Microbial Signatures in Breast Cancer: Exploring New Potentials Across Body Niches
Abstract
1. Introduction
2. Changes in the Microbiota of Different Body Niches in the Context of Breast Cancer
2.1. Gut Microbiota
2.2. Breast Tissue Microbiota
2.3. Nipple Aspirate Fluid Microbiota
2.4. Skin Microbiota
2.5. Oral Microbiota
2.6. Female Urinary Tract
2.7. Female Reproductive Tract Microbiota
2.8. Blood Microbiota
3. Utilizing the Microbiota for Breast Cancer Diagnosis, Prognosis
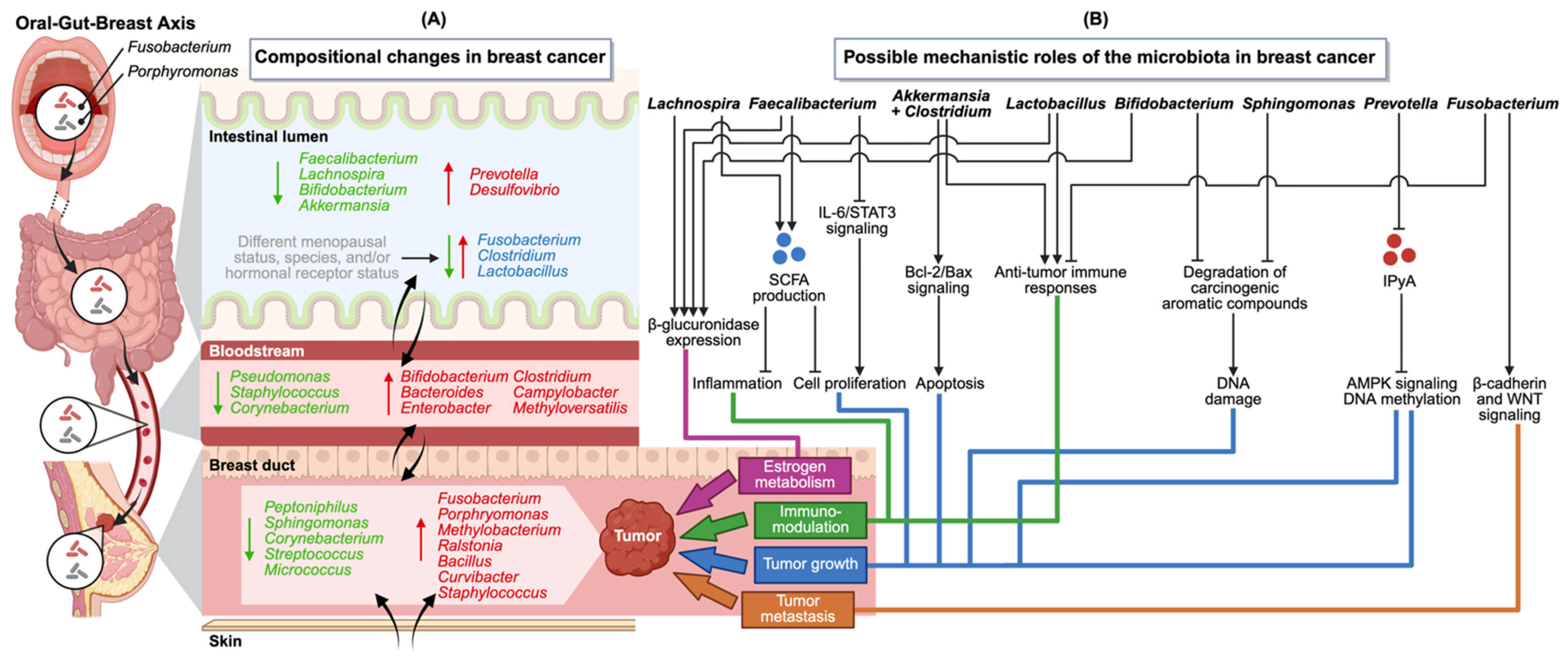
4. Emerging Mechanistic Role of the Oral–Gut–Breast Axis in Breast Cancer
5. Challenges and Prospects for Clinical Translation of Microbiota Findings
6. Conclusions
Authors Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Filho, A.M.; Laversanne, M.; Ferlay, J.; Colombet, M.; Piñeros, M.; Znaor, A.; Parkin, D.M.; Soerjomataram, I.; Bray, F. The GLOBOCAN 2022 Cancer Estimates: Data Sources, Methods, and a Snapshot of the Cancer Burden Worldwide. Int. J. Cancer 2025, 156, 1336–1346. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-S.; Zhao, Z.; Yang, Z.-N.; Xu, F.; Lu, H.-J.; Zhu, Z.-Y.; Shi, W.; Jiang, J.; Yao, P.-P.; Zhu, H.-P. Risk Factors and Preventions of Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- MetaHIT Consortium; Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Green, M.; Arora, K.; Prakash, S. Microbial Medicine: Prebiotic and Probiotic Functional Foods to Target Obesity and Metabolic Syndrome. Int. J. Mol. Sci. 2020, 21, 2890. [Google Scholar] [CrossRef]
- Chiu, L.; Bazin, T.; Truchetet, M.-E.; Schaeverbeke, T.; Delhaes, L.; Pradeu, T. Protective Microbiota: From Localized to Long-Reaching Co-Immunity. Front. Immunol. 2017, 8, 1678. [Google Scholar] [CrossRef]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and Cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef]
- Sarangi, A.N.; Goel, A.; Aggarwal, R. Methods for Studying Gut Microbiota: A Primer for Physicians. J. Clin. Exp. Hepatol. 2019, 9, 62–73. [Google Scholar] [CrossRef]
- Zhu, J.; Liao, M.; Yao, Z.; Liang, W.; Li, Q.; Liu, J.; Yang, H.; Ji, Y.; Wei, W.; Tan, A.; et al. Breast Cancer in Postmenopausal Women Is Associated with an Altered Gut Metagenome. Microbiome 2018, 6, 136. [Google Scholar] [CrossRef]
- Hou, M.-F.; Ou-Yang, F.; Li, C.-L.; Chen, F.-M.; Chuang, C.-H.; Kan, J.-Y.; Wu, C.-C.; Shih, S.-L.; Shiau, J.-P.; Kao, L.-C.; et al. Comprehensive Profiles and Diagnostic Value of Menopausal-Specific Gut Microbiota in Premenopausal Breast Cancer. Exp. Mol. Med. 2021, 53, 1636–1646. [Google Scholar] [CrossRef]
- Ma, J.; Sun, L.; Liu, Y.; Ren, H.; Shen, Y.; Bi, F.; Zhang, T.; Wang, X. Alter between Gut Bacteria and Blood Metabolites and the Anti-Tumor Effects of Faecalibacterium prausnitzii in Breast Cancer. BMC Microbiol. 2020, 20, 82. [Google Scholar] [CrossRef]
- Zeber-Lubecka, N.; Kulecka, M.; Jagiełło-Gruszfeld, A.; Dąbrowska, M.; Kluska, A.; Piątkowska, M.; Bagińska, K.; Głowienka, M.; Surynt, P.; Tenderenda, M.; et al. Breast Cancer but Not the Menopausal Status Is Associated with Small Changes of the Gut Microbiota. Front. Oncol. 2024, 14, 1279132. [Google Scholar] [CrossRef]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the Association Between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: A Population-Based Case-Control Pilot Study. JNCI J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef]
- Byrd, D.A.; Vogtmann, E.; Wu, Z.; Han, Y.; Wan, Y.; Clegg-Lamptey, J.; Yarney, J.; Wiafe-Addai, B.; Wiafe, S.; Awuah, B.; et al. Associations of Fecal Microbial Profiles with Breast Cancer and Nonmalignant Breast Disease in the Ghana Breast Health Study. Int. J. Cancer 2021, 148, 2712–2723. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, Z.; Peng, Q.; Lian, W.; Chen, D. Comparison of the Gut Microbiota in Patients with Benign and Malignant Breast Tumors: A Pilot Study. Evol. Bioinform. 2021, 17, 117693432110575. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, Y.; Ye, S.; Yin, S.; Gu, J. Changes of Intestinal Microflora of Breast Cancer in Premenopausal Women. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Tseng, C.; Vigen, C.; Yu, Y.; Cozen, W.; Garcia, A.A.; Spicer, D. Gut Microbiome Associations with Breast Cancer Risk Factors and Tumor Characteristics: A Pilot Study. Breast Cancer Res. Treat. 2020, 182, 451–463. [Google Scholar] [CrossRef]
