Investigating SMR Peptide Interactions with Breast Cancer-Associated Proteins
Abstract
1. Introduction
2. Results
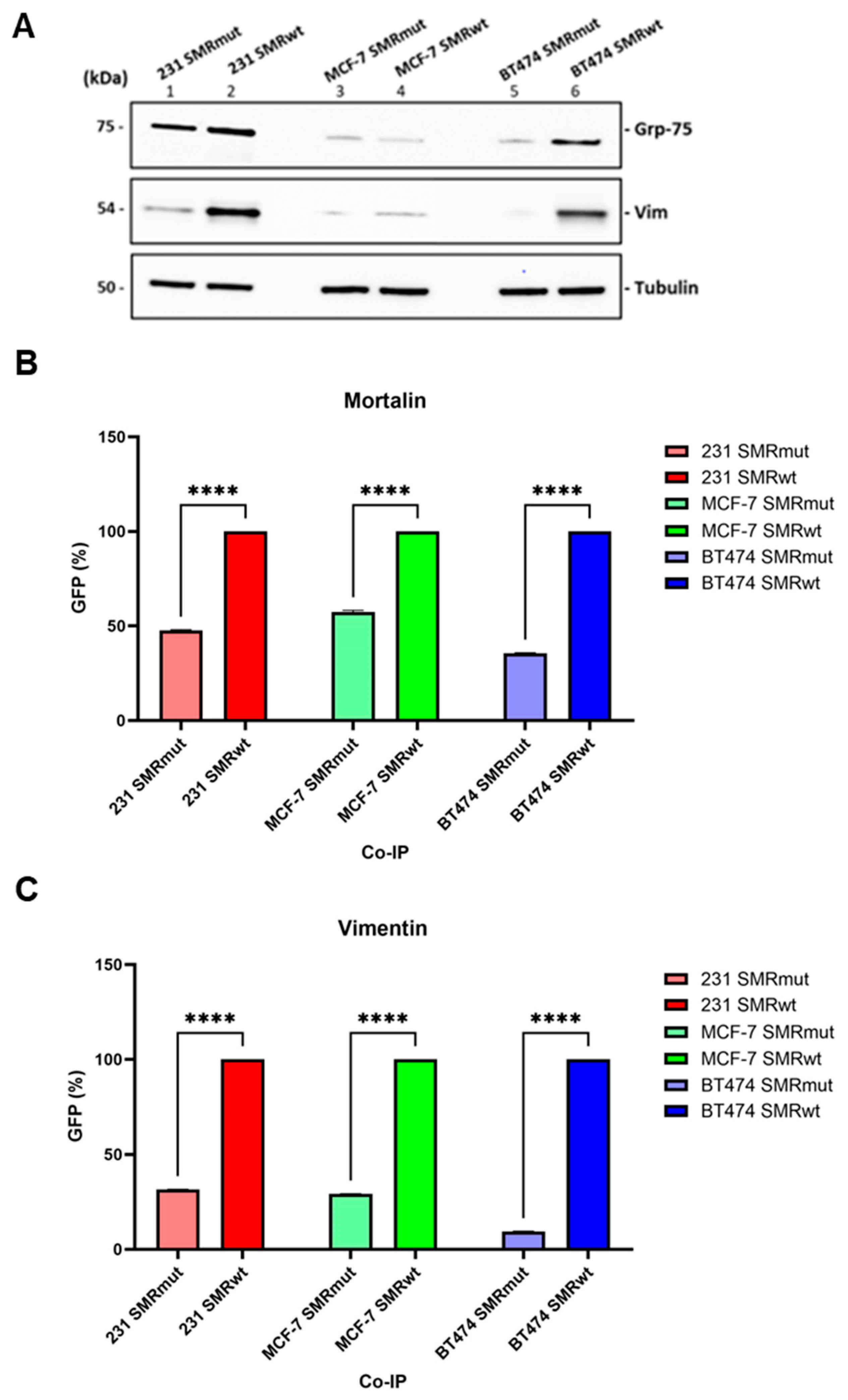
2.1. SMRwt Peptide Interacts with Host Cell Proteins Mortalin and Vimentin in BC Cell Lines
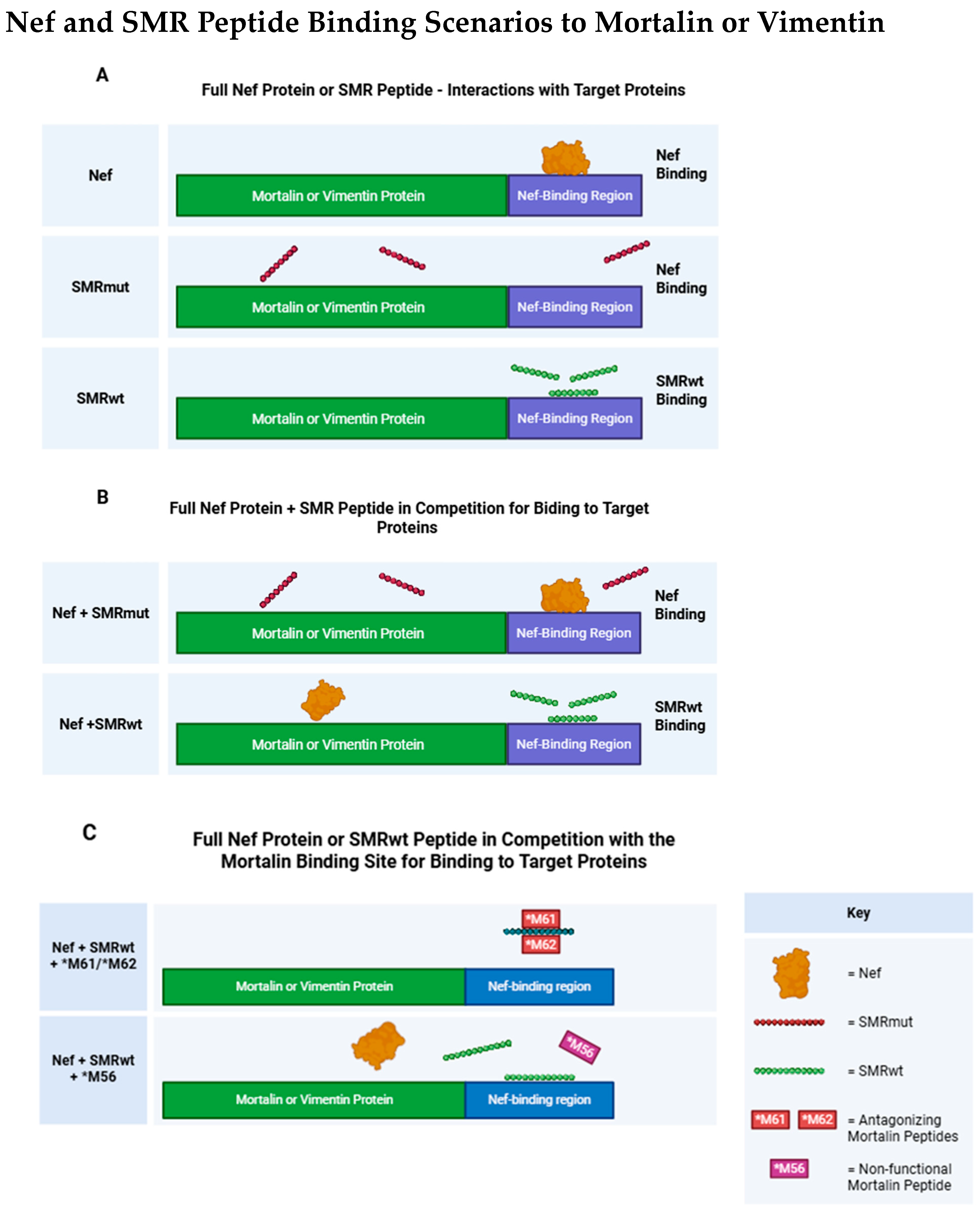
2.2. Peptide Sequence Motifs Specific to HIV Nef-SMR, Mortalin, and Vimentin: Roles in Protein Binding and Interaction
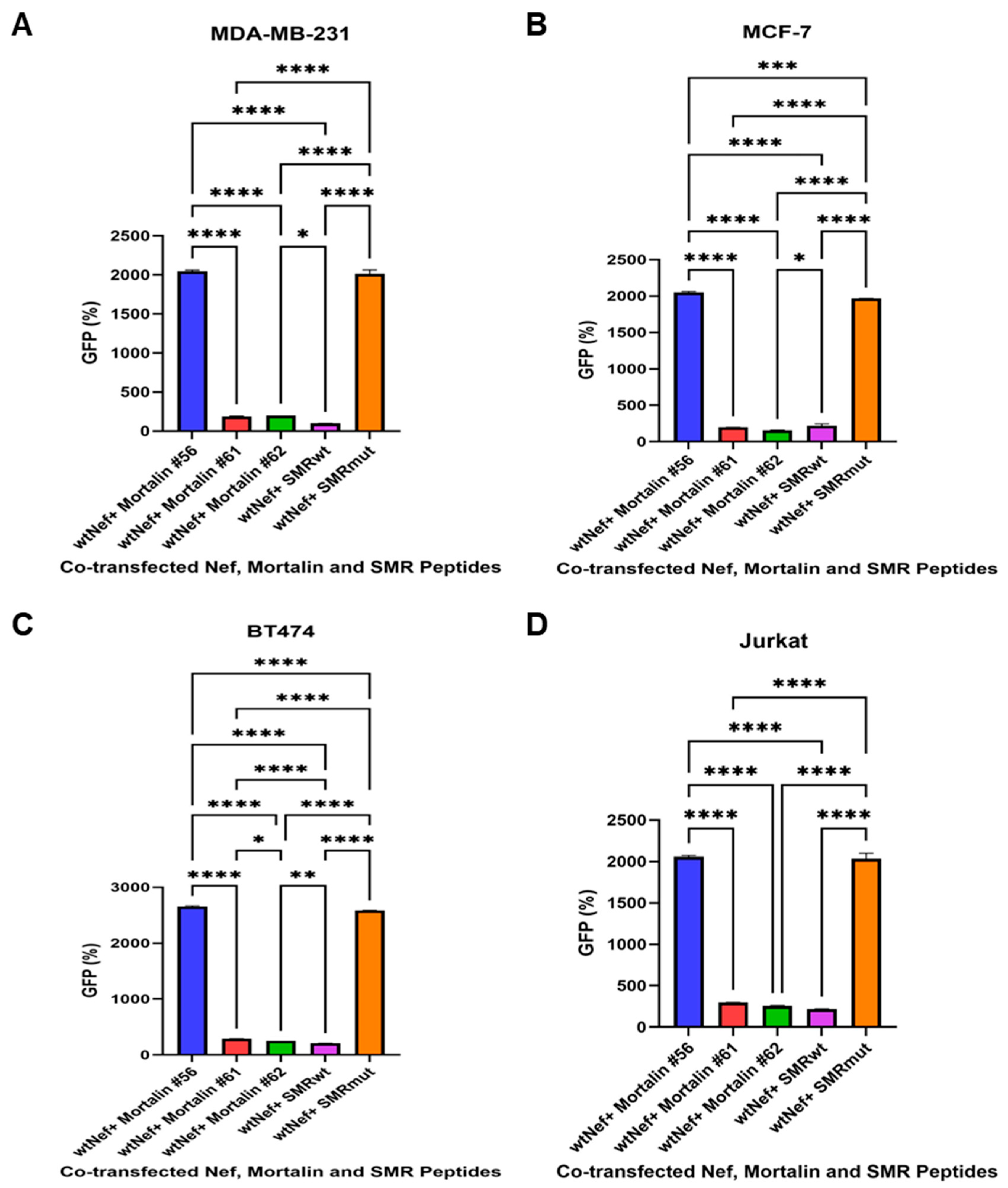
2.3. The Effects of Mortalin-Derived Peptides on the Secretion of EVs Containing Nef-GFP (exNef) on Three BC Cells and Jurkat Cells
2.4. Impact of SMR, Mortalin, and Vimentin Peptides on Exosome Secretion in MDA-MB-231 and MCF-7 BC Cells
2.5. Mortalin Peptides Suppress exNef and Tumor Exosome Release
2.5.1. Mortalin-Derived Peptides Inhibit exNef Secretion in Jurkat T Cells
2.5.2. Mortalin-Derived Peptides Suppress Tumor Exosome Secretion in MDA-MB-231 Cells
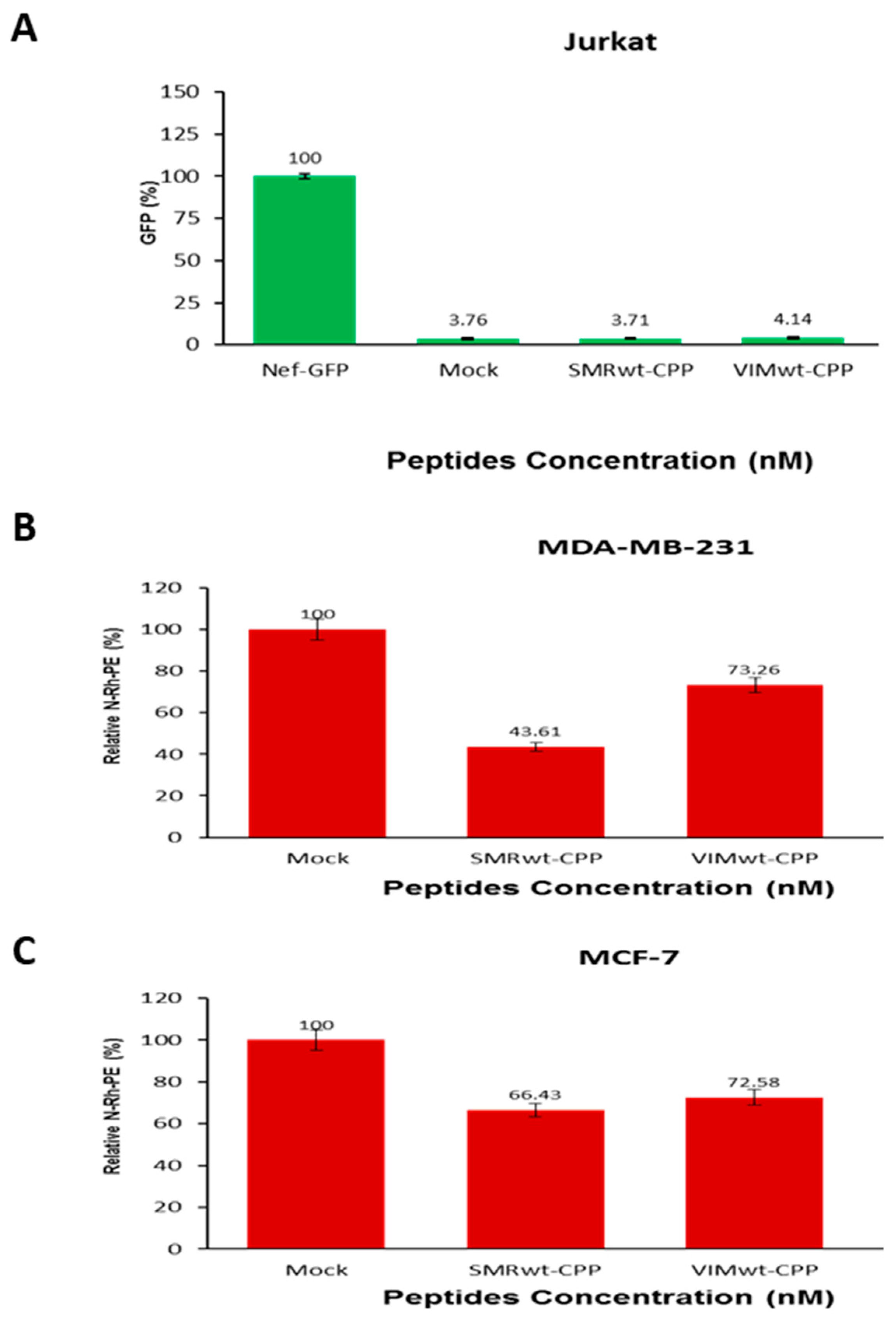
2.6. Targeted Peptides Attenuate EV Release in Jurkat Cells and Breast Cancer Cells
2.6.1. SMRwt and VIMwt Peptides Reduce Extracellular Vesicle Secretion in Jurkat T Cells
2.6.2. SMRwt and VIMwt Peptides Suppress EV Secretion in Breast Cancer Cells
2.7. Comparative Analysis of Binding Affinity Between HIV-1 Nef, SMRwt/SMRmut Peptides, and Target Proteins Mortalin and Vimentin Using SPR
2.8. SPR Kinetic Study of the Interaction Between Mortalin Peptides and Nef Reveals Differential Binding Affinity
2.9. SPR Analysis of Competition Binding Between SMR Peptide and Mortalin Peptides to Vimentin and Mortalin Proteins
2.10. Competitive Binding of Mortalin Peptides with SMRwt: Implications for Mortalin and Vimentin Interactions
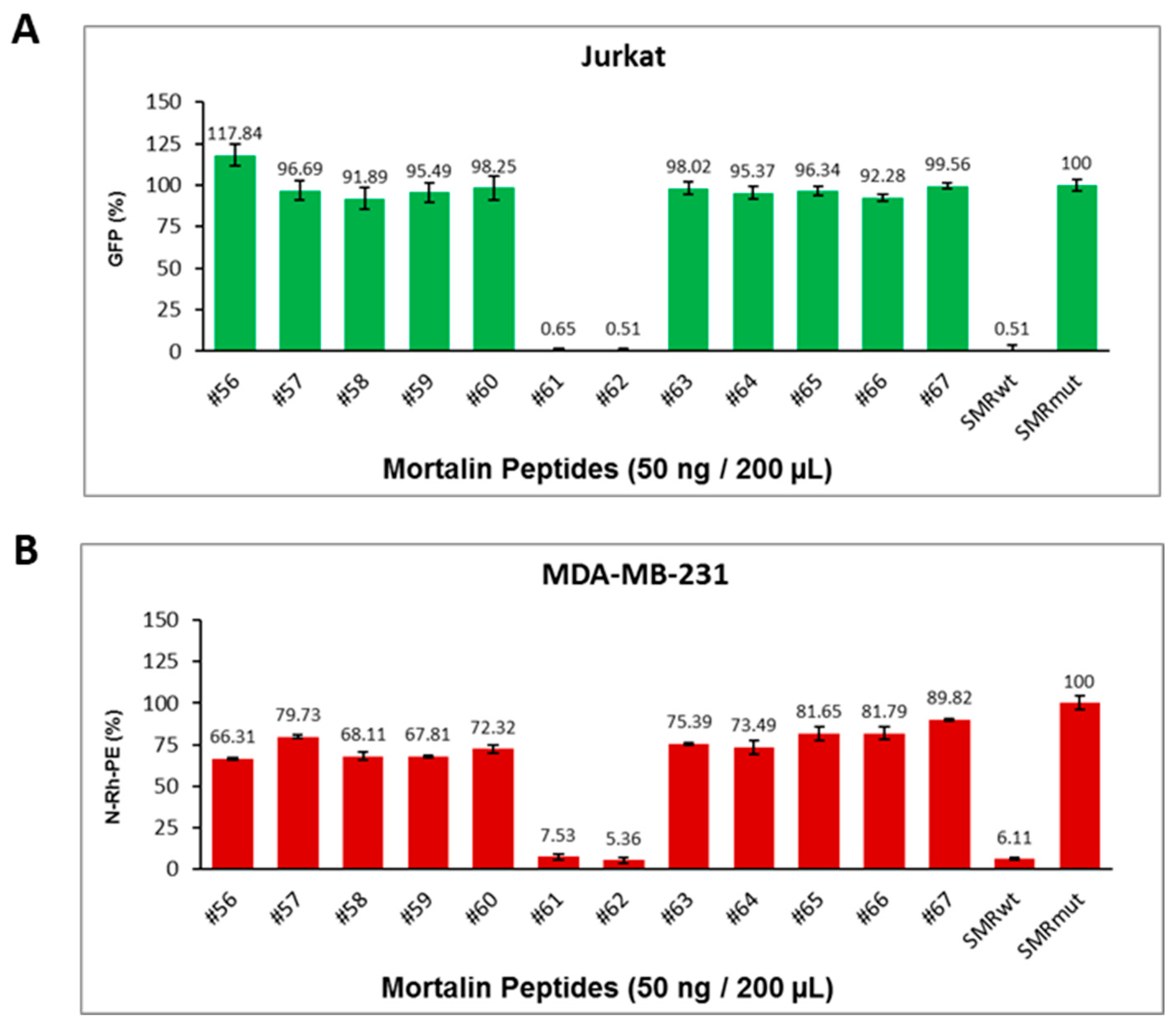
2.11. Development and Testing of Mortalin-Derived Scanning Peptides for Effects on EV Secretion
2.12. Identification of a Vimentin-Derived Peptide with Homology to the Mortalin SMR Binding Sequence
3. Discussion
Conclusions and Future Directions
4. Materials and Methods
4.1. Cell Culture
4.2. Antibodies
4.3. SMR Peptides
4.4. Proteins
4.5. Surface Plasmon Resonance (SPR)
4.6. Cell Transfection and Transfections with Peptides
