3-Hydroxy-β-ionone Suppresses Breast Cancer Progression by Inducing Apoptosis and Blocking EMT Through the TGF-β/Smad Signaling Pathway
Abstract
1. Introduction
2. Results
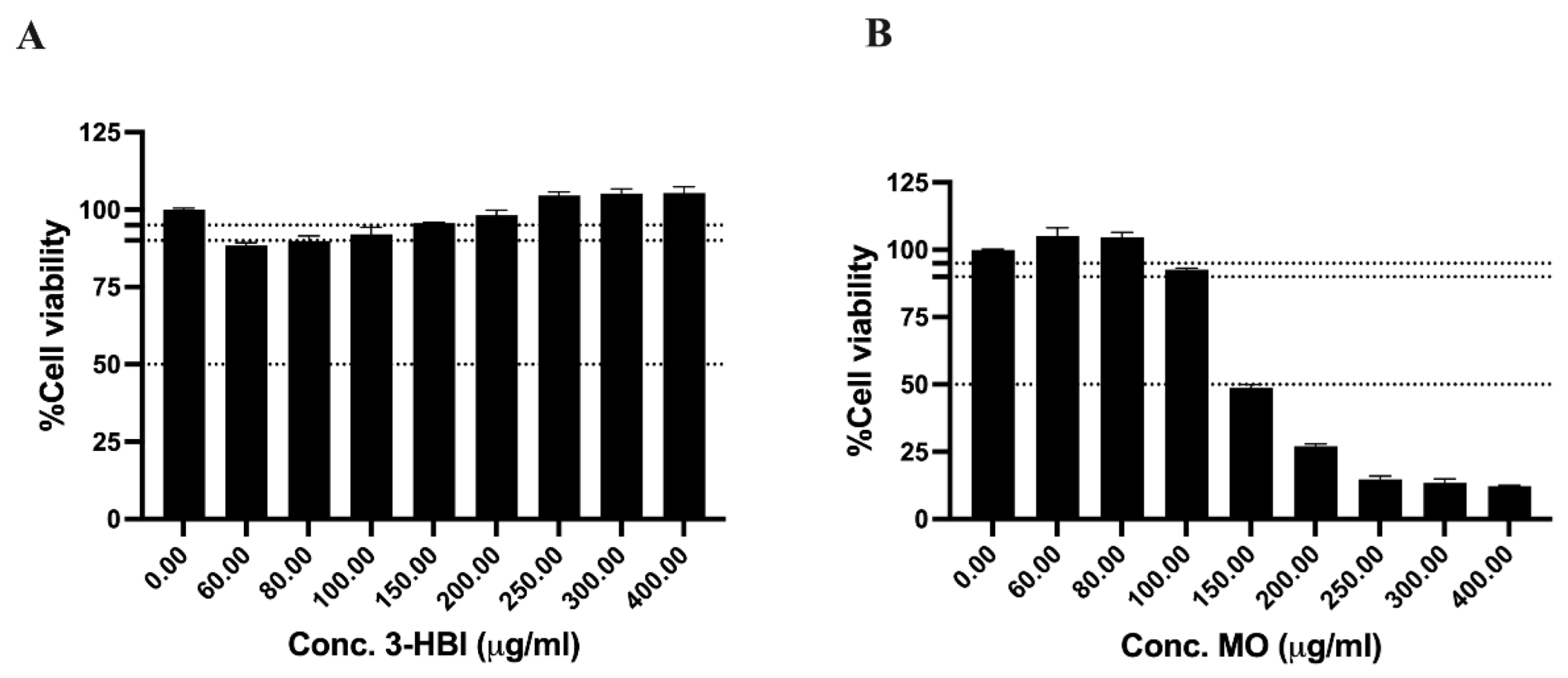
2.1. The Cytotoxicity of 3-Hydroxy-β-ionone (3-HBI)-Identified from Moringa oleifera Lam. (MO) Extract on Breast Cancer Cell Lines
2.2. The Cytotoxicity of 3-HBI on the Fibroblast Cell Line
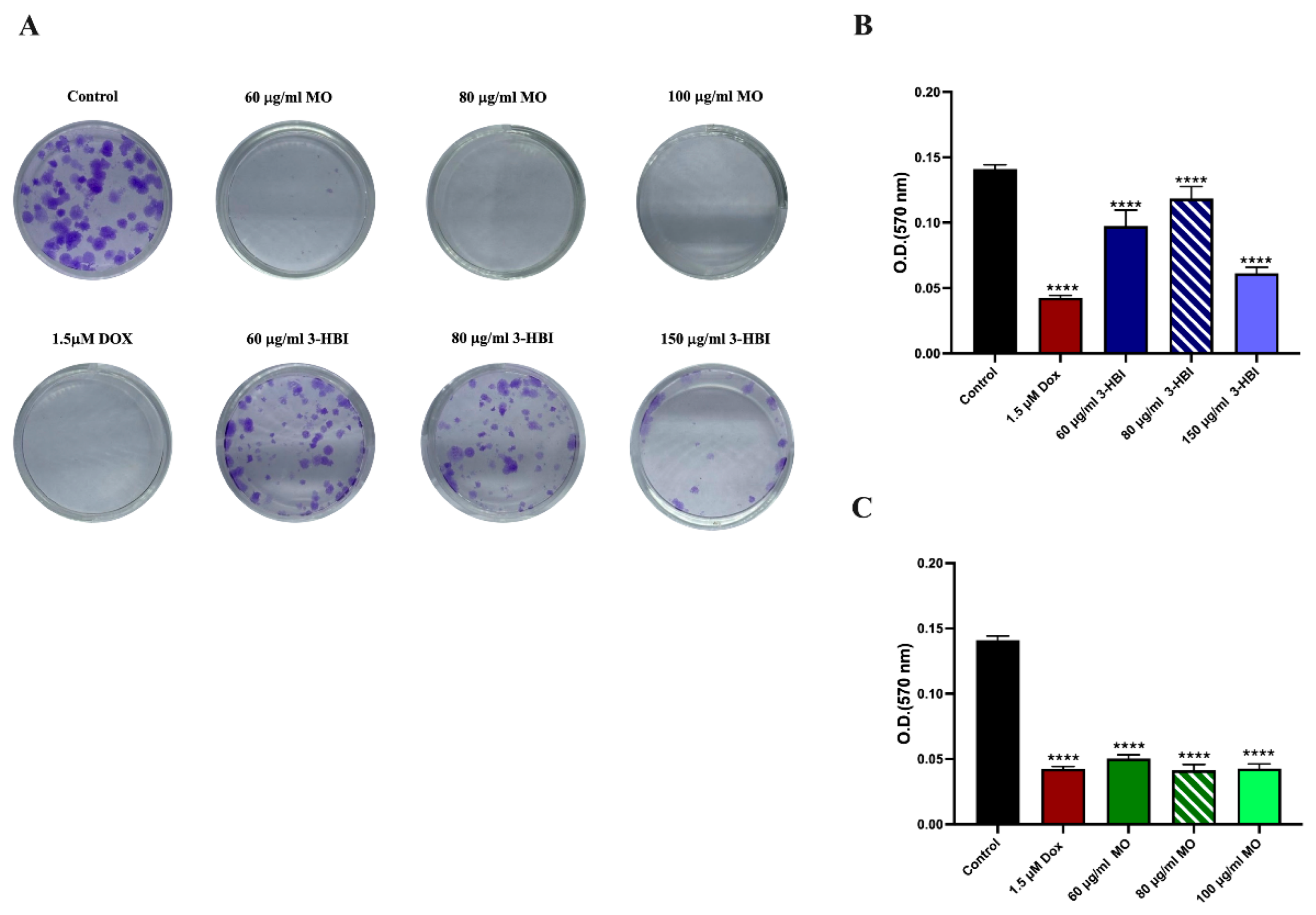
2.3. 3-HBI Inhibited TNBC Colony Formation
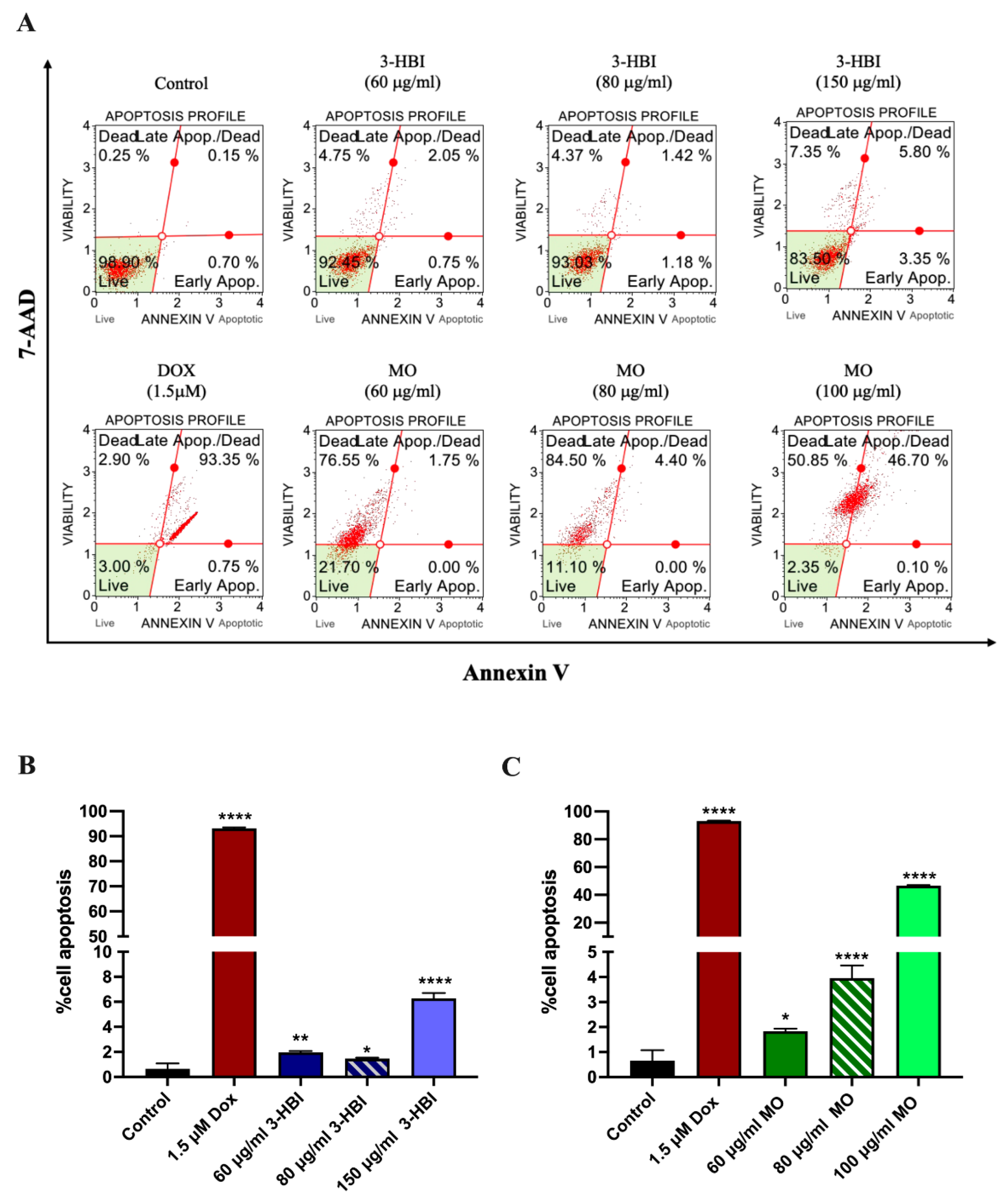
2.4. The Role of 3-HBI on Breast Cancer Cell Apoptosis
2.4.1. In Silico Binding to Apoptotic Regulators
2.4.2. 3-HBI-Induced Breast Cancer Cell Apoptosis
2.4.3. The Effect of 3-HBI on Genes and Proteins in the Apoptosis Pathway
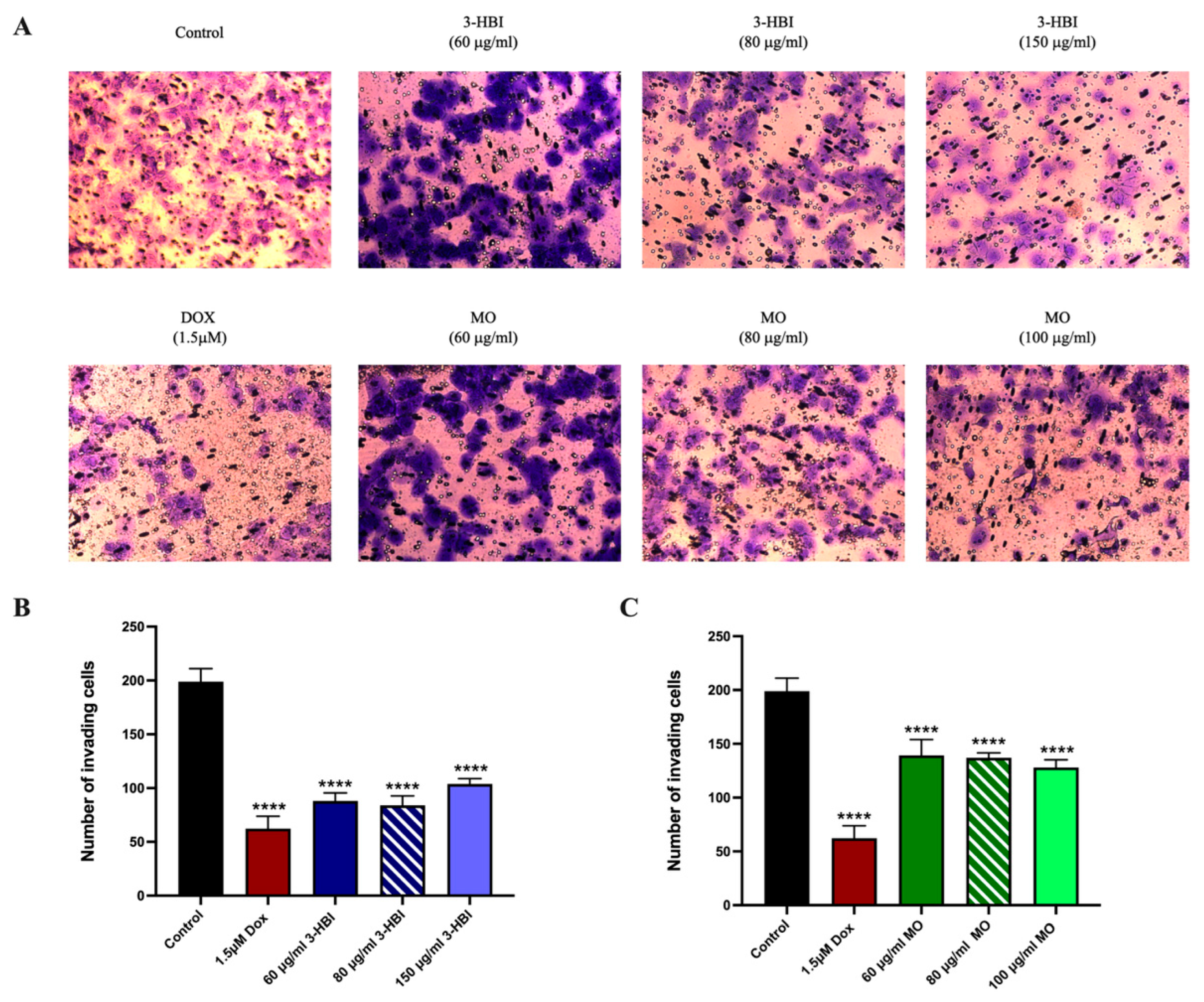
2.5. 3-HBI Inhibits TNBC Metastasis
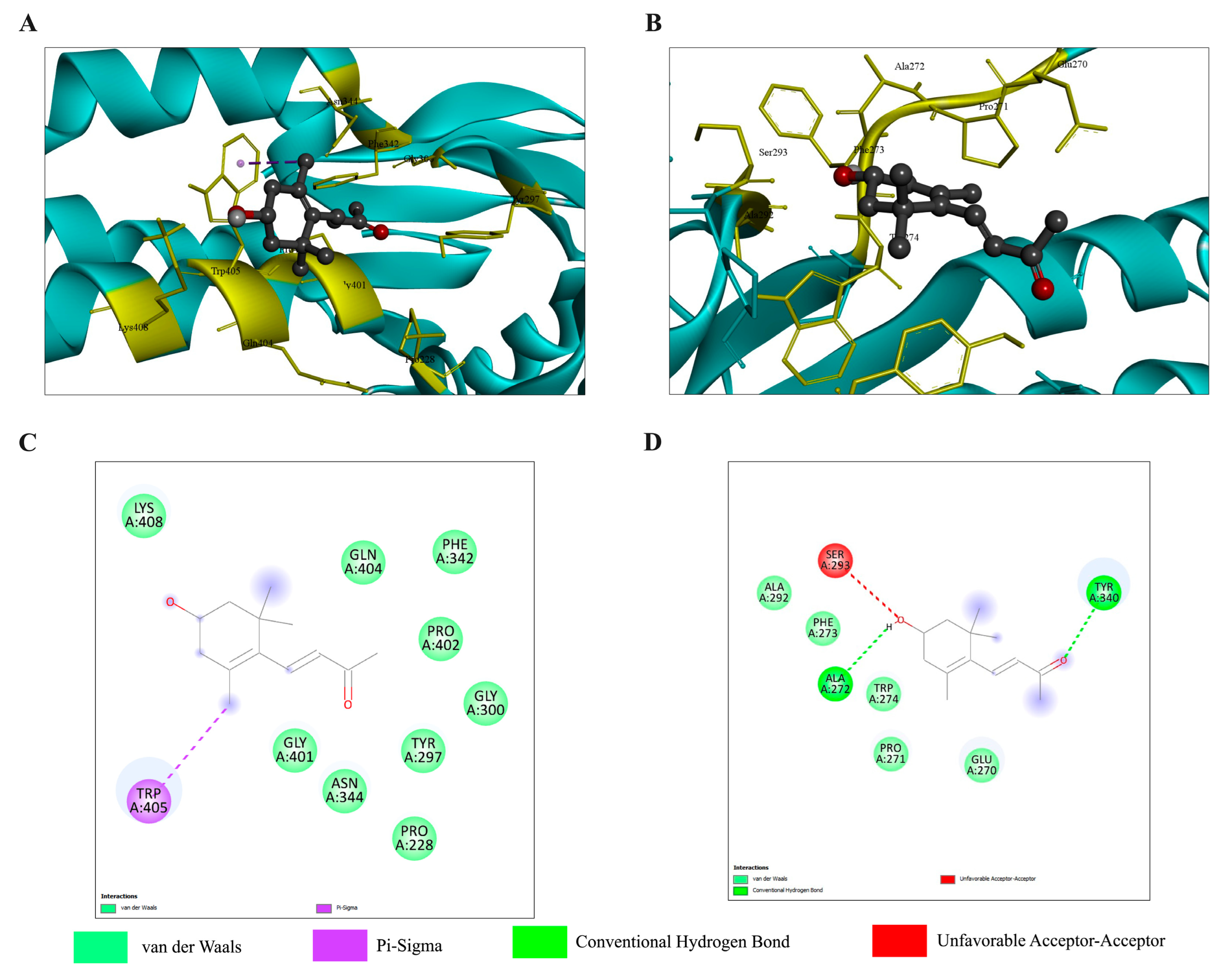
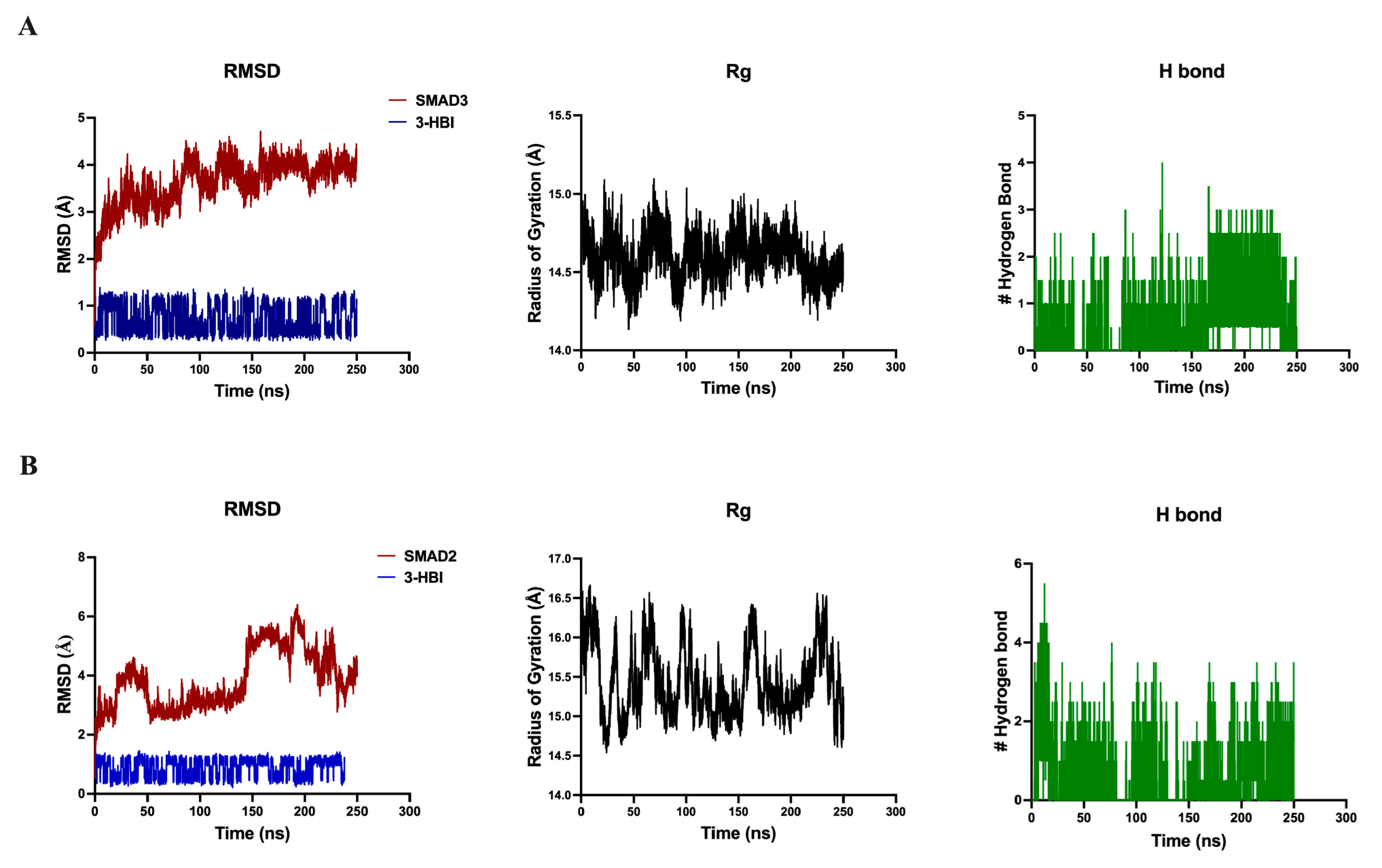
2.5.1. Molecular Docking and Molecular Dynamics Simulation with the TGF-β/Smad Pathway
2.5.2. 3-HBI Inhibited TNBC Migration and Invasion
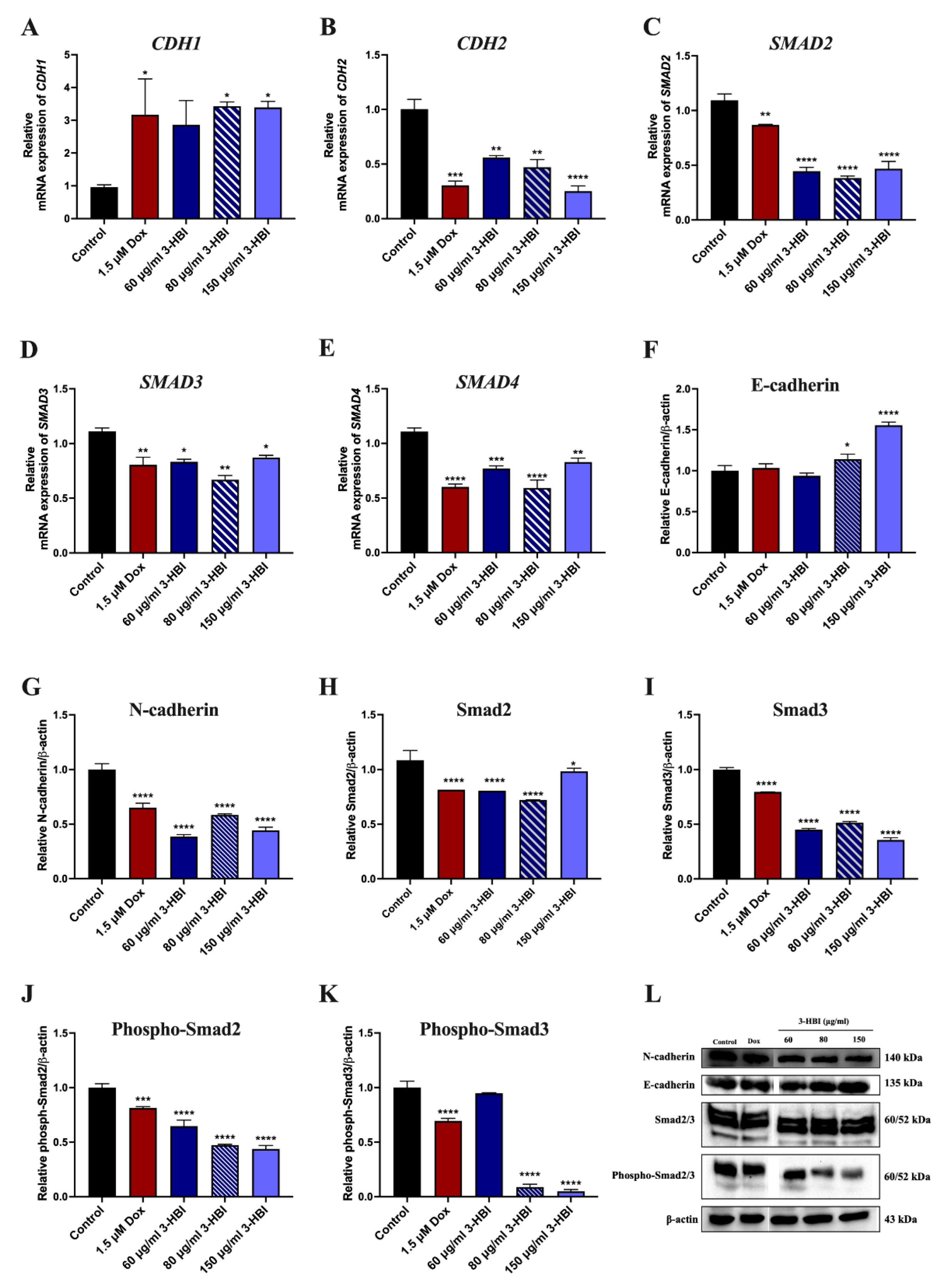
2.5.3. The Effect of 3-HBI on Genes and Proteins in the TGF-β/Smad Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Chemicals
4.2. Preparation of Moringa oleifera Lam. (MO) Extracts and Identification of Active Compounds
4.3. Cytotoxicity Assay
4.4. Colony Formation Assay
4.5. Migration Assay
4.6. Transwell Invasion Assay
4.7. Reverse Transcription-Quantitative Real-Time PCR (RT-qPCR)
4.8. Western Blot Analysis
4.9. Molecular Docking
4.10. Molecular Dynamics Simulation
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| TNBC | Triple-negative breast cancer |
| EMT | Epithelial-to-mesenchymal transition |
| 3-HBI | 3-hydroxy-β-ionone |
| MO | Moringa oleifera Lam. leaf extract |
| Dox | Doxorubicin |
| Smad | Suppressor of mothers against decapentaplegic |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef] [PubMed]