- Feng, K.; Ren, F.; Wang, X. Relationships among Breast, Gut, and Oral Microbiota across Diverse Pathological Types of Breast Cancer, a Chinese Cohort Study. Front. Mol. Biosci. 2023, 10, 1325552. [Google Scholar] [CrossRef]
- Byrd, V.; Getz, T.; Padmanabhan, R.; Arora, H.; Eng, C. The Microbiome in PTEN Hamartoma Tumor Syndrome. Endocr.-Relat. Cancer 2018, 25, 233–243. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial Dysbiosis Is Associated with Human Breast Cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Hadzega, D.; Minarik, G.; Karaba, M.; Kalavska, K.; Benca, J.; Ciernikova, S.; Sedlackova, T.; Nemcova, P.; Bohac, M.; Pindak, D.; et al. Uncovering Microbial Composition in Human Breast Cancer Primary Tumour Tissue Using Transcriptomic RNA-Seq. Int. J. Mol. Sci. 2021, 22, 9058. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Chen, B.; Yang, J.; Wang, J.; Zhu, D.; Meng, Q.; Zhang, L. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of in situ Tissue Microbiomes for Promoting Malignancy. Front. Oncol. 2018, 8, 318. [Google Scholar] [CrossRef] [PubMed]
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of Human Breast Tissue Microbiota from Core Needle Biopsies through the Analysis of Multi Hypervariable 16S-rRNA Gene Regions. Sci. Rep. 2018, 8, 16893. [Google Scholar] [CrossRef]
- German, R.; Marino, N.; Hemmerich, C.; Podicheti, R.; Rusch, D.B.; Stiemsma, L.T.; Gao, H.; Xuei, X.; Rockey, P.; Storniolo, A.M. Exploring Breast Tissue Microbial Composition and the Association with Breast Cancer Risk Factors. Breast Cancer Res. 2023, 25, 82. [Google Scholar] [CrossRef]
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.-C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human Breast Microbiome Correlates with Prognostic Features and Immunological Signatures in Breast Cancer. Genome Med. 2021, 13, 60. [Google Scholar] [CrossRef]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Hoskinson, C.; Zheng, K.; Gabel, J.; Kump, A.; German, R.; Podicheti, R.; Marino, N.; Stiemsma, L.T. Composition and Functional Potential of the Human Mammary Microbiota Prior to and Following Breast Tumor Diagnosis. mSystems 2022, 7, e01489-21. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast Tissue, Oral and Urinary Microbiomes in Breast Cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef]
- Esposito, M.V.; Fosso, B.; Nunziato, M.; Casaburi, G.; D’Argenio, V.; Calabrese, A.; D’Aiuto, M.; Botti, G.; Pesole, G.; Salvatore, F. Microbiome Composition Indicate Dysbiosis and Lower Richness in Tumor Breast Tissues Compared to Healthy Adjacent Paired Tissue, within the Same Women. BMC Cancer 2022, 22, 30. [Google Scholar] [CrossRef]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef]
- Desalegn, Z.; Smith, A.; Yohannes, M.; Cao, X.; Anberber, E.; Bekuretsion, Y.; Assefa, M.; Bauer, M.; Vetter, M.; Kantelhardt, E.J.; et al. Human Breast Tissue Microbiota Reveals Unique Microbial Signatures That Correlate with Prognostic Features in Adult Ethiopian Women with Breast Cancer. Cancers 2023, 15, 4893. [Google Scholar] [CrossRef]
- Banerjee, S.; Wei, Z.; Tian, T.; Bose, D.; Shih, N.N.C.; Feldman, M.D.; Khoury, T.; De Michele, A.; Robertson, E.S. Prognostic Correlations with the Microbiome of Breast Cancer Subtypes. Cell Death Dis. 2021, 12, 831. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the Microbiome of Nipple Aspirate Fluid of Breast Cancer Survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [PubMed]
- Jiwa, N.; Danckert, N.; Takats, Z.; Marchesi, J.; Leff, D. Exploring the Nipple Aspirate Fluid Microbiome in Breast Cancer Patients. Eur. J. Surg. Oncol. 2022, 48, e194. [Google Scholar] [CrossRef]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef]
- Thyagarajan, S.; Zhang, Y.; Thapa, S.; Allen, M.S.; Phillips, N.; Chaudhary, P.; Kashyap, M.V.; Vishwanatha, J.K. Comparative Analysis of Racial Differences in Breast Tumor Microbiome. Sci. Rep. 2020, 10, 14116. [Google Scholar] [CrossRef]
