatol. 2009, 51, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Matsuda, Y.; Naito, Z.; Ishiwata, T. CD44 in Human Glioma Correlates with Histopathological Grade and Cell Migration. Pathol. Int. 2012, 62, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Midgley, A.C.; Rogers, M.; Hallett, M.B.; Clayton, A.; Bowen, T.; Phillips, A.O.; Steadman, R. Transforming Growth Factor-Β1 (TGF-Β1)-Stimulated Fibroblast to Myofibroblast Differentiation Is Mediated by Hyaluronan (HA)-Facilitated Epidermal Growth Factor Receptor (EGFR) and CD44 Co-Localization in Lipid Rafts. J. Biol. Chem. 2013, 288, 14824–14838. [Google Scholar] [CrossRef]
- Funaro, A.; Spagnoli, G.C.; Momo, M.; Knapp, W.; Malavasi, F. Stimulation of T Cells via CD44 Requires Leukocyte-Function-Associated Antigen Interactions and Interleukin-2 Production. Hum. Immunol. 1994, 40, 267–278. [Google Scholar] [CrossRef]
- Buscher, K.; Riese, S.B.; Shakibaei, M.; Reich, C.; Dernedde, J.; Tauber, R.; Ley, K. The Transmembrane Domains of L-Selectin and CD44 Regulate Receptor Cell Surface Positioning and Leukocyte Adhesion under Flow. J. Biol. Chem. 2010, 285, 13490–13497. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Wang, H.; Guo, Q.; Liu, Y.; He, Y.; Zhang, G.; Yang, C.; Du, Y.; Gao, F. CD44s-Activated tPA/LRP1-NFκB Pathway Drives Lamellipodia Outgrowth in Luminal-Type Breast Cancer Cells. Front. Cell Dev. Biol. 2023, 11, 1224827. [Google Scholar] [CrossRef] [PubMed]
- Föger, N.; Marhaba, R.; Zöller, M. Involvement of CD44 in Cytoskeleton Rearrangement and Raft Reorganization in T Cells. J. Cell Sci. 2001, 114, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S. CD44 Intracellular Domain: A Long Tale of a Short Tail. Cancers 2023, 15, 5041. [Google Scholar] [CrossRef]
- Du, Y.; Bradshaw, W.J.; Leisner, T.M.; Annor-Gyamfi, J.K.; Qian, K.; Bashore, F.M.; Sikdar, A.; Nwogbo, F.O.; Ivanov, A.A.; Frye, S.V.; et al. Discovery of FERM Domain Protein–Protein Interaction Inhibitors for MSN and CD44 as a Potential Therapeutic Approach for Alzheimer’s Disease. J. Biol. Chem. 2023, 299, 105382. [Google Scholar] [CrossRef]
- Weng, X.; Maxwell-Warburton, S.; Hasib, A.; Ma, L.; Kang, L. The Membrane Receptor CD44: Novel Insights into Metabolism. Trends Endocrinol. Metab. 2022, 33, 318–332. [Google Scholar] [CrossRef]
- Tsukita, S.; Oishi, K.; Sato, N.; Sagara, J.; Kawai, A.; Tsukita, S. ERM Family Members as Molecular Linkers between the Cell Surface Glycoprotein CD44 and Actin-Based Cytoskeletons. J. Cell Biol. 1994, 126, 391–401. [Google Scholar] [CrossRef]
- Mori, T.; Kitano, K.; Terawaki, S.; Maesaki, R.; Fukami, Y.; Hakoshima, T. Structural Basis for CD44 Recognition by ERM Proteins. J. Biol. Chem. 2008, 283, 29602–29612. [Google Scholar] [CrossRef]
- Yonemura, S.; Hirao, M.; Doi, Y.; Takahashi, N.; Kondo, T.; Tsukita, S.; Tsukita, S. Ezrin/Radixin/Moesin (ERM) Proteins Bind to a Positively Charged Amino Acid Cluster in the Juxta-Membrane Cytoplasmic Domain of CD44, CD43, and ICAM-2. J. Cell Biol. 1998, 140, 885–895. [Google Scholar] [CrossRef]
- Tissue Expression of CD44—Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000026508-CD44/tissue (accessed on 18 July 2025).
- Ma, X.; Dighe, A.; Maziarz, J.; Neumann, E.; Erkenbrack, E.; Hei, Y.-Y.; Liu, Y.; Suhail, Y.; Pak, I.; Levchenko, A.; et al. Evolution of Higher Mesenchymal CD44 Expression in the Human Lineage. Evol. Med. Public Health 2022, 10, 447–462. [Google Scholar] [CrossRef]
- Williams, K.; Motiani, K.; Giridhar, P.V.; Kasper, S. CD44 Integrates Signaling in Normal Stem Cell, Cancer Stem Cell and (Pre)Metastatic Niches. Exp. Biol. Med. 2013, 238, 324–338. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, G.; Zhang, P.; Zhuang, X.; Hu, G. A CD44v+ Subpopulation of Breast Cancer Stem-like Cells with Enhanced Lung Metastasis Capacity. Cell Death Dis. 2017, 8, e2679. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, K.; Jiang, P.; Zhang, X.; Li, X.; Li, Z. CD44v/CD44s Expression Patterns Are Associated with the Survival of Pancreatic Carcinoma Patients. Diagn. Pathol. 2014, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Erb, U.; Megaptche, A.P.; Gu, X.; Büchler, M.W.; Zöller, M. CD44 Standard and CD44v10 Isoform Expression on Leukemia Cells Distinctly Influences Niche Embedding of Hematopoietic Stem Cells. J. Hematol. Oncol. 2014, 7, 29. [Google Scholar] [CrossRef]
- Lo, C.W.-S.; Chan, C.K.W.; Yu, J.; He, M.; Choi, C.H.J.; Lau, J.Y.W.; Wong, N. Development of CD44E/s Dual-Targeting DNA Aptamer as Nanoprobe to Deliver Treatment in Hepatocellular Carcinoma. Nanotheranostics 2022, 6, 161–174. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, Z.; Katsha, A.; Hong, J.; Belkhiri, A.; El-Rifai, W. Regulation of CD44E by DARPP-32-Dependent Activation of SRp20 Splicing Factor in Gastric Tumorigenesis. Oncogene 2016, 35, 1847–1856. [Google Scholar] [CrossRef][Green Version]
- Oliferenko, S.; Paiha, K.; Harder, T.; Gerke, V.; Schwärzler, C.; Schwarz, H.; Beug, H.; Günthert, U.; Huber, L.A. Analysis of Cd44-Containing Lipid Rafts. J. Cell Biol. 1999, 146, 843–854. [Google Scholar] [CrossRef]
- Murai, T. Lipid Raft-Mediated Regulation of Hyaluronan–CD44 Interactions in Inflammation and Cancer. Front. Immunol. 2015, 6, 420. [Google Scholar] [CrossRef]
- Sun, F.; Schroer, C.F.E.; Palacios, C.R.; Xu, L.; Luo, S.-Z.; Marrink, S.J. Molecular Mechanism for Bidirectional Regulation of CD44 for Lipid Raft Affiliation by Palmitoylations and PIP2. PLoS Comput. Biol. 2020, 16, e1007777. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Park, Y.; Hemati, H.; Liu, X. Cell Aggregation Activates Small GTPase Rac1 and Induces CD44 Cleavage by Maintaining Lipid Raft Integrity. J. Biol. Chem. 2023, 299, 105377. [Google Scholar] [CrossRef] [PubMed]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of Alternative Splicing of CD44 in Cancer. Cell. Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.K.; Carpenito, C.; Dougherty, S.T.; Hayes, G.M.; Dougherty, G.J. Identification and Characterization of CD44RC, a Novel Alternatively Spliced Soluble CD44 Isoform That Can Potentiate the Hyaluronan Binding Activity of Cell Surface CD44. Neoplasia 1999, 1, 446–452. [Google Scholar] [CrossRef][Green Version]