4.7. Co-Immunoprecipitation (Co-IP)
4.8. Western Immunoblot Analysis
4.9. Exosome Isolation and Purification
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast Cancer |
| EV | Extracellular Vesicle |
| SPR | Surface Plasmon Resonance |
| SMR | Secretion Modification Region |
| EMT | Epithelial–Mesenchymal Transition |
References
- Marino, N.; Woditschka, S.; Reed, L.T.; Nakayama, J.; Mayer, M.; Wetzel, M.; Steeg, P.S. Breast cancer metastasis: Issues for the personalization of its prevention and treatment. Am. J. Pathol. 2013, 183, 1084–1095. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, T.; Hooda, J.; Atkinson, J.M.; Whiteside, T.L.; Oesterreich, S.; Lee, A.V. Exosomes in Breast Cancer—Mechanisms of Action and Clinical Potential. Mol. Cancer Res. 2021, 19, 935–945. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, X.; Sun, C.; Huang, X.; Li, J.; Fu, Z.; Li, W.; Yin, Y. The Advancing Roles of Exosomes in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 731062. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer. 2021, 144, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Piombino, C.; Mastrolia, I.; Omarini, C.; Candini, O.; Dominici, M.; Piacentini, F.; Toss, A. The Role of Exosomes in Breast Cancer Diagnosis. Biomedicines 2021, 9, 312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Y.; Ma, Q.; Wang, T.; Xing, J.; Li, Q.; Wang, D.; Wang, G. The involvement and application potential of exosomes in breast cancer immunotherapy. Front. Immunol. 2024, 15, 1384946. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kok, V.C.; Yu, C.C. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, H.X.; Gires, O. Tumor-derived extracellular vesicles in breast cancer: From bench to bedside. Cancer Lett. 2019, 460, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Mohan, V.; Krishnaswamy, V.R.; Solomonov, I.; Sagi, I. Exosomes as a storehouse of tissue remodeling proteases and mediators of cancer progression. Cancer Metastasis Rev. 2019, 38, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Huang, W.; Tan, Q.Y.; Fan, X.Q.; Jiang, Y.G.; Liu, L.; Zhong, Y.Y.; Liang, Y.G.; Wang, R.W. Breast cancer