- Elzamly, S.; Badri, N.; Padilla, O.; Dwivedi, A.K.; Alvarado, L.A.; Hamilton, M.; Diab, N.; Rock, C.; Elfar, A.; Teleb, M.; et al. Epithelial-Mesenchymal Transition Markers in Breast Cancer and Pathological Responseafter Neoadjuvant Chemotherapy. Breast Cancer 2018, 12, 1178223418788074. [Google Scholar] [CrossRef]
- Zheng, C.; Yan, S.; Lu, L.; Yao, H.; He, G.; Chen, S.; Li, Y.; Peng, X.; Cheng, Z.; Wu, M.; et al. Lovastatin Inhibits EMT and Metastasis of Triple-Negative Breast Cancer Stem Cells Through Dysregulation of Cytoskeleton-Associated Proteins. Front. Oncol. 2021, 11, 656687. [Google Scholar] [CrossRef]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef]
- Wang, X.; Eichhorn, P.J.A.; Thiery, J.P. TGF-β, EMT, and resistance to anti-cancer treatment. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2023; Volume 97, pp. 1–11. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; He, X.; Zhang, P.; Sun, C.; Xu, X.; Lu, Y.; Li, F. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem. Biophys. Res. Commun. 2018, 502, 160–165. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; Dekker, T.J.A.; Hawinkels, L.J.A.C.; Putter, H.; Smit, V.T.H.B.M.; Kroep, J.R.; Kuppen, P.J.K.; van de Velde, C.J.H.; ten Dijke, P.; Tollenaar, R.A.E.M.; et al. The prognostic role of TGF-β signaling pathway in breast cancer patients. Ann. Oncol. 2013, 24, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, J.; Mao, K.; Deng, H.; Yang, Y.; Zhou, E.; Liu, J. Role of transforming growth factor-β1 in triple negative breast cancer patients. Int. J. Surg. 2017, 45, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Sylvin Benjamin, A.; Marie Alfrede, M.; Sadrine Tchoukouegno, N.; Job, T.; Charline Florence, A.; Dieudonne, N.; Liselotte, K. Natural Terpenoids Against Female Breast Cancer: A 5-year Recent Research. Curr. Med. Chem. 2018, 25, 3162–3213. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, S.C.; Joy, M.P.; Aiswarya, S.U.; Ajmani, A.; Keerthana, C.K.; Rayginia, T.P.; Isakov, N.; Anto, R.J. Unlocking nature’s pharmacy: An in-depth exploration of phytochemicals as potential sources of anti-cancer and anti-inflammatory molecules. Explor. Drug Sci. 2024, 2, 744–784. [Google Scholar] [CrossRef]