- Bechtel, S.; Rosenfelder, H.; Duda, A.; Schmidt, C.P.; Ernst, U.; Wellenreuther, R.; Mehrle, A.; Schuster, C.; Bahr, A.; Blöcker, H.; et al. The Full-ORF Clone Resource of the German cDNA Consortium. BMC Genom. 2007, 8, 399. [Google Scholar] [CrossRef]
- Harn, H.J.; Isola, N.; Cooper, D.L. The Multispecific Cell Adhesion Molecule CD44 Is Represented in Reticulocyte cDNA. Biochem. Biophys. Res. Commun. 1991, 178, 1127–1134. [Google Scholar] [CrossRef]
- Stamenkovic, I.; Amiot, M.; Pesando, J.M.; Seed, B. A Lymphocyte Molecule Implicated in Lymph Node Homing Is a Member of the Cartilage Link Protein Family. Cell 1989, 56, 1057–1062. [Google Scholar] [CrossRef]
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A Human Lymphocyte Homing Receptor, the Hermes Antigen, Is Related to Cartilage Proteoglycan Core and Link Proteins. Cell 1989, 56, 1063–1072. [Google Scholar] [CrossRef]
- Kozlowski, L.P. IPC—Isoelectric Point Calculator. Biol. Direct 2016, 11, 55. [Google Scholar] [CrossRef]
- Baj-Krzyworzeka, M.; Weglarczyk, K.; Szatanek, R.; Mytar, B.; Baran, J.; Siedlar, M. The Role of CD44H Molecule in the Interactions between Human Monocytes and Pancreatic Adenocarcinoma-Derived Microvesicles. Folia Histochem. Cytobiol. 2019, 57, 28–34. [Google Scholar] [CrossRef]
- Stamenkovic, I.; Yu, Q. CHAPTER 5—CD44 Meets Merlin and Ezrin: Their Interplay Mediates the Pro-Tumor Activity of CD44 and Tumor-Suppressing Effect of Merlin. In Hyaluronan in Cancer Biology; Stern, R., Ed.; Academic Press: San Diego, CA, USA, 2009; pp. 71–87. ISBN 978-0-12-374178-3. [Google Scholar]
- Bonente, D.; Bianchi, L.; De Salvo, R.; Nicoletti, C.; De Benedetto, E.; Bacci, T.; Bini, L.; Inzalaco, G.; Franci, L.; Chiariello, M.; et al. Co-Expression of Podoplanin and CD44 in Proliferative Vitreoretinopathy Epiretinal Membranes. Int. J. Mol. Sci. 2023, 24, 9728. [Google Scholar] [CrossRef]
- Perez, A.; Neskey, D.M.; Wen, J.; Pereira, L.; Reategui, E.P.; Goodwin, W.J.; Carraway, K.L.; Franzmann, E.J. CD44 Interacts with EGFR and Promotes Head and Neck Squamous Cell Carcinoma Initiation and Progression. Oral Oncol. 2013, 49, 306–313. [Google Scholar] [CrossRef]
- Liu, Z.; Chu, S.; Yao, S.; Li, Y.; Fan, S.; Sun, X.; Su, L.; Liu, X. CD74 Interacts with CD44 and Enhances Tumorigenesis and Metastasis via RHOA-Mediated Cofilin Phosphorylation in Human Breast Cancer Cells. Oncotarget 2016, 7, 68303–68313. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Sleeman, J. Chapter Nine—CD44 Is a Multidomain Signaling Platform That Integrates Extracellular Matrix Cues with Growth Factor and Cytokine Signals. In Advances in Cancer Research; Simpson, M.A., Heldin, P., Eds.; Hyaluronan Signaling and Turnover; Academic Press: San Diego, CA, USA, 2014; Volume 123, pp. 231–254. [Google Scholar]
- Cowman, M.K.; Turley, E.A. Functional Organization of Extracellular Hyaluronan, CD44, and RHAMM. Proteoglycan Res. 2023, 1, e4. [Google Scholar] [CrossRef]
- Lee, J.-L.; Wang, M.-J.; Sudhir, P.-R.; Chen, J.-Y. CD44 Engagement Promotes Matrix-Derived Survival through the CD44-SRC-Integrin Axis in Lipid Rafts. Mol. Cell. Biol. 2008, 28, 5710–5723. [Google Scholar] [CrossRef]
- Skandalis, S.S.; Kozlova, I.; Engström, U.; Hellman, U.; Heldin, P. Proteomic Identification of CD44 Interacting Proteins. IUBMB Life 2010, 62, 833–840. [Google Scholar] [CrossRef]
- Bai, Y.; Liu, Y.-J.; Wang, H.; Xu, Y.; Stamenkovic, I.; Yu, Q. Inhibition of the Hyaluronan-CD44 Interaction by Merlin Contributes to the Tumor-Suppressor Activity of Merlin. Oncogene 2007, 26, 836–850. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-N.; Tsai, M.-F.; Wu, S.-G.; Chang, T.-H.; Shih, J.-Y. CD44s and CD44v8-10 Isoforms Confer Acquired Resistance to Osimertinib by Activating the ErbB3/STAT3 Signaling Pathway. Life Sci. 2024, 336, 122345. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Leng, L.; Wang, T.; Wang, W.; Du, X.; Li, J.; McDonald, C.; Chen, Z.; Murphy, J.W.; Lolis, E.; et al. CD44 Is the Signaling Component of the Macrophage Migration Inhibitory Factor-CD74 Receptor Complex. Immunity 2006, 25, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yang, C.; Gao, F. The State of CD44 Activation in Cancer Progression and Therapeutic Targeting. FEBS J. 2022, 289, 7970–7986. [Google Scholar] [CrossRef]
- Cichy, J.; Puré, E. The Liberation of CD44. J. Cell Biol. 2003, 161, 839–843. [Google Scholar] [CrossRef]
- Bruno, P.S.; Arshad, A.; Gogu, M.-R.; Waterman, N.; Flack, R.; Dunn, K.; Darie, C.C.; Neagu, A.-N. Post-Translational Modifications of Proteins Orchestrate All Hallmarks of Cancer. Life 2025, 15, 126. [Google Scholar] [CrossRef]
- Liao, C.; Wang, Q.; An, J.; Chen, J.; Li, X.; Long, Q.; Xiao, L.; Guan, X.; Liu, J. CD44 Glycosylation as a Therapeutic Target in Oncology. Front. Oncol. 2022, 12, 883831. [Google Scholar] [CrossRef] [PubMed]
- Vuorio, J.; Škerlová, J.; Fábry, M.; Veverka, V.; Vattulainen, I.; Řezáčová, P.; Martinez-Seara, H. N-Glycosylation Can Selectively Block or Foster Different Receptor–Ligand Binding Modes. Sci. Rep. 2021, 11, 5239. [Google Scholar] [CrossRef] [PubMed]
- Leon, F.; Seshacharyulu, P.; Nimmakalaya, R.K.; Chugh, S.; Karmakar, S.; Nallasamy, P.; Vengoji, R.; Rachagani, S.; Cox, J.L.; Mallya, K.; et al. Reduction in O-Glycome Induces Differentially Glycosylated CD44 to Promote Stemness and Metastasis in Pancreatic Cancer. Oncogene 2022, 41, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.; Oh, S.; Shin, I. Ablation of CD44 Induces Glycolysis-to-Oxidative Phosphorylation Transition via Modulation of the c-Src-Akt-LKB1-AMPKα Pathway. Biochem. J. 2016, 473, 3013–3030. [Google Scholar] [CrossRef]
- Puré, E.; Camp, R.L.; Peritt, D.; Panettieri, R.A.; Lazaar, A.L.; Nayak, S. Defective Phosphorylation and Hyaluronate Binding of CD44 with Point Mutations in the Cytoplasmic Domain. J. Exp. Med. 1995, 181, 55–62. [Google Scholar] [CrossRef]
- Stamenkovic, I.; Yu, Q. Shedding Light on Proteolytic Cleavage of CD44: The Responsible Sheddase and Functional Significance of Shedding. J. Investig. Dermatol. 2009, 129, 1321–1324. [Google Scholar] [CrossRef]
- Wöhner, B.; Li, W.; Hey, S.; Drobny, A.; Werny, L.; Becker-Pauly, C.; Lucius, R.; Zunke, F.; Linder, S.; Arnold, P. Proteolysis of CD44 at the Cell Surface Controls a Downstream Protease Network. Front. Mol. Biosci. 2023, 10, 1026810. [Google Scholar] [CrossRef]