anti-estrogen resistance protein 1 (BCAR1/p130cas) in pulmonary disease tissue and serum. Mol. Diagn. Ther. 2011, 15, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- E-Kobon, T.; Thongararm, P.; Roytrakul, S.; Meesuk, L.; Chumnanpuen, P. Prediction of anticancer peptides against MCF-7 breast cancer cells from the peptidomes of Achatina fulica mucus fractions. Comput. Struct. Biotechnol. J. 2016, 14, 49–57. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marqus, S.; Pirogova, E.; Piva, T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017, 24, 21. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chu, H.L.; Yip, B.S.; Chen, K.H.; Yu, H.Y.; Chih, Y.H.; Cheng, H.T.; Chou, Y.T.; Cheng, J.W. Novel antimicrobial peptides with high anticancer activity and selectivity. PLoS ONE 2015, 10, e0126390. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chih, Y.H.; Lin, Y.S.; Yip, B.S.; Wei, H.J.; Chu, H.L.; Yu, H.Y.; Cheng, H.T.; Chou, Y.T.; Cheng, J.W. Ultrashort Antimicrobial Peptides with Antiendotoxin Properties. Antimicrob. Agents Chemother. 2015, 59, 5052–5056. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lumongga, F.; Dharmajaya, R.; Siregar, K.; Delyuzar, D.; Handjari, D.; Jusuf, N.; Munir, D.; Asrul, A. Correlation Between Intensity of Vimentin Immuno-expression in Young Women with Triple Negative Breast Cancer and Its Cliniocopathological Parameters. Med. Arch. 2022, 76, 454–457. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grasset, E.M.; Dunworth, M.; Sharma, G.; Loth, M.; Tandurella, J.; Cimino-Mathews, A.; Gentz, M.; Bracht, S.; Haynes, M.; Fertig, E.J.; et al. Triple-negative breast cancer metastasis involves complex epithelial-mesenchymal transition dynamics and requires vimentin. Sci. Transl. Med. 2022, 14, eabn7571. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, H.; Wang, Y.; Liu, C.; Li, W.; Zhou, F.; Wang, X.; Zheng, J. The Apolipoprotein C1 is involved in breast cancer progression via EMT and MAPK/JNK pathway. Pathol. Res. Pract. 2022, 229, 153746. [Google Scholar] [CrossRef] [PubMed]