- Balmaganbetova, F.K.; Amanzholkyzy, A.; Nurgaliyeva, R.E.; Kaldybayeva, A.T.; Zhexenova, A.N. Comparative Analysis of Vaginal Microbiota in Women with Breast Cancer in Kazakhstan. Asian Pac. J. Cancer Prev. 2021, 22, 1313–1318. [Google Scholar] [CrossRef]
- Peng, Y.; Gu, J.; Liu, F.; Wang, P.; Wang, X.; Si, C.; Gong, J.; Zhou, H.; Qin, A.; Song, F. Integrated Analysis of Microbiota and Gut Microbial Metabolites in Blood for Breast Cancer. mSystems 2024, 9, e0064324. [Google Scholar] [CrossRef]
- An, J.; Yang, J.; Kwon, H.; Lim, W.; Kim, Y.-K.; Moon, B.-I. Prediction of Breast Cancer Using Blood Microbiome and Identification of Foods for Breast Cancer Prevention. Sci. Rep. 2023, 13, 5110. [Google Scholar] [CrossRef]
- Shi, J.; Geng, C.; Sang, M.; Gao, W.; Li, S.; Yang, S.; Li, Z. Effect of Gastrointestinal Microbiome and Its Diversity on the Expression of Tumor-Infiltrating Lymphocytes in Breast Cancer. Oncol. Lett. 2019, 17, 5050–5056. [Google Scholar] [CrossRef]
- Klymiuk, I.; Bilgilier, C.; Mahnert, A.; Prokesch, A.; Heininger, C.; Brandl, I.; Sahbegovic, H.; Singer, C.; Fuereder, T.; Steininger, C. Chemotherapy-Associated Oral Microbiome Changes in Breast Cancer Patients. Front. Oncol. 2022, 12, 949071. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.; Gasbarrini, A.; Mele, M. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Mayneris-Perxachs, J.; Arnoriaga-Rodríguez, M.; Luque-Córdoba, D.; Priego-Capote, F.; Pérez-Brocal, V.; Moya, A.; Burokas, A.; Maldonado, R.; Fernández-Real, J.-M. Gut Microbiota Steroid Sexual Dimorphism and Its Impact on Gonadal Steroids: Influences of Obesity and Menopausal Status. Microbiome 2020, 8, 136. [Google Scholar] [CrossRef] [PubMed]
- Santos-Marcos, J.A.; Rangel-Zuñiga, O.A.; Jimenez-Lucena, R.; Quintana-Navarro, G.M.; Garcia-Carpintero, S.; Malagon, M.M.; Landa, B.B.; Tena-Sempere, M.; Perez-Martinez, P.; Lopez-Miranda, J.; et al. Influence of Gender and Menopausal Status on Gut Microbiota. Maturitas 2018, 116, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, J.; Li, X.; Sun, Q.; Qin, P.; Wang, Q. Compositional and Functional Features of the Female Premenopausal and Postmenopausal Gut Microbiota. FEBS Lett. 2019, 593, 2655–2664. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low Counts of Faecalibacterium prausnitzii in Colitis Microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Luu, T.H.; Michel, C.; Bard, J.-M.; Dravet, F.; Nazih, H.; Bobin-Dubigeon, C. Intestinal Proportion of Blautia sp. Is Associated with Clinical Stage and Histoprognostic Grade in Patients with Early-Stage Breast Cancer. Nutr. Cancer 2017, 69, 267–275. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, B.; Tang, T.; Xiao, Z.; Ye, F.; Li, X.; Wu, S.; Huang, J.-G.; Jiang, S. Gut Microbiota and Risk of Five Common Cancers: A Univariable and Multivariable Mendelian Randomization Study. Cancer Med. 2023, 12, 10393–10405. [Google Scholar] [CrossRef]
- Lee, K.; Kruper, L.; Dieli-Conwright, C.M.; Mortimer, J.E. The Impact of Obesity on Breast Cancer Diagnosis and Treatment. Curr. Oncol. Rep. 2019, 21, 41. [Google Scholar] [CrossRef]
- Hossain, F.; Majumder, S.; David, J.; Bunnell, B.A.; Miele, L. Obesity Modulates the Gut Microbiome in Triple-Negative Breast Cancer. Nutrients 2021, 13, 3656. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells 2020, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- The Endogenous Hormones And Breast Cancer Collaborative Group. Endogenous Sex Hormones and Breast Cancer in Postmenopausal Women: Reanalysis of Nine Prospective Studies. J. Natl. Cancer Inst. 2002, 94, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.N.; Falk, R.T.; Schairer, C.; Moore, S.C.; Fuhrman, B.J.; Dallal, C.M.; Bauer, D.C.; Dorgan, J.F.; Shu, X.-O.; Zheng, W.; et al. Association of Estrogen Metabolism with Breast Cancer Risk in Different Cohorts of Postmenopausal Women. Cancer Res. 2017, 77, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Pfeiffer, R.M.; Byrd, D.A.; Wan, Y.; Ansong, D.; Clegg-Lamptey, J.-N.; Wiafe-Addai, B.; Edusei, L.; Adjei, E.; Titiloye, N.; et al. Associations of Circulating Estrogens and Estrogen Metabolites with Fecal and Oral Microbiome in Postmenopausal Women in the Ghana Breast Health Study. Microbiol. Spectr. 2023, 11, e0157223. [Google Scholar] [CrossRef]
