Mechanism for Nucleotidyl Transfer in LINE-1 ORF2p Revealed by QM/MM Simulations
Abstract
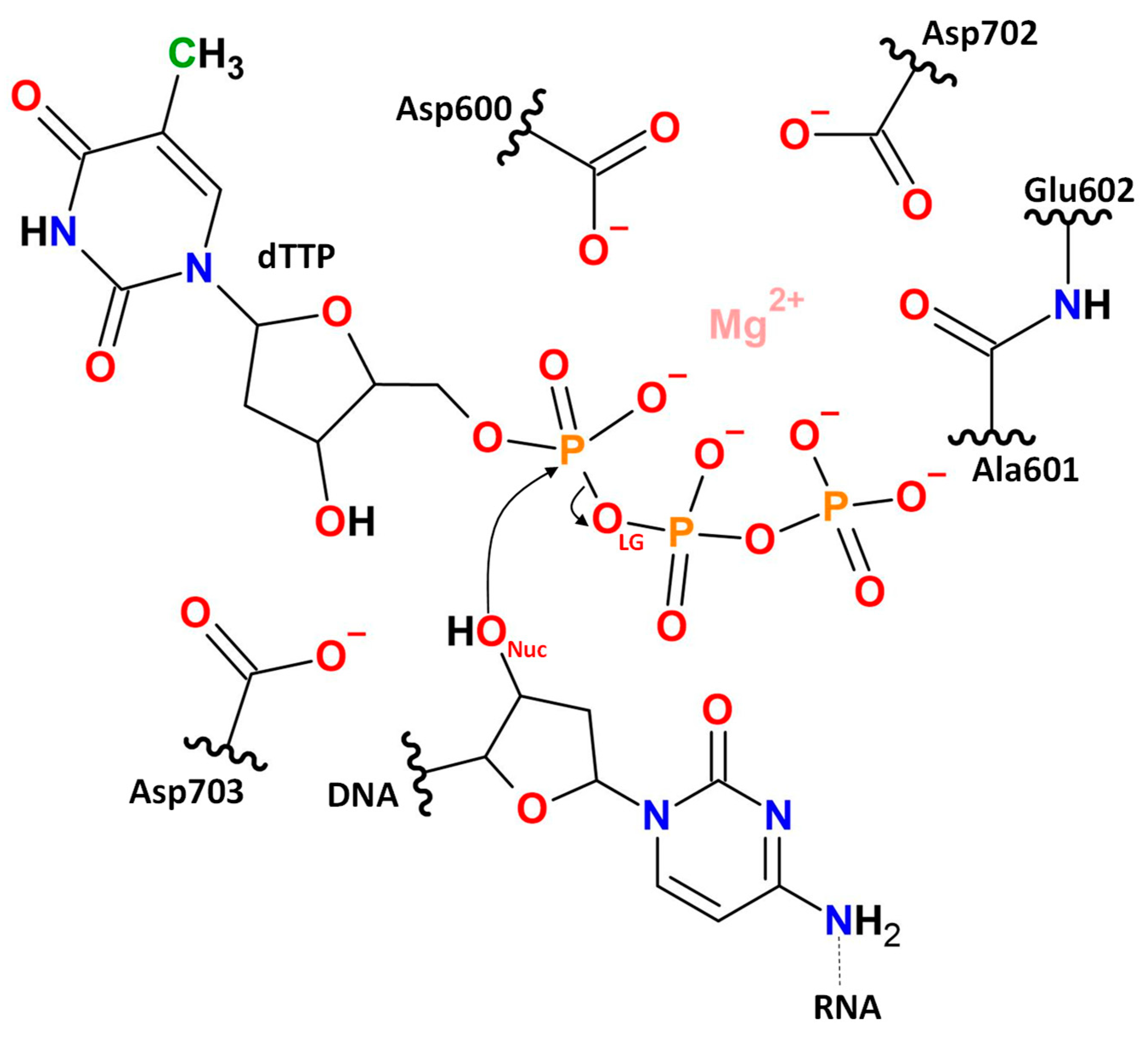
1. Introduction
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- Rodriguez-Martin, B.; Alvarez, E.G.; Baez-Ortega, A.; Zamora, J.; Supek, F.; Demeulemeester, J.; Santamarina, M.; Ju, Y.S.; Temes, J.; Garcia-Souto, D.; et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020, 52, 306–319. [Google Scholar] [CrossRef]
- Carter, V.; LaCava, J.; Taylor, M.S.; Liang, S.Y.; Mustelin, C.; Ukadike, K.C.; Bengtsson, A.; Lood, C.; Mustelin, T. High Prevalence and Disease Correlation of Autoantibodies Against p40 Encoded by Long Interspersed Nuclear Elements in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2020, 72, 89–99. [Google Scholar] [CrossRef]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Dai, L.; LaCava, J.; Taylor, M.S.; Boeke, J.D. Expression and detection of LINE-1 ORF-encoded proteins. Mob. Genet. Elem. 2014, 4, e29319. [Google Scholar] [CrossRef]
- Khazina, E.; Truffault, V.; Büttner, R.; Schmidt, S.; Coles, M.; Weichenrieder, O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011, 18, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Thawani, A.; Collins, K.; Nogales, E. Structural and biochemical studies of mobile retrotransposon proteins in action. Curr. Opin. Struct. Biol. 2025, 92, 103053. [Google Scholar] [CrossRef]
- Thawani, A.; Ariza, A.J.F.; Nogales, E.; Collins, K. Template and target-site recognition by human LINE-1 in retrotransposition. Nature 2024, 626, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.T.; van Eeuwen, T.; Hoyos, D.; Zalevsky, A.; Tchesnokov, E.P.; Sánchez, R.; Miller, B.D.; Di Stefano, L.H.; Ruiz, F.X.; Hancock, M.; et al. Structures, functions and adaptations of the human LINE-1 ORF2 protein. Nature 2024, 626, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, G.E.; Hu, H.; Boulanger, J.; Nguyen, T.H.D. Structural mechanism of LINE-1 target-primed reverse transcription. Science 2025, 388, eads8412. [Google Scholar] [CrossRef]