- Thalla, D.G.; Jung, P.; Bischoff, M.; Lautenschlager, F. Role of Extracellular Vimentin in Cancer-Cell Functionality and Its Influence on Cell Monolayer Permeability Changes Induced by SARS-CoV-2 Receptor Binding Domain. Int. J. Mol. Sci. 2021, 22, 7469. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, Z.; Fang, Z.; Ma, J. Regulatory mechanisms and clinical significance of vimentin in breast cancer. Biomed. Pharmacother. 2021, 133, 111068. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Q.; Li, Y.J.; Wei, Z.J.; Fan, Y.; Li, X.D.; Chen, Z.M.; Hou, D.Y.; Xiao, W.Y.; Ding, M.R.; Wang, H.; et al. Binding-Induced Fibrillogenesis Peptides Recognize and Block Intracellular Vimentin Skeletonization against Breast Cancer. Nano Lett. 2021, 21, 6202–6210. [Google Scholar] [CrossRef] [PubMed]
- Korsching, E.; Packeisen, J.; Liedtke, C.; Hungermann, D.; Wulfing, P.; van Diest, P.J.; Brandt, B.; Boecker, W.; Buerger, H. The origin of vimentin expression in invasive breast cancer: Epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J. Pathol. 2005, 206, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; He, Y. Challenges of Convalescent Plasma Therapy on COVID-19. J. Clin. Virol. 2020, 127, 104358. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, H.; Zhu, W.; Zhang, L.; Lei, H.; Wu, X.; Guo, L.; Chen, X.; Wang, Y.; Tang, H. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci. Rep. 2015, 5, 8421. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arrindell, J.; Desnues, B. Vimentin: From a cytoskeletal protein to a critical modulator of immune response and a target for infection. Front. Immunol. 2023, 14, 1224352. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Berr, A.L.; Wiese, K.; Dos Santos, G.; Koch, C.M.; Anekalla, K.R.; Kidd, M.; Davis, J.M.; Cheng, Y.; Hu, Y.S.; Ridge, K.M. Vimentin is required for tumor progression and metastasis in a mouse model of non-small cell lung cancer. Oncogene 2023, 42, 2074–2087. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, J.I. Editorial: The role of mortalin in biology and disease. Front. Cell Dev. Biol. 2023, 11, 1196430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Londono, C.; Osorio, C.; Gama, V.; Alzate, O. Mortalin, apoptosis, and neurodegeneration. Biomolecules 2012, 2, 143–164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Esfahanian, N.; Knoblich, C.D.; Bowman, G.A.; Rezvani, K. Mortalin: Protein partners, biological impacts, pathological roles, and therapeutic opportunities. Front. Cell Dev. Biol. 2023, 11, 1028519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yoon, A.R.; Wadhwa, R.; Kaul, S.C.; Yun, C.O. Why is Mortalin a Potential Therapeutic Target for Cancer? Front. Cell. Dev. Biol. 2022, 10, 914540. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wei, B.; Cao, J.; Tian, J.H.; Yu, C.Y.; Huang, Q.; Yu, J.J.; Ma, R.; Wang, J.; Xu, F.; Wang, L.B. Mortalin maintains breast cancer stem cells stemness via activation of Wnt/GSK3beta/beta-catenin signaling pathway. Am. J. Cancer Res. 2021, 11, 2696–2716. [Google Scholar] [PubMed] [PubMed Central]
- Kim, S.Y.; Byrn, R.; Groopman, J.; Baltimore, D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: Evidence for differential gene expression. J. Virol. 1989, 63, 3708–3713. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Campbell, T.D.; Khan, M.; Huang, M.B.; Bond, V.C.; Powell, M.D. HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethn Dis. 2008, 18 (Suppl. S2), S2-14-9. [Google Scholar] [PubMed] [PubMed Central]
- Ali, S.A.; Huang, M.B.; Campbell, P.E.; Roth, W.W.; Campbell, T.; Khan, M.; Newman, G.; Villinger, F.; Powell, M.D.; Bond, V.C. Genetic characterization of HIV type 1 Nef-induced vesicle secretion. AIDS Res. Hum. Retroviruses 2010, 26, 173–192. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, H.; Ji, M.; Chen, L.; Liu, Q.; Che, S.; Xu, M.; Lin, Z. The clinicopathological significance of Mortalin overexpression in invasive ductal carcinoma of breast. J. Exp. Clin. Cancer Res. 2016, 35, 42. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Masoud, V.; Pages, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elwakeel, A. Abrogating the Interaction Between p53 and Mortalin (Grp75/HSPA9/mtHsp70) for Cancer Therapy: The Story so far. Front. Cell Dev. Biol. 2022, 10, 879632. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rai, R.; Kennedy, A.L.; Isingizwe, Z.R.; Javadian, P.; Benbrook, D.M. Similarities and Differences of Hsp70, hsc70, Grp78 and Mortalin as Cancer Biomarkers and Drug Targets. Cells 2021, 10, 2996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, P.K.; Hong, S.K.; Starenki, D.; Oshima, K.; Shao, H.; Gestwicki, J.E.; Tsai, S.; Park, J.I. Mortalin/HSPA9 targeting selectively induces KRAS tumor cell death by perturbing mitochondrial membrane permeability. Oncogene 2020, 39, 4257–4270. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wadhwa, R.; Colgin, L.; Yaguchi, T.; Taira, K.; Reddel, R.R.; Kaul, S.C. Rhodacyanine dye MKT-077 inhibits in vitro telomerase assay but has no detectable effects on telomerase activity in vivo. Cancer Res. 2002, 62, 4434–4438. [Google Scholar] [PubMed]
- Wadhwa, R.; Sugihara, T.; Yoshida, A.; Nomura, H.; Reddel, R.R.; Simpson, R.; Maruta, H.; Kaul, S.C. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000, 60, 6818–6821. [Google Scholar] [PubMed]
- Alfadhalah, T.; Elamir, H. Exploring leadership styles in government hospitals in Kuwait. Leadersh Health Serv. 2019, 32, 458–476. [Google Scholar] [CrossRef] [PubMed]
- Barlin, M.; Erdmann-Gilmore, P.; Mudd, J.L.; Zhang, Q.; Seymour, R.W.; Guo, Z.; Miessner, J.R.; Goedegebuure, S.P.; Bi, Y.; Osorio, O.A.; et al. Proteins in Tumor-Derived Plasma Extracellular Vesicles Indicate Tumor Origin. Mol. Cell Proteom. 2023, 22, 100476. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, M.B.; Gonzalez, R.R.; Lillard, J.; Bond, V.C. Secretion modification region-derived peptide blocks exosome release and mediates cell cycle arrest in breast cancer cells. Oncotarget 2017, 8, 11302–11315. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, M.B.; Wu, J.Y.; Lillard, J.; Bond, V.C. SMR peptide antagonizes mortalin promoted release of extracellular vesicles and affects mortalin protection from complement-dependent cytotoxicity in breast cancer cells and leukemia cells. Oncotarget 2019, 10, 5419–5438. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, M.B.; Brena, D.; Wu, J.Y.; Roth, W.W.; Owusu, S.; Bond, V.C. Novel secretion modification region (SMR) peptide exhibits anti-metastatic properties in human breast cancer cells. Sci. Rep. 2022, 12, 13204. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Miller-Kleinhenz, J.; Guo, X.; Qian, W.; Zhou, H.; Bozeman, E.N.; Zhu, L.; Ji, X.; Wang, Y.A.; Styblo, T.; O’Regan, R.; et al. Dual-targeting Wnt and uPA receptors using peptide conjugated ultra-small nanoparticle drug carriers inhibited cancer stem-cell phenotype in chemo-resistant breast cancer. Biomaterials 2018, 152, 47–62. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Peess, C.; von Proff, L.; Goller, S.; Andersson, K.; Gerg, M.; Malmqvist, M.; Bossenmaier, B.; Schraml, M. Deciphering the stepwise binding mode of HRG1beta to HER3 by surface plasmon resonance and interaction map. PLoS ONE 2015, 10, e0116870. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Douzi, B. Protein-Protein Interactions: Surface Plasmon Resonance. Methods Mol. Biol. 2017, 1615, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gouw, M.; Michael, S.; Samano-Sanchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Calyseva, J.; et al. ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dinkel, H.; Van Roey, K.; Michael, S.; Kumar, M.; Uyar, B.; Altenberg, B.; Milchevskaya, V.; Schneider, M.; Kuhn, H.; Behrendt, A.; et al. ELM 2016--data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016, 44, D294–D300. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Zeke, A.; Lazar, T.; Glavina, J.; Nagy-Kanta, E.; Donagh, J.M.; Kalman, Z.E.; Pascarelli, S.; et al. ELM-the Eukaryotic Linear Motif resource-2024 update. Nucleic Acids Res. 2024, 52, D442–D455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Drescher, D.G.; Selvakumar, D.; Drescher, M.J. Analysis of Protein Interactions by Surface Plasmon Resonance. Adv. Protein Chem. Struct. Biol. 2018, 110, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Myszka, D.G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997, 8, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Hahnefeld, C.; Drewianka, S.; Herberg, F.W. Determination of kinetic data using surface plasmon resonance biosensors. Methods Mol. Med. 2004, 94, 299–320. [Google Scholar] [CrossRef] [PubMed]