- Urbaniak, C.; Burton, J.P.; Reid, G. Breast, Milk and Microbes: A Complex Relationship That Does Not End with Lactation. Women’s Health 2012, 8, 385–398. [Google Scholar] [CrossRef]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast Cancer Colonization by Fusobacterium nucleatum Accelerates Tumor Growth and Metastatic Progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- Zheng, H.-H.; Du, C.-T.; Yu, C.; Tang, X.-Y.; Huang, R.-L.; Zhang, Y.-Z.; Gao, W.; Xie, G.-H. The Relationship of Tumor Microbiome and Oral Bacteria and Intestinal Dysbiosis in Canine Mammary Tumor. Int. J. Mol. Sci. 2022, 23, 10928. [Google Scholar] [CrossRef]
- Bernardo, G.; Le Noci, V.; Ottaviano, E.; De Cecco, L.; Camisaschi, C.; Guglielmetti, S.; Di Modica, M.; Gargari, G.; Bianchi, F.; Indino, S.; et al. Reduction of Staphylococcus epidermidis in the Mammary Tumor Microbiota Induces Antitumor Immunity and Decreases Breast Cancer Aggressiveness. Cancer Lett. 2023, 555, 216041. [Google Scholar] [CrossRef]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of Human Breast Tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- Dandeu, L.N.R.; Lachovsky, J.; Sidlik, S.; Marenco, P.; Orschanski, D.; Aguilera, P.; Vázquez, M.; Carballo, M.D.P.; Fernández, E.; Penas-Steinhardt, A.; et al. Relevance of Oncobiome in Breast Cancer Evolution in an Argentine Cohort. mSphere 2025, 10, e00597-24. [Google Scholar] [CrossRef] [PubMed]
- McCall, S.A.; Lichy, J.H.; Bijwaard, K.E.; Aguilera, N.S.; Chu, W.S.; Taubenberger, J.K. Epstein-Barr Virus Detection in Ductal Carcinoma of the Breast. J. Natl. Cancer Inst. 2001, 93, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.; Glenn, W.K.; Ye, Y.; Tran, B.; Delprado, W.; Lutze-Mann, L.; Whitaker, N.J.; Lawson, J.S. Human Papilloma Virus Is Associated with Breast Cancer. Br. J. Cancer 2009, 101, 1345–1350. [Google Scholar] [CrossRef]
- Afzal, S.; Fiaz, K.; Noor, A.; Sindhu, A.S.; Hanif, A.; Bibi, A.; Asad, M.; Nawaz, S.; Zafar, S.; Ayub, S.; et al. Interrelated Oncogenic Viruses and Breast Cancer. Front. Mol. Biosci. 2022, 9, 781111. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.-M.; Li, Y.; Wang, X.; Cao, W.-M.; Liu, D.-X. Non-Invasive Biomarkers for Early Detection of Breast Cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef]
- Kennedy, M.S.; Chang, E.B. The Microbiome: Composition and Locations. Prog. Mol. Biol. Transl. Sci. 2020, 176, 1–42. [Google Scholar] [CrossRef]
- Issrani, R.; Reddy, R.J.; El-Metwally, T.H.; Prabhu, N. Periodontitis as a Risk Factor for Breast Cancer—What We Know Till Date? Asian Pac. J. Cancer Prev. 2021, 22, 3109–3114. [Google Scholar] [CrossRef]
- Hugenholtz, F.; van der Veer, C.; Terpstra, M.L.; Borgdorff, H.; van Houdt, R.; Bruisten, S.; Geerlings, S.E.; van de Wijgert, J.H.H.M. Urine and Vaginal Microbiota Compositions of Postmenopausal and Premenopausal Women Differ Regardless of Recurrent Urinary Tract Infection and Renal Transplant Status. Sci. Rep. 2022, 12, 2698. [Google Scholar] [CrossRef]
- Ammitzbøll, N.; Bau, B.P.J.; Bundgaard-Nielsen, C.; Villadsen, A.B.; Jensen, A.-M.; Leutscher, P.D.C.; Glavind, K.; Hagstrøm, S.; Arenholt, L.T.S.; Sørensen, S. Pre- and Postmenopausal Women Have Different Core Urinary Microbiota. Sci. Rep. 2021, 11, 2212. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, X.; Wei, W.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The Microbiota Continuum along the Female Reproductive Tract and Its Relation to Uterine-Related Diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef] [PubMed]