- Stamos, J.L.; Lentzsch, A.M.; Lambowitz, A.M. Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications. Mol. Cell 2017, 68, 926–939. [Google Scholar] [CrossRef]
- Baldwin, E.T.; Götte, M.; Tchesnokov, E.P.; Arnold, E.; Hagel, M.; Nichols, C.; Dossang, P.; Lamers, M.; Wan, P.; Steinbacher, S.; et al. Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K–specific inhibition. Proc. Natl. Acad. Sci. USA 2022, 119, e2200260119. [Google Scholar] [CrossRef]
- Kulakova, A.M.; Khrenova, M.G.; Zvereva, M.I.; Polyakov, I.V. Domain Mobility in the ORF2p Complex Revealed by Molecular Dynamics Simulations and Big Data Analysis. Int. J. Mol. Sci. 2024, 26, 73. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Smithline, Z.B. Crystallographic evidence for two-metal-ion catalysis in human pol η. Protein Sci. 2019, 28, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Unarta, I.C.; Goonetilleke, E.C.; Wang, D.; Huang, X. Nucleotide addition and cleavage by RNA polymerase II: Coordination of two catalytic reactions using a single active site. J. Biol. Chem. 2023, 299, 102844. [Google Scholar] [CrossRef] [PubMed]
- Steitz, T.A. DNA- and RNA-dependent DNA polymerases. Curr. Opin. Struct. Biol. 1993, 3, 31–38. [Google Scholar] [CrossRef]
- Steitz, T.A. A mechanism for all polymerases. Nature 1998, 391, 231–232. [Google Scholar] [CrossRef] [PubMed]
- Beese, L.S.; Steitz, T.A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar] [CrossRef]
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502. [Google Scholar] [CrossRef]
- Walker, A.R.; Cisneros, G.A. Computational Simulations of DNA Polymerases: Detailed Insights on Structure/Function/Mechanism from Native Proteins to Cancer Variants. Chem. Res. Toxicol. 2017, 30, 1922–1935. [Google Scholar] [CrossRef]
- Nakamura, T.; Zhao, Y.; Yamagata, Y.; Hua, Y.; Yang, W. Watching DNA polymerase η make a phosphodiester bond. Nature 2012, 487, 196–201. [Google Scholar] [CrossRef]
- Yang, W.; Weng, P.J.; Gao, Y. A new paradigm of DNA synthesis: Three-metal-ion catalysis. Cell Biosci. 2016, 6, 51, Erratum in Cell Biosci. 2017, 7, 32. https://doi.org/10.1186/s13578-017-0159-1. [Google Scholar] [CrossRef]