- Kabakov, A.E.; Gabai, V.L. HSP70s in Breast Cancer: Promoters of Tumorigenesis and Potential Targets/Tools for Therapy. Cells 2021, 10, 3446. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Na, Y.; Kaul, S.C.; Ryu, J.; Lee, J.S.; Ahn, H.M.; Kaul, Z.; Kalra, R.S.; Li, L.; Widodo, N.; Yun, C.O.; et al. Stress chaperone mortalin contributes to epithelial-mesenchymal transition and cancer metastasis. Cancer Res. 2016, 76, 2754–2765. [Google Scholar] [CrossRef] [PubMed]



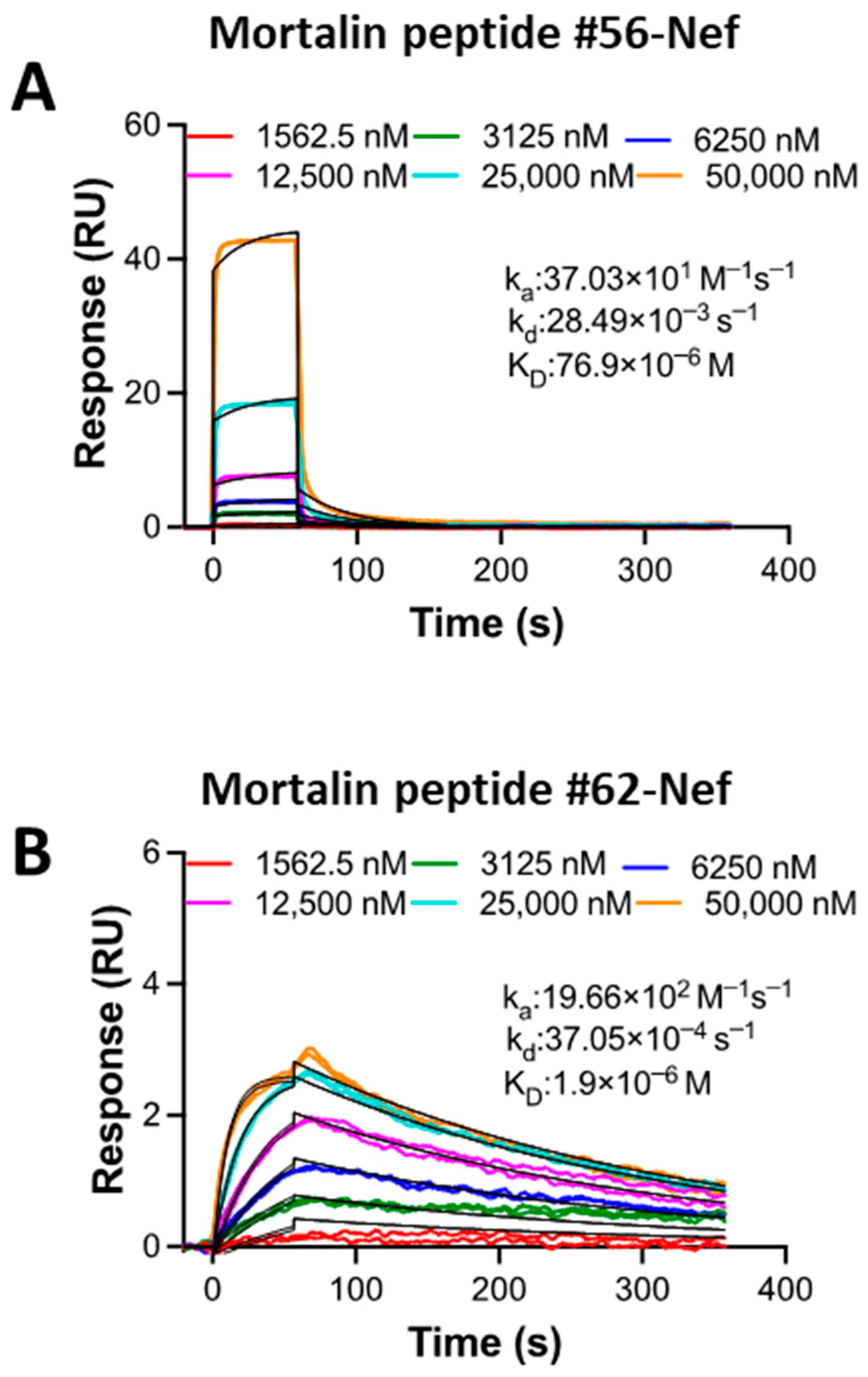
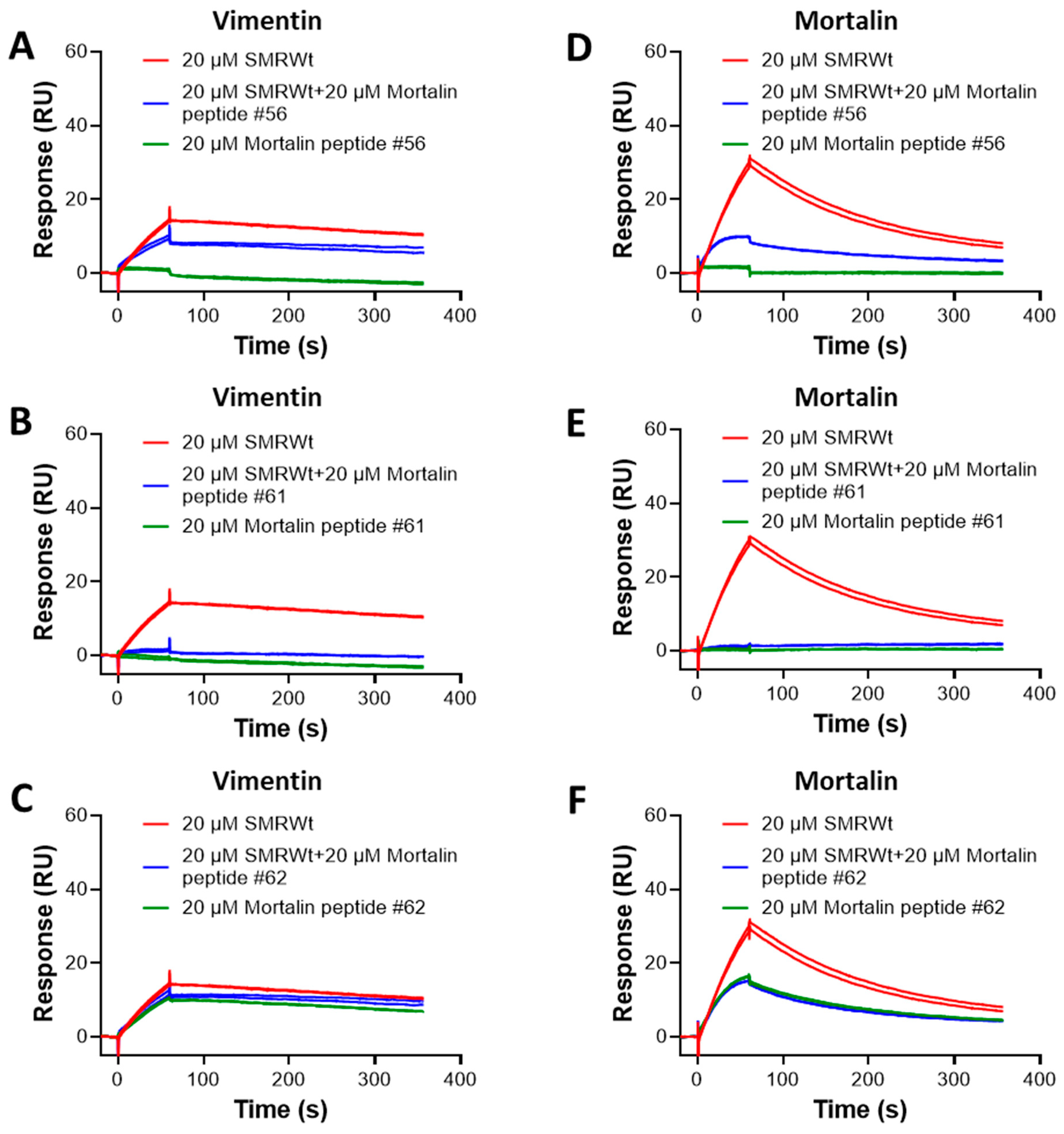
| Analyte | Ligand | |
|---|---|---|
| Vimentin KD | Mortalin KD | |
| SMRwt | 6.63 +/− 0.74 µM | 20.73 +/− 2.33 µM |
| SMRmut | 5.03 +/− 2.12 µM | NB |
| Nef | 0.75 +/− 1.1 nM | 3.16 +/− 0.03 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.-B.; Tiwari, P.B.; Üren, A.; Shelton, M.N.; Brena, D.; Wu, J.Y.; Khan, M.B.; Powell, M.D.; Stiles, J.K.; Johnson, E.L.; et al. Investigating SMR Peptide Interactions with Breast Cancer-Associated Proteins. Int. J. Mol. Sci. 2025, 26, 8848. https://doi.org/10.3390/ijms26188848
Huang M-B, Tiwari PB, Üren A, Shelton MN, Brena D, Wu JY, Khan MB, Powell MD, Stiles JK, Johnson EL, et al. Investigating SMR Peptide Interactions with Breast Cancer-Associated Proteins. International Journal of Molecular Sciences. 2025; 26(18):8848. https://doi.org/10.3390/ijms26188848
Chicago/Turabian StyleHuang, Ming-Bo, Purushottam B. Tiwari, Aykut Üren, Martin N. Shelton, Dara Brena, Jennifer Y. Wu, Mahfuz B. Khan, Michael D. Powell, Jonathan K. Stiles, Erica L. Johnson, and et al. 2025. "Investigating SMR Peptide Interactions with Breast Cancer-Associated Proteins" International Journal of Molecular Sciences 26, no. 18: 8848. https://doi.org/10.3390/ijms26188848
APA StyleHuang, M.-B., Tiwari, P. B., Üren, A., Shelton, M. N., Brena, D., Wu, J. Y., Khan, M. B., Powell, M. D., Stiles, J. K., Johnson, E. L., Yan, F., Yang, L., & Bond, V. C. (2025). Investigating SMR Peptide Interactions with Breast Cancer-Associated Proteins. International Journal of Molecular Sciences, 26(18), 8848. https://doi.org/10.3390/ijms26188848