- Marschalek, J.; Farr, A.; Marschalek, M.-L.; Domig, K.J.; Kneifel, W.; Singer, C.F.; Kiss, H.; Petricevic, L. Influence of Orally Administered Probiotic Lactobacillus Strains on Vaginal Microbiota in Women with Breast Cancer during Chemotherapy: A Randomized Placebo-Controlled Double-Blinded Pilot Study. Breast Care 2017, 12, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Prasanchit, P.; Pongchaikul, P.; Lertsittichai, P.; Tantitham, C.; Manonai, J. Vaginal Microbiomes of Breast Cancer Survivors Treated with Aromatase Inhibitors with and without Vulvovaginal Symptoms. Sci. Rep. 2024, 14, 7417. [Google Scholar] [CrossRef]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell. Infect. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef]
- Tan, C.C.S.; Ko, K.K.K.; Chen, H.; Liu, J.; Loh, M.; SG10K_Health Consortium; Chia, M.; Nagarajan, N. No Evidence for a Common Blood Microbiome Based on a Population Study of 9,770 Healthy Humans. Nat. Microbiol. 2023, 8, 973–985. [Google Scholar] [CrossRef]
- Çorbacıoğlu, Ş.K.; Aksel, G. Receiver Operating Characteristic Curve Analysis in Diagnostic Accuracy Studies: A Guide to Interpreting the Area under the Curve Value. Turk. J. Emerg. Med. 2023, 23, 195–198. [Google Scholar] [CrossRef]
- Stanton, S.E.; Disis, M.L. Clinical Significance of Tumor-Infiltrating Lymphocytes in Breast Cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef]
- Urbaniak, C.; McMillan, A.; Angelini, M.; Gloor, G.B.; Sumarah, M.; Burton, J.P.; Reid, G. Effect of Chemotherapy on the Microbiota and Metabolome of Human Milk, a Case Report. Microbiome 2014, 2, 24. [Google Scholar] [CrossRef]
- Csendes, D.; Gutlapalli, S.D.; Prakash, K.; Swarnakari, K.M.; Bai, M.; Manoharan, M.P.; Raja, R.; Jamil, A.; Desai, A.; Desai, D.M.; et al. Gastrointestinal Microbiota and Breast Cancer Chemotherapy Interactions: A Systematic Review. Cureus 2022, 14, e31648. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Yu, Y.; Jung, K.S.; Kim, Y.H.; Kim, J.-J. Tumor Microenvironment Can Predict Chemotherapy Response of Patients with Triple-Negative Breast Cancer Receiving Neoadjuvant Chemotherapy. Cancer Res. Treat. 2024, 56, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, D.; Wu, D.; Hong, W.; Kang, Y.; Tang, L.; Yang, Q.; Wang, X.; Li, B.; Li, R.; et al. Oral Combined Probiotics Clostridium butyricum and Akkermansia muciniphila Inhibits the Progression of 4T1 Breast Cancer by Activating Bcl-2/Bax Pathway. Cancer Med. 2025, 14, e70987. [Google Scholar] [CrossRef]
- Lakritz, J.R.; Poutahidis, T.; Levkovich, T.; Varian, B.J.; Ibrahim, Y.M.; Chatzigiagkos, A.; Mirabal, S.; Alm, E.J.; Erdman, S.E. Beneficial Bacteria Stimulate Host Immune Cells to Counteract Dietary and Genetic Predisposition to Mammary Cancer in Mice. Int. J. Cancer 2014, 135, 529–540. [Google Scholar] [CrossRef]
- Banerjee, S.; Gupta, N.; Pramanik, K.; Gope, M.; GhoshThakur, R.; Karmakar, A.; Gogoi, N.; Hoque, R.R.; Mandal, N.C.; Balachandran, S. Microbes and Microbial Strategies in Carcinogenic Polycyclic Aromatic Hydrocarbons Remediation: A Systematic Review. Environ. Sci. Pollut. Res. 2023, 31, 1811–1840. [Google Scholar] [CrossRef]
- Reddy, B.S.; Rivenson, A. Inhibitory Effect of Bifidobacterium longum on Colon, Mammary, and Liver Carcinogenesis Induced by 2-Amino-3-Methylimidazo[4,5-f]Quinoline, a Food Mutagen. Cancer Res. 1993, 53, 3914–3918. [Google Scholar]
- Fredrickson, J.K.; Balkwill, D.L.; Drake, G.R.; Romine, M.F.; Ringelberg, D.B.; White, D.C. Aromatic-Degrading Sphingomonas Isolates from the Deep Subsurface. Appl. Environ. Microbiol. 1995, 61, 1917–1922. [Google Scholar] [CrossRef]
- Zylstra, G.J.; Kim, E. Aromatic Hydrocarbon Degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 1997, 19, 408–414. [Google Scholar] [CrossRef]
- Su, J.; Li, D.; Chen, Q.; Li, M.; Su, L.; Luo, T.; Liang, D.; Lai, G.; Shuai, O.; Jiao, C.; et al. Anti-Breast Cancer Enhancement of a Polysaccharide From Spore of Ganoderma lucidum With Paclitaxel: Suppression on Tumor Metabolism With Gut Microbiota Reshaping. Front. Microbiol. 2018, 9, 3099. [Google Scholar] [CrossRef]
- Little, A.; Tangney, M.; Tunney, M.M.; Buckley, N.E. Fusobacterium nucleatum: A Novel Immune Modulator in Breast Cancer? Expert Rev. Mol. Med. 2023, 25, e15. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. The Microbiome–Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef]