- Ravindranath, A.K.; Kaur, S.; Wernyj, R.P.; Kumaran, M.N.; Miletti-Gonzalez, K.E.; Chan, R.; Lim, E.; Madura, K.; Rodriguez-Rodriguez, L. CD44 Promotes Multi-Drug Resistance by Protecting P-Glycoprotein from FBXO21-Mediated Ubiquitination. Oncotarget 2015, 6, 26308–26321. [Google Scholar] [CrossRef]
- Xu, Y.; Bai, Z.; Lan, T.; Fu, C.; Cheng, P. CD44 and Its Implication in Neoplastic Diseases. MedComm 2024, 5, e554. [Google Scholar] [CrossRef]
- Yung, S.; Chan, T.M. The Role of Hyaluronan and CD44 in the Pathogenesis of Lupus Nephritis. Autoimmune Dis. 2012, 2012, 207190. [Google Scholar] [CrossRef]
- Yi, P.; Cao, P.; Yang, M.; Xiong, F.; Jiang, J.; Mei, Y.; Xin, Y.; Zhao, M.; Wu, H.; Lu, Q. Overexpressed CD44 Is Associated with B-Cell Activation via the HA-CD44-AIM2 Pathway in Lupus B Cells. Clin. Immunol. 2023, 255, 109710. [Google Scholar] [CrossRef] [PubMed]
- Norris, P.A.A.; Kaur, G.; Khan, R.; Zhu, G.; Ni, H.; Lazarus, A.H. Anti-Inflammatory Activity of CD44 Antibodies in Murine Immune Thrombocytopenia Is Mediated by Fcγ Receptor Inhibition. Blood 2021, 137, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- Tremmel, M.; Matzke, A.; Albrecht, I.; Laib, A.M.; Olaku, V.; Ballmer-Hofer, K.; Christofori, G.; Héroult, M.; Augustin, H.G.; Ponta, H.; et al. A CD44v6 Peptide Reveals a Role of CD44 in VEGFR-2 Signaling and Angiogenesis. Blood 2009, 114, 5236–5244. [Google Scholar] [CrossRef] [PubMed]
- Ziranu, P.; Pretta, A.; Aimola, V.; Cau, F.; Mariani, S.; D’Agata, A.P.; Codipietro, C.; Rizzo, D.; Dell’Utri, V.; Sanna, G.; et al. CD44: A New Prognostic Marker in Colorectal Cancer? Cancers 2024, 16, 1569. [Google Scholar] [CrossRef]
- Inoue, A.; Ohnishi, T.; Nishikawa, M.; Ohtsuka, Y.; Kusakabe, K.; Yano, H.; Tanaka, J.; Kunieda, T. A Narrative Review on CD44’s Role in Glioblastoma Invasion, Proliferation, and Tumor Recurrence. Cancers 2023, 15, 4898. [Google Scholar] [CrossRef]
- Mehner, L.-M.; Munoz-Sagredo, L.; Sonnentag, S.J.; Treffert, S.M.; Orian-Rousseau, V. Targeting CD44 and Other Pleiotropic Co-Receptors as a Means for Broad Inhibition of Tumor Growth and Metastasis. Clin. Exp. Metastasis 2024, 41, 599–611. [Google Scholar] [CrossRef]
- Johnson, P.; Ruffell, B. CD44 and Its Role in Inflammation and Inflammatory Diseases. Inflamm. Allergy Drug Targets 2009, 8, 208–220. [Google Scholar] [CrossRef]
- Puré, E.; Cuff, C.A. A Crucial Role for CD44 in Inflammation. Trends Mol. Med. 2001, 7, 213–221. [Google Scholar] [CrossRef]
- Krolikoski, M.; Monslow, J.; Puré, E. The CD44-HA Axis and Inflammation in Atherosclerosis: A Temporal Perspective. Matrix Biol. 2019, 78–79, 201–218. [Google Scholar] [CrossRef]
- Salathia, S.; Gigliobianco, M.R.; Casadidio, C.; Di Martino, P.; Censi, R. Hyaluronic Acid-Based Nanosystems for CD44 Mediated Anti-Inflammatory and Antinociceptive Activity. Int. J. Mol. Sci. 2023, 24, 7286. [Google Scholar] [CrossRef]
- Wong, C.C.Y.; Gao, L.Y.; Xu, Y.; Chau, M.K.M.; Zhang, D.; Yap, D.Y.H.; Ying, S.K.Y.; Lee, C.K.; Yung, S.; Chan, T.M. Cluster of Differentiation-44 as a Novel Biomarker of Lupus Nephritis and Its Role in Kidney Inflammation and Fibrosis. Front. Immunol. 2024, 15, 1443153. [Google Scholar] [CrossRef]
- Yung, S.; Gao, L.; Chan, T.M. LSO-014 Clinico-Pathological Association of Serum CD44 Level in Lupus Nephritis Patients. Lupus Sci. Med. 2023, 10. [Google Scholar] [CrossRef]
- Fromont Hankard, G.; Cezard, J.P.; Aigrain, Y.; Navarro, J.; Peuchmaur, M. CD44 Variant Expression in Inflammatory Colonic Mucosa Is Not Disease Specific but Associated with Increased Crypt Cell Proliferation. Histopathology 1998, 32, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Franić, I.; Režić-Mužinić, N.; Markotić, A.; Živković, P.M.; Vilović, M.; Rušić, D.; Božić, J. Expression of CD44 in Leukocyte Subpopulations in Patients with Inflammatory Bowel Diseases. Diagnostics 2022, 12, 2014. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cai, J.; Gu, Y.; Li, X.; He, W.; Chen, Y.; Wang, Z.; Li, K.; Qin, G.; Gu, X.; et al. Novel pH-Responsive and CD44-Targeting Silica Nanoparticles for Inflammatory Bowel Disease Therapy. Chem. Eng. J. 2025, 513, 163017. [Google Scholar] [CrossRef]
- Wittig, B.M.; Sabat, R.; Holzlöhner, P.; Witte-Händel, E.; Heilmann, K.; Witte, K.; Triebus, J.; Tzankov, A.; Laman, J.D.; Bokemeyer, B.; et al. Absence of Specific Alternatively Spliced Exon of CD44 in Macrophages Prevents Colitis. Mucosal Immunol. 2018, 11, 846–860. [Google Scholar] [CrossRef]
- Kitano, A.; Oshitani, N.; Matsumoto, T.; Kobayashi, K. CD44 Variants in Ulcerative Colitis and Crohn’s Disease. Lancet 1996, 348, 266–267. [Google Scholar] [CrossRef]
- Collins, C.B.; Ho, J.; Wilson, T.E.; Wermers, J.D.; Tlaxca, J.L.; Lawrence, M.B.; Solga, M.; Lannigan, J.; Rivera–Nieves, J. CD44 Deficiency Attenuates Chronic Murine Ileitis. Gastroenterology 2008, 135, 1993–2002. [Google Scholar] [CrossRef]
- Vadhan, A.; Hou, M.-F.; Vijayaraghavan, P.; Wu, Y.-C.; Hu, S.C.-S.; Wang, Y.-M.; Cheng, T.-L.; Wang, Y.-Y.; Yuan, S.-S.F. CD44 Promotes Breast Cancer Metastasis through AKT-Mediated Downregulation of Nuclear FOXA2. Biomedicines 2022, 10, 2488. [Google Scholar] [CrossRef]
- Louderbough, J.M.V.; Schroeder, J.A. Understanding the Dual Nature of CD44 in Breast Cancer Progression. Mol. Cancer Res. 2011, 9, 1573–1586. [Google Scholar] [CrossRef]
- Li, X.-P.; Zhang, X.-W.; Zheng, L.-Z.; Guo, W.-J. Expression of CD44 in Pancreatic Cancer and Its Significance. Int. J. Clin. Exp. Pathol. 2015, 8, 6724–6731. [Google Scholar] [PubMed]
- Zhao, S.; Chen, C.; Chang, K.; Karnad, A.; Jagirdar, J.; Kumar, A.P.; Freeman, J.W. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness and Response to Therapy. Clin. Cancer Res. 2016, 22, 5592–5604. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Zhao, X.; Cao, L.; Karnad, A.; Kumar, A.P.; Freeman, J.W. Gemcitabine Resistance of Pancreatic Cancer Cells Is Mediated by IGF1R Dependent Upregulation of CD44 Expression and Isoform Switching. Cell Death Dis. 2022, 13, 682. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Vadhan, A.; Chen, P.-H.; Lee, Y.-L.; Chao, C.-Y.; Cheng, K.-H.; Chang, Y.-C.; Hu, S.C.-S.; Yuan, S.-S.F. CD44 Promotes Lung Cancer Cell Metastasis through ERK–ZEB1 Signaling. Cancers 2021, 13, 4057. [Google Scholar] [CrossRef]