- Weaver, T.M.; Washington, M.T.; Freudenthal, B.D. New insights into DNA polymerase mechanisms provided by time-lapse crystallography. Curr. Opin. Struct. Biol. 2022, 77, 102465. [Google Scholar] [CrossRef] [PubMed]
- Pata, J.D.; Yin, Y.W.; Lahiri, I. Editorial: Nucleic Acid Polymerases: The Two-Metal-Ion Mechanism and Beyond. Front. Mol. Biosci. 2022, 9, 948326. [Google Scholar] [CrossRef] [PubMed]
- Raper, A.T.; Reed, A.J.; Suo, Z. Kinetic Mechanism of DNA Polymerases: Contributions of Conformational Dynamics and a Third Divalent Metal Ion. Chem. Rev. 2018, 118, 6000–6025. [Google Scholar] [CrossRef]
- Geronimo, I.; Vidossich, P.; Donati, E.; De Vivo, M. Computational investigations of polymerase enzymes: Structure, function, inhibition, and biotechnology. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1534. [Google Scholar] [CrossRef]
- Wu, W.-J.; Yang, W.; Tsai, M.-D. How DNA polymerases catalyse replication and repair with contrasting fidelity. Nat. Rev. Chem. 2017, 1, 0068. [Google Scholar] [CrossRef]
- Khrenova, M.G.; Mulashkina, T.I.; Kulakova, A.M.; Polyakov, I.V.; Nemukhin, A.V. Computer Modeling of the Mechanisms of Enzymatic Reactions: Lessons from 20 Years of Practice. Moscow Univ. Chem. Bull. 2024, 79, 86–92. [Google Scholar] [CrossRef]
- Lin, P.; Pedersen, L.C.; Batra, V.K.; Beard, W.A.; Wilson, S.H.; Pedersen, L.G. Energy analysis of chemistry for correct insertion by DNA polymerase β. Proc. Natl. Acad. Sci. USA 2006, 103, 13294–13299. [Google Scholar] [CrossRef]
- Bojin, M.D.; Schlick, T. A Quantum Mechanical Investigation of Possible Mechanisms for the Nucleotidyl Transfer Reaction Catalyzed by DNA Polymerase β. J. Phys. Chem. B 2007, 111, 11244–11252. [Google Scholar] [CrossRef]
- Cisneros, G.A.; Perera, L.; García-Díaz, M.; Bebenek, K.; Kunkel, T.A.; Pedersen, L.G. Catalytic mechanism of human DNA polymerase λ with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. DNA Repair 2008, 7, 1824–1834. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schlick, T. Quantum Mechanics/Molecular Mechanics Investigation of the Chemical Reaction in Dpo4 Reveals Water-Dependent Pathways and Requirements for Active Site Reorganization. J. Am. Chem. Soc. 2008, 130, 13240–13250. [Google Scholar] [CrossRef]