- Wali, A.F.; Pillai, J.R.; Talath, S.; Shivappa, P.; Sridhar, S.B.; El-Tanani, M.; Rangraze, I.R.; Mohamed, O.I.; Al Ani, N.N. Phytochemicals in Breast Cancer Prevention and Treatment: A Comprehensive Review. Curr. Issues Mol. Biol. 2025, 47, 30. [Google Scholar] [CrossRef]
- Luetragoon, T.; Pankla Sranujit, R.; Noysang, C.; Thongsri, Y.; Potup, P.; Suphrom, N.; Nuengchamnong, N.; Usuwanthim, K. Bioactive Compounds in Moringa oleifera Lam. Leaves Inhibit the Pro-Inflammatory Mediators in Lipopolysaccharide-Induced Human Monocyte-Derived Macrophages. Molecules 2020, 25, 191. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Salam, M.A.; Suenaga, K. Isolation and Identification of Potent Allelopathic Substances in a Traditional Bangladeshi Rice Cultivar Kartikshail. Plant Prod. Sci. 2011, 14, 128–134. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Seki, T.; Shigemori, H. Allelopathy and allelopathic substance in the moss Rhynchostegium pallidifolium. J. Plant Physiol. 2010, 167, 468–471. [Google Scholar] [CrossRef]
- Luetragoon, T.; Pankla Sranujit, R.; Noysang, C.; Thongsri, Y.; Potup, P.; Suphrom, N.; Nuengchamnong, N.; Usuwanthim, K. Anti-Cancer Effect of 3-Hydroxy-β-Ionone Identified from Moringa oleifera Lam. Leaf on Human Squamous Cell Carcinoma 15 Cell Line. Molecules 2020, 25, 3563. [Google Scholar] [CrossRef]
- de Moura Espíndola, R.; Mazzantini, R.P.; Ong, T.P.; de Conti, A.; Heidor, R.; Moreno, F.S. Geranylgeraniol and β-ionone inhibit hepatic preneoplastic lesions, cell proliferation, total plasma cholesterol and DNA damage during the initial phases of hepatocarcinogenesis, but only the former inhibits NF-κB activation. Carcinogenesis 2005, 26, 1091–1099. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, Y.S.; Jang, W.J.; Rakib, A.M.; Oh, T.W.; Kim, B.H.; Kim, S.Y.; Kim, J.O.; Ha, Y.L. Anti-proliferative Effects of β-ionone on Human Lung Cancer A-549 Cells. J. Life Sci. 2013, 23, 1351–1359. [Google Scholar] [CrossRef][Green Version]