- Dorgan, J.F.; Longcope, C.; Stephenson, H.E.; Falk, R.T.; Miller, R.; Franz, C.; Kahle, L.; Campbell, W.S.; Tangrea, J.A.; Schatzkin, A. Relation of Prediagnostic Serum Estrogen and Androgen Levels to Breast Cancer Risk. Cancer Epidemiol. Biomark. Prev. 1996, 5, 533–539. [Google Scholar]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of Beta-Glucosidase and Beta-Glucuronidase Activity and of Beta-Glucuronidase Gene Gus in Human Colonic Bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Q.; Zhang, W.; Kang, M.; Ma, J.; Zhao, L. Gut Microbial Beta-Glucuronidase: A Vital Regulator in Female Estrogen Metabolism. Gut Microbes 2023, 15, 2236749. [Google Scholar] [CrossRef] [PubMed]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The Gut Microbiome Switches Mutant P53 from Tumour-Suppressive to Oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- McGuinness, A.J.; Stinson, L.F.; Snelson, M.; Loughman, A.; Stringer, A.; Hannan, A.J.; Cowan, C.S.M.; Jama, H.A.; Caparros-Martin, J.A.; West, M.L.; et al. From Hype to Hope: Considerations in Conducting Robust Microbiome Science. Brain Behav. Immun. 2024, 115, 120–130. [Google Scholar] [CrossRef]
- Filardo, S.; Di Pietro, M.; Sessa, R. Current Progresses and Challenges for Microbiome Research in Human Health: A Perspective. Front. Cell. Infect. Microbiol. 2024, 14, 1377012. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Peters, B.A.; Shapiro, J.A.; Church, T.R.; Miller, G.; Trinh-Shevrin, C.; Yuen, E.; Friedlander, C.; Hayes, R.B.; Ahn, J. A Taxonomic Signature of Obesity in a Large Study of American Adults. Sci. Rep. 2018, 8, 9749. [Google Scholar] [CrossRef]
- Thu, M.S.; Chotirosniramit, K.; Nopsopon, T.; Hirankarn, N.; Pongpirul, K. Human Gut, Breast, and Oral Microbiome in Breast Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2023, 13, 1144021. [Google Scholar] [CrossRef]
- Luan, B.; Ge, F.; Lu, X.; Li, Z.; Zhang, H.; Wu, J.; Yang, Q.; Chen, L.; Zhang, W.; Chen, W. Changes in the Fecal Microbiota of Breast Cancer Patients Based on 16S rRNA Gene Sequencing: A Systematic Review and Meta-Analysis. Clin. Transl. Oncol. 2024, 26, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Li, X.; Xia, Q.; Yang, S.; Zhang, H.; Yang, H. Potential Values of Formalin-Fixed Paraffin-Embedded Tissues for Intratumoral Microbiome Analysis in Breast Cancer. Heliyon 2023, 9, e16267. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Díaz, J.; Álvarez-Mercado, A.I.; Ruiz-Marín, C.M.; Reina-Pérez, I.; Pérez-Alonso, A.J.; Sánchez-Andujar, M.B.; Torné, P.; Gallart-Aragón, T.; Sánchez-Barrón, M.T.; Reyes Lartategui, S.; et al. Association of Breast and Gut Microbiota Dysbiosis and the Risk of Breast Cancer: A Case-Control Clinical Study. BMC Cancer 2019, 19, 495. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Marín, C.M.; Isabel Álvarez-Mercado, A.; Plaza-Díaz, J.; Rodríguez-Lara, A.; Gallart-Aragón, T.; Sánchez-Barrón, M.T.; de Reyes Lartategui, S.; Alcaide-Lucena, M.; Fernández, M.F.; Fontana, L. A Clustering Study of Sociodemographic Data, Dietary Patterns, and Gut Microbiota in Healthy and Breast Cancer Women Participating in the MICROMA Study. Mol. Nutr. Food Res. 2024, 68, e2400253. [Google Scholar] [CrossRef]
- Hülpüsch, C.; Neumann, A.U.; Reiger, M.; Fischer, J.C.; De Tomassi, A.; Hammel, G.; Gülzow, C.; Fleming, M.; Dapper, H.; Mayinger, M.; et al. Association of Skin Microbiome Dynamics With Radiodermatitis in Patients with Breast Cancer. JAMA Oncol. 2024, 10, 516. [Google Scholar] [CrossRef]
- Patuleia, S.I.S.; Moelans, C.B.; Koopman, J.; van Steenhoven, J.E.C.; van Dalen, T.; van der Pol, C.C.; Jager, A.; Ausems, M.G.E.M.; van Diest, P.J.; van der Wall, E.; et al. Patient-Centered Research: How Do Women Tolerate Nipple Fluid Aspiration as a Potential Screening Tool for Breast Cancer? BMC Cancer 2022, 22, 705. [Google Scholar] [CrossRef]
- de Groot, J.S.; Moelans, C.B.; Elias, S.G.; Hennink, A.; Verolme, B.; Suijkerbuijk, K.P.M.; Jager, A.; Seynaeve, C.; Bos, P.; Witkamp, A.J.; et al. Repeated Nipple Fluid Aspiration: Compliance and Feasibility Results from a Prospective Multicenter Study. PLoS ONE 2015, 10, e0127895. [Google Scholar] [CrossRef][Green Version]