- Wang, L.; Broyde, S.; Zhang, Y. Polymerase-Tailored Variations in the Water-Mediated and Substrate-Assisted Mechanism for Nucleotidyl Transfer: Insights from a Study of T7 DNA Polymerase. J. Mol. Biol. 2009, 389, 787–796. [Google Scholar] [CrossRef]
- Rungrotmongkol, T.; Mulholland, A.J.; Hannongbua, S. QM/MM simulations indicate that Asp185 is the likely catalytic base in the enzymatic reaction of HIV-1 reverse transcriptase. Medchemcomm 2014, 5, 593. [Google Scholar] [CrossRef]
- Wang, H.; Huang, N.; Dangerfield, T.; Johnson, K.A.; Gao, J.; Elber, R. Exploring the Reaction Mechanism of HIV Reverse Transcriptase with a Nucleotide Substrate. J. Phys. Chem. B 2020, 124, 4270–4283. [Google Scholar] [CrossRef]
- Aranda, J.; Wieczór, M.; Terrazas, M.; Brun-Heath, I.; Orozco, M. Mechanism of reaction of RNA-dependent RNA polymerase from SARS-CoV-2. Chem Catal. 2022, 2, 1084–1099. [Google Scholar] [CrossRef]
- Maghsoud, Y.; Roy, A.; Leddin, E.M.; Cisneros, G.A. Effects of the Y432S Cancer-Associated Variant on the Reaction Mechanism of Human DNA Polymerase κ. J. Chem. Inf. Model. 2024, 64, 4231–4249. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Smidansky, E.; Maksimchuk, K.R.; Arnold, J.J.; Korneeva, V.S.; Götte, M.; Konigsberg, W.; Cameron, C.E. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. USA 2007, 104, 4267–4272. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.C. GTP hydrolysis mechanism of Ras-like GTPases. J. Mol. Biol. 2004, 340, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Sobti, M.; Ueno, H.; Noji, H.; Stewart, A.G. The six steps of the complete F1-ATPase rotary catalytic cycle. Nat. Commun. 2021, 12, 4690. [Google Scholar] [CrossRef]
- Mishra, A.K.; Lambright, D.G. Small GTPases and their GAPs. Biopolymers 2016, 105, 431–448. [Google Scholar] [CrossRef]