- Fridlender, M.; Kapulnik, Y.; Koltai, H. Plant derived substances with anti-cancer activity: From folklore to practice. Front. Plant Sci. 2015, 6, 799. [Google Scholar] [CrossRef] [PubMed]
- Juthani, R.; Punatar, S.; Mittra, I. New light on chemotherapy toxicity and its prevention. BJC Rep. 2024, 2, 41. [Google Scholar] [CrossRef]
- Pilco-Ferreto, N.; Calaf, G.M. Influence of doxorubicin on apoptosis and oxidative stress in breast cancer cell lines. Int. J. Oncol. 2016, 49, 753–762. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Chung, J.Y.-F.; Chan, M.K.-K.; Li, J.S.-F.; Chan, A.S.-W.; Tang, P.C.-T.; Leung, K.-T.; To, K.-F.; Lan, H.-Y.; Tang, P.M.-K. TGF-β Signaling: From Tissue Fibrosis to Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 7575. [Google Scholar] [CrossRef]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Luetragoon, T.; Daowtak, K.; Thongsri, Y.; Potup, P.; Calder, P.C.; Usuwanthim, K. Anti-Inflammatory Potential of 3-Hydroxy-beta-Ionone from Moringa oleifera: Decreased Transendothelial Migration of Monocytes Through an Inflamed Human Endothelial Cell Monolayer by Inhibiting the IkappaB-alpha/NF-kappaB Signaling Pathway. Molecules 2024, 29, 5873. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2024, 53, D1516–D1525. [Google Scholar] [CrossRef] [PubMed]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Bustamante Munguira, E.; Andrés Juan, C.; Pérez-Lebeña, E. Michael Acceptors as Anti-Cancer Compounds: Coincidence or Causality? Int. J. Mol. Sci. 2024, 25, 6099. [Google Scholar] [CrossRef]
- Oussou, K.F.; Guclu, G.; Sevindik, O.; Kelebek, H.; Starowicz, M.; Selli, S. GC-MS-Olfactometric Characterization of Volatile and Key Odorants in Moringa (Moringa oleifera) and Kinkeliba (Combretum micranthum G. Don) Herbal Tea Infusions Prepared from Cold and Hot Brewing. Separations 2023, 10, 10. [Google Scholar] [CrossRef]