- Hu, B.; Ma, Y.; Yang, Y.; Zhang, L.; Han, H.; Chen, J. CD44 Promotes Cell Proliferation in Non-Small Cell Lung Cancer. Oncol. Lett. 2018, 15, 5627–5633. [Google Scholar] [CrossRef]
- Alaei, E.; Farahani, N.; Orouei, S.; Alimohammadi, M.; Daneshi, S.; Mousavi, T.; Mahmoodieh, B.; Taheriazam, A.; Rahimzadeh, P.; Hashemi, M. The Clinicopathologic and Prognostic Value of CD44 Expression in Patients with Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Mol. Cell. Probes 2025, 81, 102028. [Google Scholar] [CrossRef]
- Grisar, J.; Munk, M.; Steiner, C.W.; Amoyo-Minar, L.; Tohidast-Akrad, M.; Zenz, P.; Steiner, G.; Smolen, J.S. Expression Patterns of CD44 and CD44 Splice Variants in Patients with Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2012, 30, 64–72. [Google Scholar]
- Naor, D.; Nedvetzki, S. CD44 in Rheumatoid Arthritis. Arthritis Res. Ther. 2003, 5, 105–115. [Google Scholar] [CrossRef]
- Gorantla, S.; Gorantla, G.; Saha, R.N.; Singhvi, G. CD44 Receptor-Targeted Novel Drug Delivery Strategies for Rheumatoid Arthritis Therapy. Expert Opin. Drug Deliv. 2021, 18, 1553–1557. [Google Scholar] [CrossRef]
- Wittig, B.; Schwärzler, C.; Föhr, N.; Günthert, U.; Zöller, M. Cutting Edge: Curative Treatment of an Experimentally Induced Colitis by a CD44 Variant V7-Specific Antibody1. J. Immunol. 1998, 161, 1069–1073. [Google Scholar] [CrossRef]
- Schuster, R.; Younesi, F.; Ezzo, M.; Hinz, B. The Role of Myofibroblasts in Physiological and Pathological Tissue Repair. Cold Spring Harb. Perspect. Biol. 2023, 15, a041231. [Google Scholar] [CrossRef] [PubMed]
- Cialdai, F.; Risaliti, C.; Monici, M. Role of Fibroblasts in Wound Healing and Tissue Remodeling on Earth and in Space. Front. Bioeng. Biotechnol. 2022, 10, 958381. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Wang, X.; Sinha, S.; Forte, E.; Thompson, S.M.; Herzog, E.L.; Driskell, R.R.; Rosenthal, N.; Biernaskie, J.; Horsley, V. Fibroblasts: Origins, Definitions, and Functions in Health and Disease. Cell 2021, 184, 3852–3872. [Google Scholar] [CrossRef] [PubMed]
- Guler, Z.; Roovers, J.P. Role of Fibroblasts and Myofibroblasts on the Pathogenesis and Treatment of Pelvic Organ Prolapse. Biomolecules 2022, 12, 94. [Google Scholar] [CrossRef]
- Ito, T.; Williams, J.D.; Fraser, D.J.; Phillips, A.O. Hyaluronan Regulates Transforming Growth Factor-Beta1 Receptor Compartmentalization. J. Biol. Chem. 2004, 279, 25326–25332. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Singleton, P.A.; Zhu, H.; Zhou, B. Hyaluronan Promotes Signaling Interaction between CD44 and the Transforming Growth Factor Beta Receptor I in Metastatic Breast Tumor Cells. J. Biol. Chem. 2002, 277, 39703–39712. [Google Scholar] [CrossRef]
- Li, L.; Qi, L.; Liang, Z.; Song, W.; Liu, Y.; Wang, Y.; Sun, B.; Zhang, B.; Cao, W. Transforming Growth Factor-Β1 Induces EMT by the Transactivation of Epidermal Growth Factor Signaling through HA/CD44 in Lung and Breast Cancer Cells. Int. J. Mol. Med. 2015, 36, 113–122. [Google Scholar] [CrossRef]
- Wang, Y.; Mack, J.A.; Maytin, E.V. CD44 Inhibits α-SMA Gene Expression via a Novel G-Actin/MRTF-Mediated Pathway That Intersects with TGFβR/p38MAPK Signaling in Murine Skin Fibroblasts. J. Biol. Chem. 2019, 294, 12779–12794. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, K.; Hackert, T.; Zöller, M. CD44/CD44v6 a Reliable Companion in Cancer-Initiating Cell Maintenance and Tumor Progression. Front. Cell Dev. Biol. 2018, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Itakura, Y.; Toyoda, M. Sialylation Regulates Myofibroblast Differentiation of Human Skin Fibroblasts. Stem Cell Res. Ther. 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Morris, N.G.; Woods, E.L.; Dally, J.; Midgley, A.C.; Steadman, R.; Moseley, R. Dysfunctional Pericellular Hyaluronan Deposition Contributes to Attenuated CD44/EGFR Co-Localization and Impaired Myofibroblast Differentiation in Chronic Wound Fibroblasts. Exp. Cell Res. 2025, 450, 114646. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.-H.; Tsai, F.-C.; Chang, G.-J.; Lai, Y.-J.; Chang, S.-H.; Chen, W.-J.; Yeh, Y.-H. CD44 Regulates Epac1-Mediated β-Adrenergic-Receptor-Induced Ca2+-Handling Abnormalities: Implication in Cardiac Arrhythmias. J. Biomed. Sci. 2023, 30, 55. [Google Scholar] [CrossRef]
- Mishra, J.P.; Mishra, S.; Gee, K.; Kumar, A. Differential Involvement of Calmodulin-Dependent Protein Kinase II-Activated AP-1 and c-Jun N-Terminal Kinase-Activated EGR-1 Signaling Pathways in Tumor Necrosis Factor-α and Lipopolysaccharide-Induced CD44 Expression in Human Monocytic Cells. J. Biol. Chem. 2005, 280, 26825–26837. [Google Scholar] [CrossRef]
- Udabage, L.; Brownlee, G.R.; Nilsson, S.K.; Brown, T.J. The Over-Expression of HAS2, Hyal-2 and CD44 Is Implicated in the Invasiveness of Breast Cancer. Exp. Cell Res. 2005, 310, 205–217. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, D.; Liang, J.; Meltzer, E.B.; Gray, A.; Miura, R.; Wogensen, L.; Yamaguchi, Y.; Noble, P.W. Severe Lung Fibrosis Requires an Invasive Fibroblast Phenotype Regulated by Hyaluronan and CD44. J. Exp. Med. 2011, 208, 1459–1471. [Google Scholar] [CrossRef]
- Suchankova, M.; Zsemlye, E.; Urban, J.; Baráth, P.; Kohútová, L.; Siváková, B.; Ganovska, M.; Tibenska, E.; Szaboova, K.; Tedlova, E.; et al. The Bronchoalveolar Lavage Fluid CD44 as a Marker for Pulmonary Fibrosis in Diffuse Parenchymal Lung Diseases. Front. Immunol. 2025, 15, 1479458. [Google Scholar] [CrossRef]
- Xia, H.; Herrera, J.; Smith, K.; Yang, L.; Gilbertsen, A.; Benyumov, A.; Racila, E.; Bitterman, P.B.; Henke, C.A. Hyaluronan/CD44 Axis Regulates S100A4-Mediated Mesenchymal Progenitor Cell Fibrogenicity in Idiopathic Pulmonary Fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L926–L941. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Jiang, H.; Wu, X.; Hao, Y.; Su, Y.; Zou, Y.; Xian, W.; Wang, F.; Du, Q. PPARG/SPP1/CD44 Signaling Pathway in Alveolar Macrophages: Mechanisms of Lipid Dysregulation and Therapeutic Targets in Idiopathic Pulmonary Fibrosis. Heliyon 2025, 11, e41628. [Google Scholar] [CrossRef]