- Nemukhin, A.V.; Grigorenko, B.L.; Lushchekina, S.V.; Varfolomeev, S.D. Quantum chemical modelling in the research of molecular mechanisms of enzymatic catalysis. Russ. Chem. Rev. 2012, 81, 1011. [Google Scholar] [CrossRef]
- Khrenova, M.G.; Mulashkina, T.I.; Stepanyuk, R.A.; Nemukhin, A.V. Modeling of enzyme-catalyzed P–O bond cleavage in the adenosine triphosphate molecule. Mendeleev Commun. 2024, 34, 1–7. [Google Scholar] [CrossRef]
- Berta, D.; Gehrke, S.; Nyíri, K.; Vértessy, B.G.; Rosta, E. Mechanism-Based Redesign of GAP to Activate Oncogenic Ras. J. Am. Chem. Soc. 2023, 145, 20302–20310. [Google Scholar] [CrossRef] [PubMed]
- Khrenova, M.G.; Grigorenko, B.L.; Kolomeisky, A.B.; Nemukhin, A.V. Hydrolysis of guanosine triphosphate (GTP) by the Ras-GAP protein complex: Reaction mechanism and kinetic scheme. J. Phys. Chem. B 2015, 119, 12838–12845. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Kots, E.D.; Nemukhin, A.V. Diversity of mechanisms in Ras–GAP catalysis of guanosine triphosphate hydrolysis revealed by molecular modeling. Org. Biomol. Chem. 2019, 17, 4879–4891. [Google Scholar] [CrossRef] [PubMed]
- Kiani, F.A.; Fischer, S. Advances in quantum simulations of ATPase catalysis in the myosin motor. Curr. Opin. Struct. Biol. 2015, 31, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D.; Khanppnavar, B.; Kantarci, I.; Mehta, V.; Korkhov, V.M. Structural insights into membrane adenylyl cyclases, initiators of cAMP signaling. Trends Biochem. Sci. 2024, 49, 156–168. [Google Scholar] [CrossRef]
- Grigorenko, B.; Polyakov, I.; Nemukhin, A. Mechanisms of ATP to cAMP Conversion Catalyzed by the Mammalian Adenylyl Cyclase: A Role of Magnesium Coordination Shells and Proton Wires. J. Phys. Chem. B 2020, 124, 451–460. [Google Scholar] [CrossRef]
- van Keulen, S.C.; Martin, J.; Colizzi, F.; Frezza, E.; Trpevski, D.; Diaz, N.C.; Vidossich, P.; Rothlisberger, U.; Hellgren Kotaleski, J.; Wade, R.C.; et al. Multiscale molecular simulations to investigate adenylyl cyclase-based signaling in the brain. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2023, 13, e1623. [Google Scholar] [CrossRef]