- Shangary, S.; Johnson, D.E. Peptides Derived from BH3 Domains of Bcl-2 Family Members: A Comparative Analysis of Inhibition of Bcl-2, Bcl-xL and Bax Oligomerization, Induction of Cytochrome c Release, and Activation of Cell Death. Biochemistry 2002, 41, 9485–9495. [Google Scholar] [CrossRef] [PubMed]
- Westphal, D.; Dewson, G.; Czabotar, P.E.; Kluck, R.M. Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2011, 1813, 521–531. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Westphal, D.; Dewson, G.; Ma, S.; Hockings, C.; Fairlie, W.D.; Lee, E.F.; Yao, S.; Robin, A.Y.; Smith, B.J.; et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 2013, 152, 519–531. [Google Scholar] [CrossRef]
- Parikh, N.; Koshy, C.; Dhayabaran, V.; Perumalsamy, L.R.; Sowdhamini, R.; Sarin, A. The N-terminus and alpha-5, alpha-6 helices of the pro-apoptotic protein Bax, modulate functional interactions with the anti-apoptotic protein Bcl-xL. BMC Cell Biol. 2007, 8, 16. [Google Scholar] [CrossRef]
- Rössig, L.; Fichtlscherer, B.; Breitschopf, K.; Haendeler, J.; Zeiher, A.M.; Mülsch, A.; Dimmeler, S. Nitric Oxide Inhibits Caspase-3 by S-Nitrosationin Vivo. J. Biol. Chem. 1999, 274, 6823–6826. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Olson, A.J. Automated docking of substrates to proteins by simulated annealing. Proteins 1990, 8, 195–202. [Google Scholar] [CrossRef]
- Khamto, N.; Chaichuang, L.; Rithchumpon, P.; Phupong, W.; Bhoopong, P.; Tateing, S.; Pompimon, W.; Semakul, N.; Chomsri, N.-o.; Meepowpan, P. Synthesis, cytotoxicity evaluation and molecular docking studies on 2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone derivatives. RSC Adv. 2021, 11, 31433–31447. [Google Scholar] [CrossRef]
- Porter, J.; Payne, A.; de Candole, B.; Ford, D.; Hutchinson, B.; Trevitt, G.; Turner, J.; Edwards, C.; Watkins, C.; Whitcombe, I.; et al. Tetrahydroisoquinoline amide substituted phenyl pyrazoles as selective Bcl-2 inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 230–233. [Google Scholar] [CrossRef]