- Yasaka, N.; Furue, M.; Tamaki, K. CD44 Expression in Normal Human Skin and Skin Tumors. J. Dermatol. 1995, 22, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.W.; Ramez, M.; Gilad, E.; Singleton, P.A.; Man, M.-Q.; Crumrine, D.A.; Elias, P.M.; Feingold, K.R. Hyaluronan–CD44 Interaction Stimulates Keratinocyte Differentiation, Lamellar Body Formation/Secretion, and Permeability Barrier Homeostasis. J. Investig. Dermatol. 2006, 126, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, T.; Zhang, W.; Yu, K.; Xu, X.; Li, W.; Song, L.; Gu, X.; Cao, R.; Cui, S. Inhibition of CD44 Suppresses the Formation of Fibrotic Scar after Spinal Cord Injury via the JAK2/STAT3 Signaling Pathway. iScience 2024, 27, 108935. [Google Scholar] [CrossRef]
- Weng, X.; Yue, W.; Shang, L.; Wang, D.; Xu, Y.; Chen, Y.; Ge, J. Inhibition of CD44 Attenuates Pressure Overload-Induced Cardiac and Lung Inflammation, Fibrosis, and Heart Failure Progression. Eur. Heart J. 2020, 41, ehaa946.0878. [Google Scholar] [CrossRef]
- Osawa, Y.; Kawai, H.; Tsunoda, T.; Komatsu, H.; Okawara, M.; Tsutsui, Y.; Yoshida, Y.; Yoshikawa, S.; Mori, T.; Yamazoe, T.; et al. Cluster of Differentiation 44 Promotes Liver Fibrosis and Serves as a Biomarker in Congestive Hepatopathy. Hepatol. Commun. 2021, 5, 1437–1447. [Google Scholar] [CrossRef]
- Han, J.; Lee, C.; Jung, Y. Current Evidence and Perspectives of Cluster of Differentiation 44 in the Liver’s Physiology and Pathology. Int. J. Mol. Sci. 2024, 25, 4749. [Google Scholar] [CrossRef]
- Petukhov, D.; Richter-Dayan, M.; Fridlender, Z.; Breuer, R.; Wallach-Dayan, S.B. Increased Regeneration Following Stress-Induced Lung Injury in Bleomycin-Treated Chimeric Mice with CD44 Knockout Mesenchymal Cells. Cells 2019, 8, 1211. [Google Scholar] [CrossRef]
- Fernández-Tabanera, E.; Melero-Fernández de Mera, R.M.; Alonso, J. CD44 In Sarcomas: A Comprehensive Review and Future Perspectives. Front. Oncol. 2022, 12, 909450. [Google Scholar] [CrossRef]
- Hiraga, T.; Ito, S.; Nakamura, H. Cancer Stem-like Cell Marker CD44 Promotes Bone Metastases by Enhancing Tumorigenicity, Cell Motility, and Hyaluronan Production. Cancer Res. 2013, 73, 4112–4122. [Google Scholar] [CrossRef]
- Menko, A.S.; Romisher, A.; Walker, J.L. The Pro-Fibrotic Response of Mesenchymal Leader Cells to Lens Wounding Involves Hyaluronic Acid, Its Receptor RHAMM, and Vimentin. Front. Cell Dev. Biol. 2022, 10, 862423. [Google Scholar] [CrossRef]
- Lin, C.-H.; Hung, P.-H.; Chen, Y.-J. CD44 Is Associated with the Aggressive Phenotype of Nasopharyngeal Carcinoma through Redox Regulation. Int. J. Mol. Sci. 2013, 14, 13266–13281. [Google Scholar] [CrossRef]
- Paulis, Y.W.J.; Huijbers, E.J.M.; van der Schaft, D.W.J.; Soetekouw, P.M.M.B.; Pauwels, P.; Tjan-Heijnen, V.C.G.; Griffioen, A.W. CD44 Enhances Tumor Aggressiveness by Promoting Tumor Cell Plasticity. Oncotarget 2015, 6, 19634–19646. [Google Scholar] [CrossRef] [PubMed]
- Suleiman, M.; Abdulrahman, N.; Yalcin, H.; Mraiche, F. The Role of CD44, Hyaluronan and NHE1 in Cardiac Remodeling. Life Sci. 2018, 209, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wang, A.; Leng, P. Aberrant N-cadherin expression in cancer. Biomed. Pharmacother. 2019, 118, 109320. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Cai, Q.; Zhang, M.; Zhu, S.; Ma, Y.; Sun, L.; Jiang, N.; Tian, J.; Niu, X.; Chen, J.; et al. A Switch from CD44+ Cell to EMT Cell Drives the Metastasis of Prostate Cancer. Oncotarget 2014, 6, 1202–1216. [Google Scholar] [CrossRef]
- Yusupov, M.; Privat-Maldonado, A.; Cordeiro, R.M.; Verswyvel, H.; Shaw, P.; Razzokov, J.; Smits, E.; Bogaerts, A. Oxidative Damage to Hyaluronan–CD44 Interactions as an Underlying Mechanism of Action of Oxidative Stress-Inducing Cancer Therapy. Redox Biol. 2021, 43, 101968. [Google Scholar] [CrossRef]
- Nurwidya, F.; Takahashi, F.; Kato, M.; Baskoro, H.; Hidayat, M.; Wirawan, A.; Takahashi, K. CD44 Silencing Decreases the Expression of Stem Cell-Related Factors Induced by Transforming Growth Factor Β1 and Tumor Necrosis Factor α in Lung Cancer: Preliminary Findings. Bosn. J. Basic Med. Sci. 2017, 17, 228–234. [Google Scholar] [CrossRef]
- Tirella, A.; Kloc-Muniak, K.; Good, L.; Ridden, J.; Ashford, M.; Puri, S.; Tirelli, N. CD44 Targeted Delivery of siRNA by Using HA-Decorated Nanotechnologies for KRAS Silencing in Cancer Treatment. Int. J. Pharm. 2019, 561, 114–123. [Google Scholar] [CrossRef]
- Gul-Uludağ, H.; Valencia-Serna, J.; Kucharski, C.; Marquez-Curtis, L.A.; Jiang, X.; Larratt, L.; Janowska-Wieczorek, A.; Uludağ, H. Polymeric Nanoparticle-Mediated Silencing of CD44 Receptor in CD34+ Acute Myeloid Leukemia Cells. Leuk. Res. 2014, 38, 1299–1308. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Khalili, N.; Razi, S.; Keshavarz-Fathi, M.; Khalili, N.; Rezaei, N. Effects of Lead and Cadmium on the Immune System and Cancer Progression. J. Environ. Health Sci. Eng. 2020, 18, 335–343. [Google Scholar] [CrossRef]
- Ju, H.; Arumugam, P.; Lee, J.; Song, J.M. Impact of Environmental Pollutant Cadmium on the Establishment of a Cancer Stem Cell Population in Breast and Hepatic Cancer. ACS Omega 2017, 2, 563–572. [Google Scholar] [CrossRef]
- Li, J.; Jiang, H.; Zhu, Y.; Ma, Z.; Li, B.; Dong, J.; Xiao, C.; Hu, A. Fine Particulate Matter (PM2.5) Induces the Stem Cell-like Properties of Hepatocellular Carcinoma by Activating ROS/Nrf2/Keap1-Mediated Autophagy. Ecotoxicol. Environ. Saf. 2024, 272, 116052. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Liang, F.; Cheng, W.; Zhou, R.; Wu, X.; Feng, Y.; Wang, Y. The Mechanisms for Lung Cancer Risk of PM2.5: Induction of Epithelial-Mesenchymal Transition and Cancer Stem Cell Properties in Human Non-Small Cell Lung Cancer Cells. Environ. Toxicol. 2017, 32, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Alexis, N.E.; Diaz-Sanchez, D.; Neas, L.M.; Harder, S.; Herbst, M.C.; Cascio, W.E.; Buse, J.B.; Peters, A.; Devlin, R.B. Ambient PM2.5 Exposure Up-Regulates the Expression of Costimulatory Receptors on Circulating Monocytes in Diabetic Individuals. Environ. Health Perspect. 2011, 119, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Figure 3. From LncRNA NEAT1 Contributes to the Acquisition of a Tumor Like-Phenotype Induced by PM 2.5 in Lung Bronchial Epithelial Cells via HIF-1α Activation. Available online: https://media.springernature.com/full/springer-static/image/art%3A10.1007%2Fs11356-021-13735-7/MediaObjects/11356_2021_13735_Fig3_HTML.png (accessed on 18 July 2025).