- Mulashkina, T.I.; Kulakova, A.M.; Khrenova, M.G. Enzymatic P–O Bond Cleavage: Criteria of Dissociative and Associative Mechanisms. J. Chem. Inf. Model. 2025, 65, 8181–8193. [Google Scholar] [CrossRef]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a Covalently Trapped Catalytic Complex of HIV-1 Reverse Transcriptase: Implications for Drug Resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Peletskaya, E.N.; Kogon, A.A.; Tuske, S.; Arnold, E.; Hughes, S.H. Nonnucleoside Inhibitor Binding Affects the Interactions of the Fingers Subdomain of Human Immunodeficiency Virus Type 1 Reverse Transcriptase with DNA. J. Virol. 2004, 78, 3387–3397. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Martinez, S.E.; Arnold, E. Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef]
- Guo, L.-T.; Olson, S.; Patel, S.; Graveley, B.R.; Pyle, A.M. Direct tracking of reverse-transcriptase speed and template sensitivity: Implications for sequencing and analysis of long RNA molecules. Nucleic Acids Res. 2022, 50, 6980–6989. [Google Scholar] [CrossRef]
- Vergara, S.; Zhou, X.; Santiago, U.; Alaoui-El-Azher, M.; Conway, J.F.; Sluis-Cremer, N.; Calero, G. Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription. Nat. Commun. 2024, 15, 10553. [Google Scholar] [CrossRef]
- Pauling, L. The Nature of the Chemical Bond, 3rd ed.; Cornell University Press: New York, NY, USA, 1960. [Google Scholar]
- Adamo, C.; Barone, V. Toward reliable adiabatic connection models free from adjustable parameters. Chem. Phys. Lett. 1997, 274, 242–250. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Clemente, C.M.; Capece, L.; Martí, M.A. Best Practices on QM/MM Simulations of Biological Systems. J. Chem. Inf. Model. 2023, 63, 2609–2627. [Google Scholar] [CrossRef]