- Garner, T.P.; Reyna, D.E.; Priyadarshi, A.; Chen, H.C.; Li, S.; Wu, Y.; Ganesan, Y.T.; Malashkevich, V.N.; Cheng, E.H.; Gavathiotis, E. An Autoinhibited Dimeric Form of BAX Regulates the BAX Activation Pathway. Mol. Cell 2016, 63, 485–497. [Google Scholar] [CrossRef]
- Renatus, M.; Stennicke, H.R.; Scott, F.L.; Liddington, R.C.; Salvesen, G.S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. USA 2001, 98, 14250–14255. [Google Scholar] [CrossRef]
- Fang, B.; Fu, G.; Agniswamy, J.; Harrison, R.W.; Weber, I.T. Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling. Apoptosis 2009, 14, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.-I.; Ito, T.; Fukatsu, Y.; Wada, H.; Kurisaki, A.; Tanokura, M. Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-β signaling. Sci. Signal 2020, 13, eabb9043. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.Y.; Lam, S.S.; Correia, J.J.; Lin, K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes. Dev. 2002, 16, 1950–1963. [Google Scholar] [CrossRef] [PubMed]
- Martin-Malpartida, P.; Batet, M.; Kaczmarska, Z.; Freier, R.; Gomes, T.; Aragón, E.; Zou, Y.; Wang, Q.; Xi, Q.; Ruiz, L.; et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat. Commun. 2017, 8, 2070. [Google Scholar] [CrossRef]
- Nardone, V.; Lucarelli, A.P.; Dalle Vedove, A.; Fanelli, R.; Tomassetti, A.; Belvisi, L.; Civera, M.; Parisini, E. Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction. J. Med. Chem. 2016, 59, 5089–5094. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2023, 52, D368–D375. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Khamto, N.; Utama, K.; Boontawee, P.; Janthong, A.; Tatieng, S.; Arthan, S.; Choommongkol, V.; Sangthong, P.; Yenjai, C.; Suree, N.; et al. Inhibitory Activity of Flavonoid Scaffolds on SARS-CoV-2 3CL(pro): Insights from the Computational and Experimental Investigations. J. Chem. Inf. Model. 2024, 64, 874–891. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]


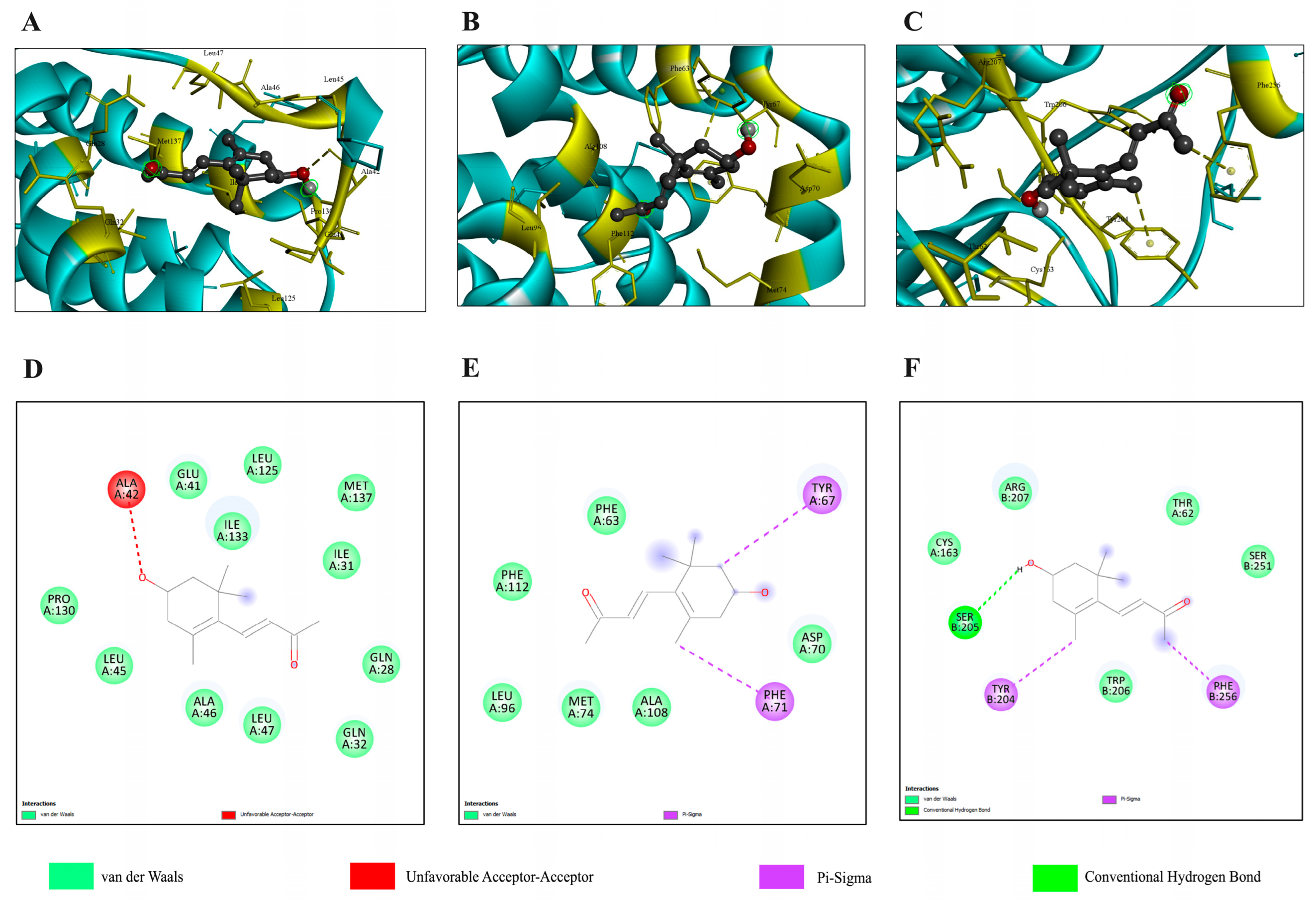