- Su, Y.; Gao, J.; Kaur, P.; Wang, Z. Neutrophils and Macrophages as Targets for Development of Nanotherapeutics in Inflammatory Diseases. Pharmaceutics 2020, 12, 1222. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, S.Y.; Wang, T.-Y.; Bai, Y.; Zhong, M.; Wang, A.; Lippmann, M.; Chen, L.-C.; Rajagopalan, S.; Sun, Q. Inflammatory Response to Fine Particulate Air Pollution Exposure: Neutrophil versus Monocyte. PLoS ONE 2013, 8, e71414. [Google Scholar] [CrossRef]
- Figure 4. From LncRNA NEAT1 Contributes to the Acquisition of a Tumor Like-Phenotype Induced by PM 2.5 in Lung Bronchial Epithelial Cells via HIF-1α Activation. Available online: https://media.springernature.com/full/springer-static/image/art%3A10.1007%2Fs11356-021-13735-7/MediaObjects/11356_2021_13735_Fig4_HTML.png?as=webp (accessed on 18 July 2025).
- Pan, J.; Liu, P.; Yu, X.; Zhang, Z.; Liu, J. The Adverse Role of Endocrine Disrupting Chemicals in the Reproductive System. Front. Endocrinol. 2024, 14, 1324993. [Google Scholar] [CrossRef]
- Singh, R.D.; Koshta, K.; Tiwari, R.; Khan, H.; Sharma, V.; Srivastava, V. Developmental Exposure to Endocrine Disrupting Chemicals and Its Impact on Cardio-Metabolic-Renal Health. Front. Toxicol. 2021, 3, 663372. [Google Scholar] [CrossRef]
- Endocrine-Disrupting Chemicals’ (EDCs) Effects on Tumour Microenvironment and Cancer Progression: Emerging Contribution of RACK1. Available online: https://www.mdpi.com/1422-0067/21/23/9229 (accessed on 18 July 2025).
- Erkekoglu, P.; Kocer-Gumusel, B.; Erkekoglu, P.; Kocer-Gumusel, B. Environmental Effects of Endocrine-Disrupting Chemicals: A Special Focus on Phthalates and Bisphenol A. In Environmental Health Risk—Hazardous Factors to Living Species; IntechOpen: London, UK, 2016; ISBN 978-953-51-2402-3. [Google Scholar]
- Primeaux, M.; Gowrikumar, S.; Dhawan, P. Role of CD44 Isoforms in Epithelial-Mesenchymal Plasticity and Metastasis. Clin. Exp. Metastasis 2022, 39, 391–406. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The Role of CD44 in Epithelial–Mesenchymal Transition and Cancer Development. Onco Targets Ther. 2015, 8, 3783–3792. [Google Scholar] [CrossRef]
- Pesonen, M.; Vähäkangas, K. Contribution of Common Plastic-Related Endocrine Disruptors to Epithelial-Mesenchymal Transition (EMT) and Tumor Progression. Chemosphere 2022, 309, 136560. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, F.; Wu, J.; Liu, L.; Li, Y.; Yu, G.; Duan, H.; Jiang, Y.; Liu, S.; He, Y.; et al. Associations of Phthalates with NAFLD and Liver Fibrosis: A Nationally Representative Cross-Sectional Study from NHANES 2017 to 2018. Front. Nutr. 2022, 9, 1059675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhen, H.; Cheng, H.; Hu, F.; Jia, Y.; Huang, B.; Jiang, M. Di-(2-Ethylhexyl) Phthalate Exposure Induces Liver Injury by Promoting Ferroptosis via Downregulation of GPX4 in Pregnant Mice. Front. Cell Dev. Biol. 2022, 10, 1014243. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Kodama, K.; Wei, K.; Tolentino, L.L.; Choi, O.; Engleman, E.G.; Butte, A.J.; McLaughlin, T. The Receptor CD44 Is Associated with Systemic Insulin Resistance and Proinflammatory Macrophages in Human Adipose Tissue. Diabetologia 2015, 58, 1579–1586. [Google Scholar] [CrossRef]
- Kodama, K.; Toda, K.; Morinaga, S.; Yamada, S.; Butte, A.J. Anti-CD44 Antibody Treatment Lowers Hyperglycemia and Improves Insulin Resistance, Adipose Inflammation, and Hepatic Steatosis in Diet-Induced Obese Mice. Diabetes 2015, 64, 867–875. [Google Scholar] [CrossRef]
- Khan, N.G.; Correia, J.; Adiga, D.; Rai, P.S.; Dsouza, H.S.; Chakrabarty, S.; Kabekkodu, S.P. A Comprehensive Review on the Carcinogenic Potential of Bisphenol A: Clues and Evidence. Environ. Sci. Pollut. Res. Int. 2021, 28, 19643–19663. [Google Scholar] [CrossRef]
- Ryszawy, D.; Pudełek, M.; Kochanowski, P.; Janik-Olchawa, N.; Bogusz, J.; Rąpała, M.; Koczurkiewicz, P.; Mikołajczyk, J.; Borek, I.; Kędracka-Krok, S.; et al. High Bisphenol A Concentrations Augment the Invasiveness of Tumor Cells through Snail-1/Cx43/ERRγ-Dependent Epithelial-Mesenchymal Transition. Toxicol. Vitr. 2020, 62, 104676. [Google Scholar] [CrossRef]
- Bora, S.S.; Gogoi, R.; Sharma, M.R.; Anshu; Borah, M.P.; Deka, P.; Bora, J.; Naorem, R.S.; Das, J.; Teli, A.B. Microplastics and Human Health: Unveiling the Gut Microbiome Disruption and Chronic Disease Risks. Front. Cell. Infect. Microbiol. 2024, 14, 1492759. [Google Scholar] [CrossRef]
- Aliya, S.; Alhammadi, M.; Ilangovan, S.; Han, S.; Tamang, S.; Son, B.; Lee, H.U.; Huh, Y.S. Microplastics: An Emerging Environmental Risk Factor for Gut Microbiota Dysbiosis and Cancer Development? Environ. Chem. Ecotoxicol. 2025, 7, 706–728. [Google Scholar] [CrossRef]
- Sofield, C.E.; Anderton, R.S.; Gorecki, A.M. Mind over Microplastics: Exploring Microplastic-Induced Gut Disruption and Gut-Brain-Axis Consequences. Curr. Issues Mol. Biol. 2024, 46, 4186–4202. [Google Scholar] [CrossRef]
- Yang, W.; Jannatun, N.; Zeng, Y.; Liu, T.; Zhang, G.; Chen, C.; Li, Y. Impacts of Microplastics on Immunity. Front. Toxicol. 2022, 4, 956885. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Hou, J.; Liao, Y.; Wei, F.; Xing, B. Polyethylene Microplastics Impede the Innate Immune Response by Disrupting the Extracellular Matrix and Signaling Transduction. iScience 2023, 26, 107390. [Google Scholar] [CrossRef] [PubMed]
- Limonta, G.; Mancia, A.; Benkhalqui, A.; Bertolucci, C.; Abelli, L.; Fossi, M.C.; Panti, C. Microplastics Induce Transcriptional Changes, Immune Response and Behavioral Alterations in Adult Zebrafish. Sci. Rep. 2019, 9, 15775. [Google Scholar] [CrossRef] [PubMed]
- Zhi, L.; Li, Z.; Su, Z.; Wang, J. Immunotoxicity of Microplastics: Carrying Pathogens and Destroying the Immune System. TrAC Trends Anal. Chem. 2024, 177, 117817. [Google Scholar] [CrossRef]
- Cheng, Y.; Yang, Y.; Bai, L.; Cui, J. Microplastics: An Often-Overlooked Issue in the Transition from Chronic Inflammation to Cancer. J. Transl. Med. 2024, 22, 959. [Google Scholar] [CrossRef]