- Pitman, S.J.; Evans, A.K.; Ireland, R.T.; Lempriere, F.; McKemmish, L.K. Benchmarking Basis Sets for Density Functional Theory Thermochemistry Calculations: Why Unpolarized Basis Sets and the Polarized 6-311G Family Should Be Avoided. J. Phys. Chem. A 2023, 127, 10295–10306. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Croitoru, A.; Park, S.-J.; Kumar, A.; Lee, J.; Im, W.; MacKerell, A.D.; Aleksandrov, A. Additive CHARMM36 Force Field for Nonstandard Amino Acids. J. Chem. Theory Comput. 2021, 17, 3554–3570. [Google Scholar] [CrossRef]
- Denning, E.J.; Priyakumar, U.D.; Nilsson, L.; Mackerell, A.D. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 2011, 32, 1929–1943. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Seritan, S.; Bannwarth, C.; Fales, B.S.; Hohenstein, E.G.; Isborn, C.M.; Kokkila-Schumacher, S.I.L.; Li, X.; Liu, F.; Luehr, N.; Snyder, J.W.; et al. TeraChem : A graphical processing unit-accelerated electronic structure package for large-scale ab initio molecular dynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1494. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Bernardi, R.C.; Rudack, T.; Scheurer, M.; Riplinger, C.; Phillips, J.C.; Maia, J.D.C.; Rocha, G.B.; Ribeiro, J.V.; Stone, J.E.; et al. NAMD goes quantum: An integrative suite for hybrid simulations. Nat. Methods 2018, 15, 351–354. [Google Scholar] [CrossRef]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Singer, K.; Smith, W. Path integral simulations of condensed phase Lennard-Jones systems. Mol. Phys. 1988, 64, 1215–1231. [Google Scholar] [CrossRef]
- Souaille, M.; Roux, B. Extension to the weighted histogram analysis method: Combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 2001, 135, 40–57. [Google Scholar] [CrossRef]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Kästner, J.; Thiel, W. Bridging the gap between thermodynamic integration and umbrella sampling provides a novel analysis method: “Umbrella integration”. J. Chem. Phys. 2005, 123, 144104. [Google Scholar] [CrossRef]
- Lu, Y.; Farrow, M.R.; Fayon, P.; Logsdail, A.J.; Sokol, A.A.; Catlow, C.R.A.; Sherwood, P.; Keal, T.W. Open-Source, Python-Based Redevelopment of the ChemShell Multiscale QM/MM Environment. J. Chem. Theory Comput. 2019, 15, 1317–1328. [Google Scholar] [CrossRef]