| Target Proteins | Estimate ΔG (kcal/mol) | Number of H-Bound | Amino Acid Interaction |
|---|---|---|---|
| Bcl-2 | −5.9 | 0 | Phe63, Tyr67, Asp70, Phe71, Met74, Lue96, Ala108, Phe11 |
| Bax | −6.5 | 0 | Gln28, Ile31, Gln32, Glu41, Ala42, Lue45, Ala46, Lue47, Lue125, Pro130, Ile133, Met137 |
| Caspase-9 | −5.2 | 3 | Arg177, Arg179, His237, Gln283, Cys285, Lys290, Val338, Ser339, Trp340, Arg341 |
| Caspase-3 | −5.4 | 1 | Thr62, Cys163, Tyr204, Ser205, Trp206, Arg207, Ser251, Phe256 |
| Target Proteins | Estimate ΔG (kcal/mol) | Number of H-Bound | Amino Acid Interaction |
|---|---|---|---|
| Smad2 | −5.1 | 2 | Glu270, Pro271, Ala272, Phe273, Trp274, Ala292, Ser293, Tyr340 |
| Smad3 | −5.4 | 0 | Pro228, Tyr297, Gly300, Phe342, Asn344, Gly401, Pro402, Gln404, Trp405, Lys408 |
| Smad4 | −4.6 | 1 | Asp52, Glu53, Ser56, Lys70, Cys71, Gln116, Tyr117, His132 |
| E-cadherin | −4.0 | 1 | Ile4, Pro5, Pro6, Ile7, Ser8, Leu21, Val22 |
| N-cadherin | −4.6 | 1 | His412, Asn417, Thr435, Pro437, Asn440, Gln467 |
| Gene Code | Target Protein | Forward (5′ → 3′) | Reverse (5′ → 3′) |
|---|---|---|---|
| BCL2 | B-cell lymphoma 2 | GATGTGATGCCTCTGCGAAG | CATGCTGATGTCTCTGGAATCT |
| BAX | Bcl-2–associated X | GGTTGTCGCCCTTTTCTA | TGTTTCCCTGAGGTTTGC |
| CASP3 | Caspase-3 | GCTATTGTAGGCGGTTGT | TGTTTCCCTGAGGTTTGC |
| SMAD2 | Suppressor of mothers against decapentaplegic 2 | TGCTCTGAAATTTGGGGACTGA | GACGACCATCAAGAGACCTGG |
| SMAD3 | Suppressor of mothers against decapentaplegic 3 | ATCGTGAAGCGCCTGCTG | CATCCAGGGACCTGGGGA |
| SMAD4 | Suppressor of mothers against decapentaplegic 4 | GCCCGAGCCCAGGTTATC | ACAATGCTCAGACAGGCATCA |
| CDH1 | E-cadherin | GAAATCACATCCTACACTGCCC | GTAGCAACTGGAGAACCATTGTC |
| CDH2 | N-cadherin | AGAAGACCAGGACTATGACTTGAG | CACCACTACTTGAGGAATTAAGGG |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | ATGACATCAAGAAGGTGGTG | CATACCAGGAAATGAGCTTG |
| Target Protein | PDB ID/UniProt | Center x, y, z | References |
|---|---|---|---|
| Bcl-2 | 2W3L | x = 38.905, y = 26.949, z = −12.545 | [40] |