- Dzierżyński, E.; Gawlik, P.J.; Puźniak, D.; Flieger, W.; Jóźwik, K.; Teresiński, G.; Forma, A.; Wdowiak, P.; Baj, J.; Flieger, J. Microplastics in the Human Body: Exposure, Detection, and Risk of Carcinogenesis: A State-of-the-Art Review. Cancers 2024, 16, 3703. [Google Scholar] [CrossRef]
- Osman, A.I.; Hosny, M.; Eltaweil, A.S.; Omar, S.; Elgarahy, A.M.; Farghali, M.; Yap, P.-S.; Wu, Y.-S.; Nagandran, S.; Batumalaie, K.; et al. Microplastic Sources, Formation, Toxicity and Remediation: A Review. Environ. Chem. Lett. 2023, 21, 2129–2169. [Google Scholar] [CrossRef]
- Li, S.; Li, C.; Zhang, Y.; He, X.; Chen, X.; Zeng, X.; Liu, F.; Chen, Y.; Chen, J. Targeting Mechanics-Induced Fibroblast Activation through CD44-RhoA-YAP Pathway Ameliorates Crystalline Silica-Induced Silicosis. Theranostics 2019, 9, 4993–5008. [Google Scholar] [CrossRef]

| Variant (dbSNP ID) | Amino Acid Change | Structural Location | Functional Description and Biological Relevance |
|---|---|---|---|
| VAR_006490 (rs3694738421) | Arg46Pro (arginine → proline) | Extracellular domain (N-terminal) | This variant underlies the Indian (In) blood group system and differentiates Ina and Inb antigens on erythrocytes. The proline-coding allele determines the rare Ina antigen, which may elicit alloimmune transfusion reactions. It is not pathogenic in systemic health but has significant clinical relevance in immunohematology. |
| VAR_030325 (rs11607491) | Substitution at position 393 | Extracellular domain | Although its functional consequences are not fully characterized, the variant is located in the ligand-binding region and may affect CD44’s interaction with hyaluronan or extracellular matrix components. |
| VAR_021147 (rs96666074) | Substitution at position 417 | Extracellular domain | Located near known glycosylation sites, this substitution may alter CD44 glycosylation patterns, potentially impacting receptor-ligand interactions or immune recognition. |
| VAR_030326 (rs14675589) | Substitution at position 479 | Juxtamembrane region (extracellular–transmembrane junction) | This variant may affect the membrane topology of CD44 and its susceptibility to proteolytic cleavage (shedding), thereby influencing the levels of soluble CD44 (sCD44) and downstream signaling. |
| VAR_030327 (rs122733971) | Substitution at position 494 | Transmembrane domain or adjacent region | This mutation may modulate the anchoring of CD44 in the membrane and alter its interactions with neighboring receptors or lipid rafts, influencing receptor clustering and signaling efficiency. |
| Ligand Type | Ligand Name | Description and Biological Functions |
|---|---|---|
| Glycosaminoglycans (GAGs) | Hyaluronic acid (HA) | Primary ligand of CD44; regulates adhesion, migration, proliferation, differentiation, and inflammatory response |
| Heparan sulfate (HS) | Facilitates the binding of growth factors, present in proteoglycan forms of CD44 | |
| Chondroitin sulfate (CS) | Associated with the CD44v isoform; influences interactions with the ECM | |
| Extracellular matrix proteins | Osteopontin (OPN, SPP1) | Modulation of cell migration and adhesion; expression in inflammation and tumorigenesis |
| Fibronectin (FN) | Adhesion and signaling; interacts with integrins and CD44 | |
| Laminin | Supports epithelial cell interactions with the basement membrane | |
| Collagen type I, II, III, IV | Effect on cell adhesion, migration, and invasion | |
| Adhesive proteins | Selectins (L-, E-selectin)—via HCELL form | Participation in leukocyte rolling is meaningful in the immune response |
| ICAM-1 (ang. intercellular adhesion molecule 1) | Supporting role in lymphocyte transmigration | |
| Signaling proteins and growth factors | TGF-β, HGF, VEGF, EGF—indirectly through complexes with HA or heparan | CD44 enables local presentation and concentration of growth signals |
| Proteases | Matrix metalloproteinases (MMP-2, MMP-9) | Interactions with CD44 promote ECM degradation and cell migration. |
| Acute phase proteins | Pentraxin 3 (PTX3) | Regulation of inflammatory response, interaction with HA and CD44 |
| Other | Serpins, complement proteins | Less frequently described ligands: immunological and proteolytic significance |
| Receptors and co-receptors (e.g., EGFR, TGF-βR)—signaling complexes | They cooperate with CD44 in the activation of PI3K, MAPK, and Rho-GTPase pathways |
| Feature | Ezrin | Radixin | Moesin |
|---|---|---|---|
| Gene symbol (human) | EZR | RDX | MSN |
| Subcellular localization | Cell membrane, microvilli, surface of cellular projections | Intercellular junctions, cortical cytoplasm | Microvilli, lamellipodia, and ECM contact zones |
| Biological function | Links the actin cytoskeleton to the plasma membrane; involved in shaping microvilli and cell adhesion | Stabilizes the plasma membrane and intercellular junctions; regulates membrane elasticity | Regulates cell shape, leukocyte transmigration, and inflammatory response |
| Interactions with CD44 | Direct interaction with CD44’s cytoplasmic domain; involved in targeting CD44 to microvilli and lipid rafts | Anchors CD44 at sites of intercellular contact and cytoskeletal reorganization | Stabilizes CD44 in migrating immune cells; involved in Rho/Rac pathway activation |
| Functional significance of CD44–ERM | Integrates extracellular signals with intracellular responses; mechanical coupling of adhesion and signaling | Maintains cell polarity and membrane tension; supports CD44 signaling | Enhances pro-inflammatory response; involved in cytoskeletal reorganization during migration |
| Diseases associated with dysfunction | Cancers (e.g., gastric cancer, leukemias), kidney diseases, viral infections (e.g., HIV, EBV) | Hearing loss (RDX mutations), liver and gallbladder cancers | Chronic inflammatory conditions, lymphomas, head and neck cancers |
| Segment | Amino Acid Range | Function |
|---|---|---|
| Extracellular domain | 21–649 | Binding to hyaluronic acid (HA) and other ECM ligands; glycosylation modifications. |
| Transmembrane segment | 650–670 | Anchoring in the membrane, defining N-/C-terminal orientation |
| Cytoplasmic domain | 671–742 | Interactions with ERM proteins, signal transduction, and regulatory phosphorylation |