- Mirzayi, C.; Renson, A.; Genomic Standards Consortium; Massive Analysis and Quality Control Society; Furlanello, C.; Sansone, S.-A.; Zohra, F.; Elsafoury, S.; Geistlinger, L.; Kasselman, L.J.; et al. Reporting Guidelines for Human Microbiome Research: The STORMS Checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef]
| Study (Year) | Methodology for Sequencing/Taxonomic Assignment | Cohort | Niche(s) | Samples | Study Design |
|---|---|---|---|---|---|
| Zhu et al. (2018) [10] | Shotgun metagenomic sequencing/IGC by bowtie2 | China | Gut (stool) | 62 breast cancer patients (18 pre-, 44 postmenopausal), 71 control patients (25 pre-, 46 postmenopausal) | Analysis of gut microbiota in pre- and postmenopausal women |
| Hou et al. (2021) [11] | 16S rRNA sequencing (V3–V4)/OTUs Greengenes 13.8 and BaseSpace RDP | China | Gut (stool) | 200 breast cancer patients (100 pre-, 100 postmenopausal), 67 age-matched controls (50 pre-, 17 postmenopausal) | Profiling menopausal-specific gut microbiota in breast cancer |
| Ma et al. (2020) [12] | 16S rRNA (variable region unspecified)/Shanghai Applied Protein Technology | China | Gut (stool) | 25 breast cancer, 25 benign breast disease patients | Comparative analysis of gut bacteria and blood metabolites |
| Zeber-Lubecka et al. (2024) [13] | Shotgun metagenomic sequencing/MetaPhlAn3 v 3.0.13 | Poland | Gut (stool) | 88 breast cancer patients (47 pre-/peri-, and 41 postmenopausal), 86 controls (51 pre-/peri-, 35 postmenopausal) | Association between breast cancer and gut microbiota |
| Goedert et al. (2015) [14] | 16S rRNA gene sequencing (V3–V4)/OTUs to RDP using the QIIME pipeline | USA | Gut (stool) | 48 postmenopausal breast cancer, 48 controls; 85.4% non-Hispanic white in both groups | Case–control study |
| Byrd et al. (2021) [15] | 16S rRNA sequencing (V4)/ASVs to the SILVA database using the DADA2 pipeline 1.2.1 | Ghana | Gut (stool) | 379 breast cancer patients, 102 benign disease patients, 414 population-based controls | Comparative analysis of fecal microbial profiles |
| Yang et al. (2021) [16] | 16S rRNA gene sequencing (V4)/OTUs to the Greengene database via the QIIME 1.9.1 pipeline | China | Gut (stool) | 83 malignant, 19 benign breast tumor patients | Comparative analysis of gut microbiota |
| He et al. (2021) [17] | 16S rRNA sequencing (V3–V4)/unknown pipeline | China | Gut (stool) | 54 premenopausal breast cancer patients, 28 healthy controls | Analysis of intestinal microflora changes in comparison to healthy controls |
| Wu et al. (2020) [18] | 16S rRNA sequencing (V3–V4)/OTUs to the RDP using the QIIME pipeline | USA | Gut (stool) | 37 breast cancer patients; 73% Hispanic, 75% overweight or obese | Associations of gut microbiomes of breast cancer patients with risk factors and tumor characteristics |
| Feng et al. (2023) [19] | 16S rRNA gene sequencing (V3, V4, V3–V4, and V4–V5)/ASVs using the QIIME 2 pipeline | China | Breast tissue (fresh frozen tissues acquired by fine needle aspiration or core needle biopsy), gut (stool), and oral (saliva) | 98 patients with different breast cancer statuses (51 luminal A, 17 luminal B, 18 HER2, and 11 triple-negative), 46 patients with benign breast disease | Comparative study of the microbiota across different sites and breast cancer subtypes |
| Byrd et al. (2018) [20] | 16S rRNA gene sequencing (V3–V4)/OTUs using the QIIME 1.9 pipeline to the RDP classifier and Greengenes 13.8 database | USA | Gut (stool), urine, oral (saline wash) | 32 PTEN Hamartoma Tumor Syndrome patients, of which 17 have cancer history, and 15 had no cancer history. 87–100% white cohort | Microbiome analysis in PTEN Hamartoma Tumor Syndrome patients |
| Xuan et al. (2014) [21] | 16S rRNA pyrosequencing (V4)/OTUs using the mothur pipeline Bayesian classifier to Greengenes database | USA | Breast tissue (FFPE) | 20 breast cancer patients with paired normal adjacent and tumor tissue; 23 healthy patients undergoing reduction mammoplasty | Comparative analysis of tumor and normal adjacent tissue from the same individual, and healthy breast tissue |