| Bax | 4S0O | x = 20.576, y = −0.177, z = 16.313 | [41] |
| Caspase-9 | 1JXQ | x = 22.835, y = 39.722, z = −16.681 | [42] |
| Caspase-3 | 3GJQ | x = 40.441, y = 33.843, z = 57.987 | [43] |
| Smad2 | 6M64 | x = −27.673, y = 5.506, z = −9.959 | [44] |
| Smad3 | 1MK2 | x = 21.095, y = −3.433, z = 0.421 | [45] |
| Smad4 | 5MEY | x = −2.39, y = −16.454, z = 6.261 | [46] |
| E-cadherin | 4ZTE | x = 8.506, y = 13.076, z = 21.823 | [47] |
| N-cadherin | A0A024RC42 | x = −2.949, y = −2.211, z = −4.454 | [48,49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutana, P.; Luetragoon, T.; Buakaew, W.; Daotak, K.; Nilsri, N.; Thongsri, Y.; Potup, P.; Léon, C.; Usuwanthim, K. 3-Hydroxy-β-ionone Suppresses Breast Cancer Progression by Inducing Apoptosis and Blocking EMT Through the TGF-β/Smad Signaling Pathway. Int. J. Mol. Sci. 2025, 26, 8771. https://doi.org/10.3390/ijms26188771
Sutana P, Luetragoon T, Buakaew W, Daotak K, Nilsri N, Thongsri Y, Potup P, Léon C, Usuwanthim K. 3-Hydroxy-β-ionone Suppresses Breast Cancer Progression by Inducing Apoptosis and Blocking EMT Through the TGF-β/Smad Signaling Pathway. International Journal of Molecular Sciences. 2025; 26(18):8771. https://doi.org/10.3390/ijms26188771
Chicago/Turabian StyleSutana, Pornsuda, Thitiya Luetragoon, Watunyoo Buakaew, Krai Daotak, Nungruthai Nilsri, Yordhathai Thongsri, Pachuen Potup, Catherine Léon, and Kanchana Usuwanthim. 2025. "3-Hydroxy-β-ionone Suppresses Breast Cancer Progression by Inducing Apoptosis and Blocking EMT Through the TGF-β/Smad Signaling Pathway" International Journal of Molecular Sciences 26, no. 18: 8771. https://doi.org/10.3390/ijms26188771
APA StyleSutana, P., Luetragoon, T., Buakaew, W., Daotak, K., Nilsri, N., Thongsri, Y., Potup, P., Léon, C., & Usuwanthim, K. (2025). 3-Hydroxy-β-ionone Suppresses Breast Cancer Progression by Inducing Apoptosis and Blocking EMT Through the TGF-β/Smad Signaling Pathway. International Journal of Molecular Sciences, 26(18), 8771. https://doi.org/10.3390/ijms26188771