| Type of Modification | Location/Target Residues | Functional Description | Biological Significance |
|---|---|---|---|
| N-glycosylation | Asn within the Asn-X-Ser/Thr motif (e.g., in the LINK domain) | Attachment of N-glycans | Essential for proper folding, stability, and affinity for HA |
| O-glycosylation | Ser/Thr (especially in variable splice regions, e.g., Thr-637/638) | Addition of short sugar chains and glycosaminoglycans (e.g., chondroitin sulfate) | Modulates ligand recognition, protects from proteolysis, and affects cell migration |
| Phosphorylation | Ser-672, Ser-706; other Ser/Thr residues in the C-terminal tail | Regulates receptor activation status and interactions with adaptor proteins (e.g., ERM) | Alters adhesion and migration signaling, influences cytoskeletal organization |
| Proteolysis (shedding) | Near the transmembrane domain (extracellular side) | Cleavage of the extracellular domain by MMPs and other proteases | Reduces surface CD44 expression, generates soluble form (sCD44), and is potentially further cleaved by γ-secretase and nuclear signaling |
| Ubiquitination | Lys-704, Lys-715 (in the cytoplasmic tail) | Covalent modification affecting intracellular trafficking and degradation | May regulate receptor abundance and endosomal sorting |
| Organ | Experimental Model | Key Observations | Functional Conclusions |
|---|---|---|---|
| Lungs (IPF) | Bleomycin-induced pulmonary fibrosis in mice; HAS2 overexpression; CD44 knockout |
| The HA–CD44 axis is essential for the pro-fibrotic phenotype in the lungs; CD44 blockade reduces fibrosis and fibroblast invasiveness. |
| Skin (SSc) | Patient-derived fibroblasts from systemic sclerosis; sCD44 concentration analysis; wound healing in CD44 KO mice |
| CD44 may support both wound healing and pathological skin sclerosis; sCD44 potentially exerts protective effects |
| Heart | Ang II-induced fibrosis; pressure overload model; CD44 KO |
| CD44 mediates cardiac remodeling by integrating inflammatory and fibrotic signals; its inhibition halts fibrosis progression |
| Liver | Hepatic congestion model (IVC ligation); CD44 and HA immunohistochemistry; CD44 neutralization (IM7) |
| CD44 activates stellate cells and promotes fibrosis independently of inflammation; it represents a promising therapeutic target and biomarker |
| Type of Pollutant | Impact on CD44 and HA | Effects in Connective Tissue | Molecular Mechanisms/Experimental Data |
|---|---|---|---|
| Heavy metals (lead, cadmium) | • Decreased expression of CD44 and CD58 on erythrocytes (e.g., in children exposed to e-waste) • Increased CD44 expression on macrophages and neutrophils at metal accumulation sites | • Impaired reparative and immune functions • Chronic inflammation • Enhanced organ fibrosis | • Lead reduces cell adhesion molecule expression (CD44/CD58), impairing ECM-cell interactions • HA fragmentation acts as a DAMP, activating immune responses via CD44 |
| Particulate matter (PM2.5, silica dust) | • CD44 mediates the recruitment of inflammatory cells to the lungs in response to ozone • CD44 blockade in silicosis models reduces collagen deposition | • Chronic lung inflammation • Pulmonary fibrosis (e.g., silicosis, fibrosing alveolitis) • Bronchial hyperreactivity | • CD44 loss protects against ozone-induced response (despite HA increase) • CD44 is essential for fibroblast activation and inflammatory cell migration • PM nanoparticles → oxidative stress, Th2 polarization, epigenetic activation of pro-fibrotic genes |
| Endocrine-disrupting chemicals (EDCs) (bisphenol A, phthalates, pesticides) | • Induction of CD44 expression in epithelial cells undergoing EMT • Increased CD44 on tissue macrophages (e.g., in adipose tissue) | • EMT and mesenchymal transition • Chronic inflammation • Fibrosis (e.g., in liver, lung, adipose tissue) | • BPA activates ERRγ → EMT in A549 cells • Phthalates increase collagen deposition and leukocyte infiltration in the liver • EDCs enhance HA and osteopontin production—both CD44 ligands |
| Microplastics | • Induce CD44 expression on macrophages and fibroblasts surrounding particles • Indirect CD44 activation via DAMPs and cytokines | • Granuloma formation around microplastics • Chronic inflammation and fibrosis in lungs, liver, intestines (“plasticosis”) | • CD44- and HA-dependent migration and activation of myofibroblasts • Microplastic particles act as toxin carriers (e.g., metals, PCBs), amplifying inflammatory response • Mechanism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedrycz-Wieczorska, A.; Chylińska-Wrzos, P.; Grzywacz, A.; Zieliński, E.; Bartosiński, A.; Kędziora-Kornatowska, K.; Lis-Sochocka, M.; Mertowska, P.; Mertowski, S.; Bojarski, K.; et al. CD44 as a Central Integrator of Inflammation and Fibrosis: From Molecular Signaling to Environmental Modulation. Int. J. Mol. Sci. 2025, 26, 8870. https://doi.org/10.3390/ijms26188870
Pedrycz-Wieczorska A, Chylińska-Wrzos P, Grzywacz A, Zieliński E, Bartosiński A, Kędziora-Kornatowska K, Lis-Sochocka M, Mertowska P, Mertowski S, Bojarski K, et al. CD44 as a Central Integrator of Inflammation and Fibrosis: From Molecular Signaling to Environmental Modulation. International Journal of Molecular Sciences. 2025; 26(18):8870. https://doi.org/10.3390/ijms26188870
Chicago/Turabian StylePedrycz-Wieczorska, Agnieszka, Patrycja Chylińska-Wrzos, Anna Grzywacz, Ewa Zieliński, Andrzej Bartosiński, Kornelia Kędziora-Kornatowska, Marta Lis-Sochocka, Paulina Mertowska, Sebastian Mertowski, Krzysztof Bojarski, and et al. 2025. "CD44 as a Central Integrator of Inflammation and Fibrosis: From Molecular Signaling to Environmental Modulation" International Journal of Molecular Sciences 26, no. 18: 8870. https://doi.org/10.3390/ijms26188870
APA StylePedrycz-Wieczorska, A., Chylińska-Wrzos, P., Grzywacz, A., Zieliński, E., Bartosiński, A., Kędziora-Kornatowska, K., Lis-Sochocka, M., Mertowska, P., Mertowski, S., Bojarski, K., Rahnama-Hezavah, M., Urbanowicz, T., & Grywalska, E. (2025). CD44 as a Central Integrator of Inflammation and Fibrosis: From Molecular Signaling to Environmental Modulation. International Journal of Molecular Sciences, 26(18), 8870. https://doi.org/10.3390/ijms26188870