- Kästner, J.; Carr, J.M.; Keal, T.W.; Thiel, W.; Wander, A.; Sherwood, P. DL-FIND: An Open-Source Geometry Optimizer for Atomistic Simulations. J. Phys. Chem. A 2009, 113, 11856–11865. [Google Scholar] [CrossRef]
- Balasubramani, S.G.; Chen, G.P.; Coriani, S.; Diedenhofen, M.; Frank, M.S.; Franzke, Y.J.; Furche, F.; Grotjahn, R.; Harding, M.E.; Hättig, C.; et al. TURBOMOLE: Modular program suite for ab initio quantum-chemical and condensed-matter simulations. J. Chem. Phys. 2020, 152, 184107. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
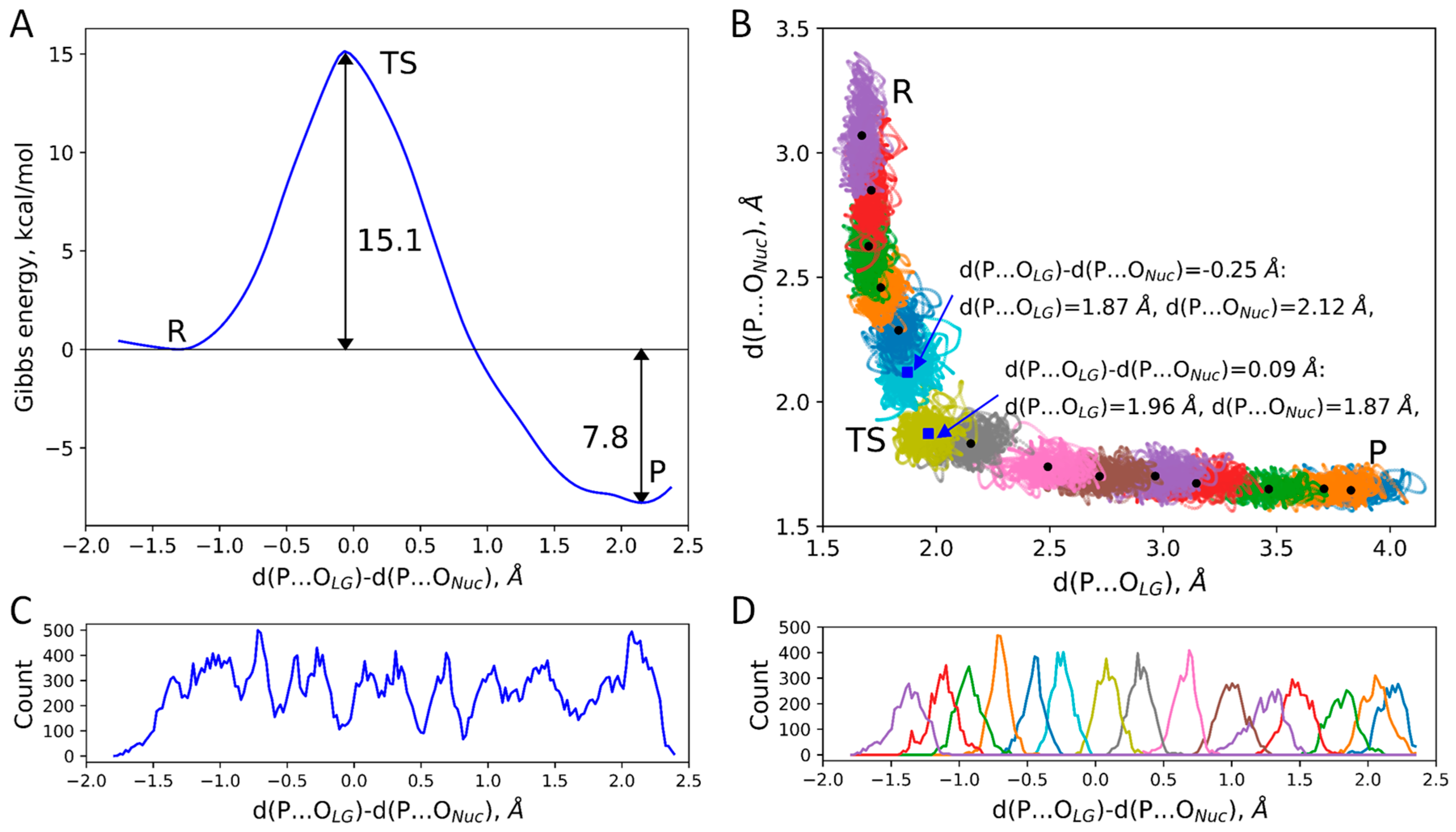
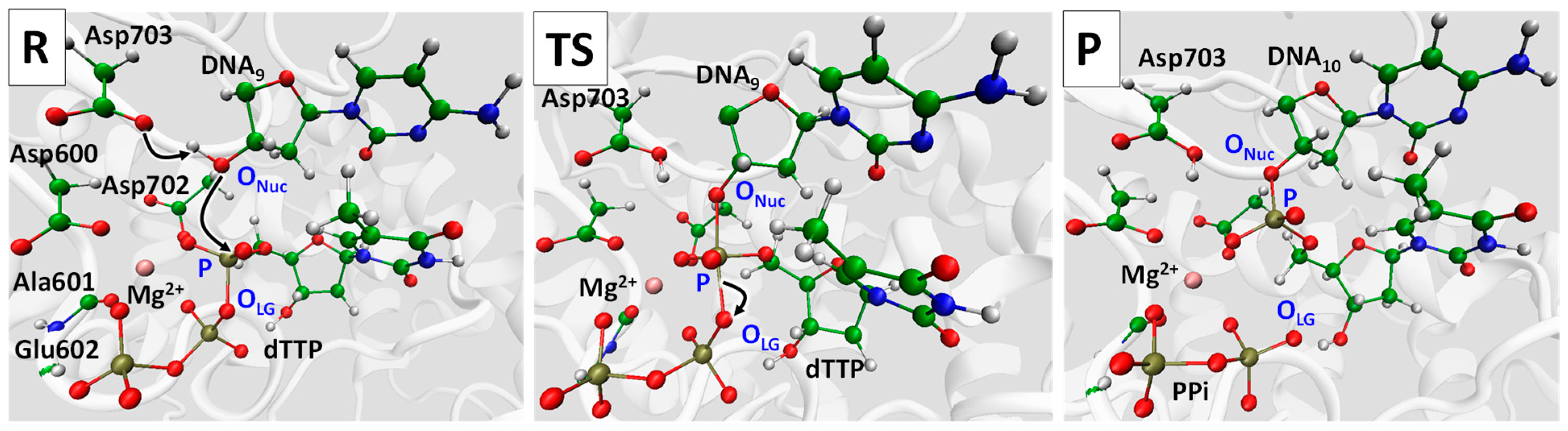
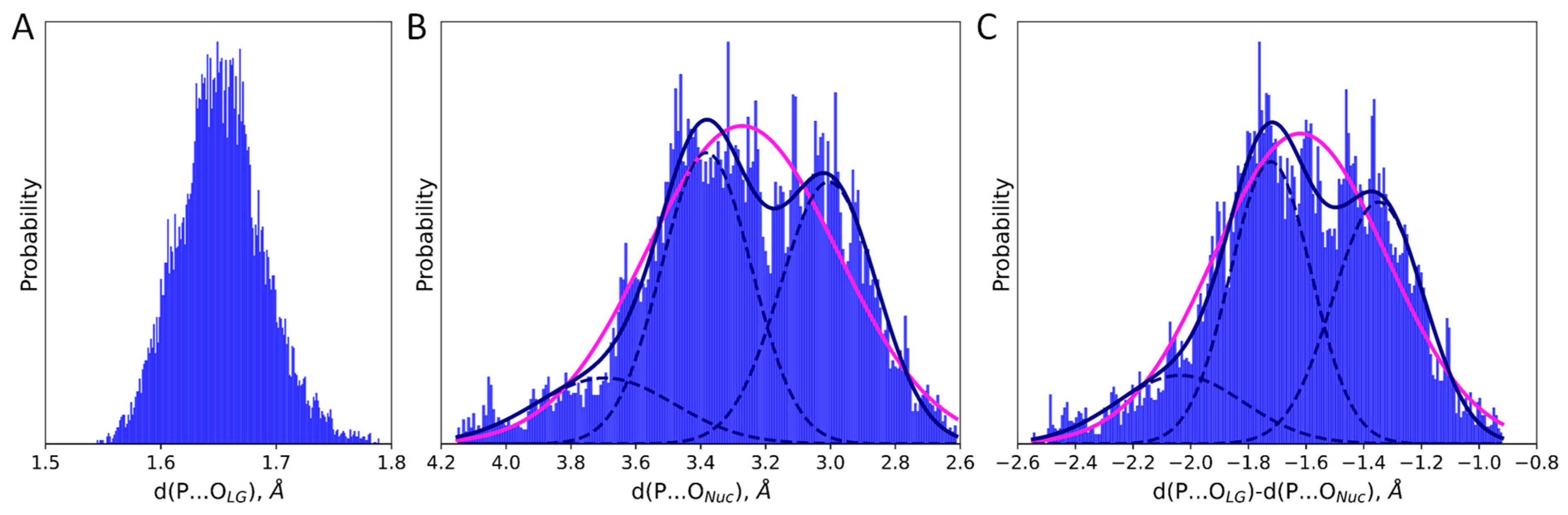
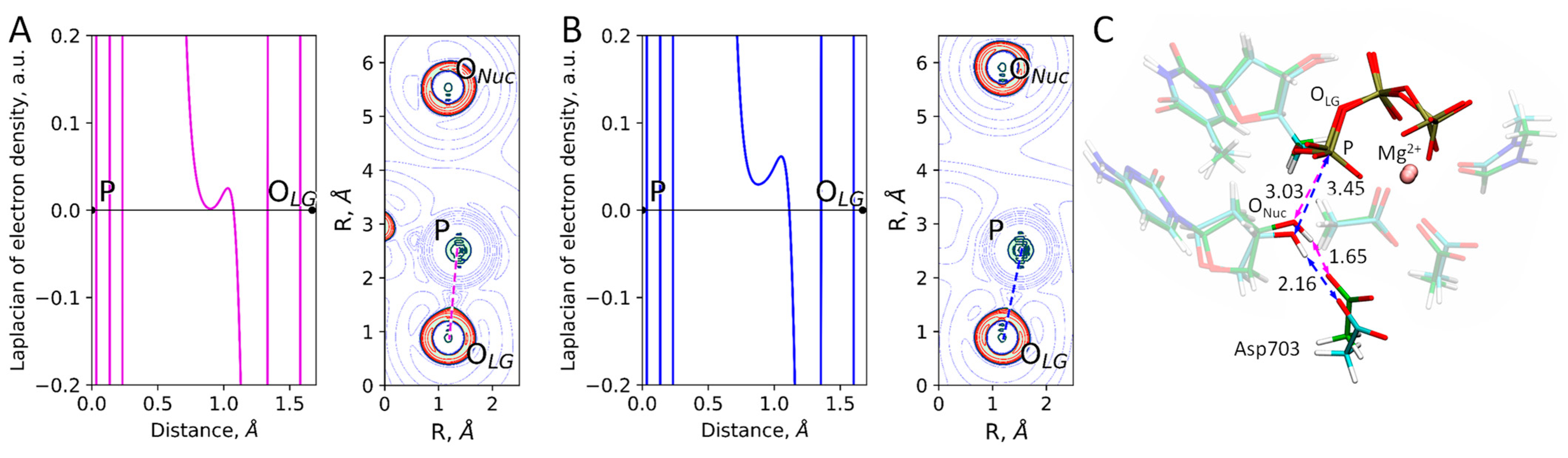
| d(P…OLG), Å | d(P…ONuc), Å | d(P…OLG)-d(P…ONuc), Å |
|---|---|---|
| 1.653 ± 0.001 (1) | 3.01 ± 0.02 (0.42) | −1.34 ± 0.02 (0.41) |
| 3.38 ± 0.02 (0.43) | −1.72 ± 0.02 (0.43) | |
| 3.70 ± 0.05 (0.15) | −2.04 ± 0.05 (0.16) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polyakov, I.V.; Miroshnichenko, K.D.; Mulashkina, T.I.; Kulakova, A.M.; Khrenova, M.G. Mechanism for Nucleotidyl Transfer in LINE-1 ORF2p Revealed by QM/MM Simulations. Int. J. Mol. Sci. 2025, 26, 8661. https://doi.org/10.3390/ijms26178661
Polyakov IV, Miroshnichenko KD, Mulashkina TI, Kulakova AM, Khrenova MG. Mechanism for Nucleotidyl Transfer in LINE-1 ORF2p Revealed by QM/MM Simulations. International Journal of Molecular Sciences. 2025; 26(17):8661. https://doi.org/10.3390/ijms26178661
Chicago/Turabian StylePolyakov, Igor V., Kirill D. Miroshnichenko, Tatiana I. Mulashkina, Anna M. Kulakova, and Maria G. Khrenova. 2025. "Mechanism for Nucleotidyl Transfer in LINE-1 ORF2p Revealed by QM/MM Simulations" International Journal of Molecular Sciences 26, no. 17: 8661. https://doi.org/10.3390/ijms26178661
APA StylePolyakov, I. V., Miroshnichenko, K. D., Mulashkina, T. I., Kulakova, A. M., & Khrenova, M. G. (2025). Mechanism for Nucleotidyl Transfer in LINE-1 ORF2p Revealed by QM/MM Simulations. International Journal of Molecular Sciences, 26(17), 8661. https://doi.org/10.3390/ijms26178661