| Hadzega et al. (2021) [22] | RNA sequencing/Kraken2 and MetaPhlan3 | Slovakia and China | Breast tissue (fresh frozen tissues) | 18 breast cancer patients, 5 healthy patients undergoing breast cosmesis surgery for Slovakian cohort; Database-downloaded data of 73 triple-negative patients and 18 healthy donor samples for Chinese cohort | Comparative analysis of primary tumor tissues of different breast cancer characteristics |
| Meng et al. (2018) [23] | 16S rRNA gene sequencing (V1–V2)/OTUs using RDP classifier with Greengenes 13.8 reference database within QIIME pipeline | China | Breast tissue (fresh frozen tissues acquired by percutaneous needle biopsy) | 22 benign, 72 malignant breast cancer patients | Comparative analysis between benign and malignant breast cancer tissues |
| Costantini et al. (2018) [24] | 16S rRNA gene sequencing (V2, V3, V4, V6+V7, V8, and V9)/OTUs using RDP classifier v. 2.11 | Italy | Breast tissue (fresh tissues obtained by core needle biopsy and/or surgical excision biopsy) | 12 core needle biopsy, 7 surgical excision biopsy tumors and healthy adjacent tissues from 16 breast patients | Characterization of microbiota in core needle biopsies versus surgical excision biopsies, comparison of breast tumor tissues with healthy adjacent tissues |
| German et al. (2023) [25] | 16S rRNA gene sequencing (V1–V2, V2–V3, V3–V4, V4–V5, V5–V7, V7–V9)/ASVs by alignment to the SILVA 138.1 SSU database via VSEARCH within the QIIME2 2021.4 pipeline | USA | Breast tissue (fresh frozen tissue cores) | 403 healthy control women, 76 breast cancer patients that donated one or more tissues from tumor biopsies, normal adjacent tissue, or distant metastatic tissues | Identification of optimal 16S rRNA gene variable region, comparative analysis of breast tissue microbial composition and association of microbial dysbiosis to breast cancer risk factors |
| Tzeng et al. (2021) [26] | 16S rRNA gene sequencing (V3–V4, V7–V9)/ASVs by the DADA2 taxonomy classifier to the SILVA database | USA | Breast tissue (fresh frozen tissues) | 221 breast cancer patients, 69 patients without breast cancer, and 18 patients without breast cancer that were categorized as high risk | Correlation study between microbiome and prognostic features |
| Urbaniak et al. (2016) [27] | 16S rRNA gene sequencing (V6)/verified OTUs with Greengenes database | Canada | Breast tissue (fresh frozen tissues) | 45 breast cancer patients, 13 benign tumor patients, and 23 disease-free patients | Comparative analysis of breast tissue microbiota |
| Hoskinson et al. (2022) [28] | 16S rRNA gene sequencing (V3–V4)/ASVs to the SILVA reference database using the DADA2 pipeline | USA | Breast tissue (fresh frozen tissues) | 50 healthy women, 15 “prediagnostic” women who were healthy at sampling and went on to be diagnosed with breast cancer later, 76 breast cancer patients that donated adjacent normal and/or tumor tissue | Comparative analysis of breast tissue microbiota from healthy, prediagnostic, malignant and adjacent normal breast tissue |
| Wang et al. (2017) [29] | 16S rRNA gene sequencing (V3–V4)/OTUs against Greengenes 13.8 database using UCLUST | USA | Breast tissue (fresh frozen tissues), oral (saline rinse), and urine | 57 breast cancer patients (tumor and adjacent normal tissue), and 21 healthy women (two tissue samples, one from each breast) | Comparison of breast tissue, oral, and urinary microbiota with breast cancer and clinical-pathologic features |
| Esposito et al. (2022) [30] | 16S rRNA gene sequencing (V4–V6)/ASVs in BioMaS against the RDP 11.5 database | Italy | Breast tissue (fresh frozen tissues) | Tumoral and adjacent non-tumoral tissue from 34 women with breast cancer | Comparison of microbiota composition of paired tumoral and adjacent non-tumoral tissue |
| Banerjee et al. (2018) [31] | PathoChip Array | USA | Breast tissue (FFPE) | Breast tissue from different breast cancer subtypes (50 ER+ or PgR+, 34 HER2+, 24 ER+ PgR+ HER2+, and 40 triple-negative), and 20 normal breast tissue controls | Study of microbial (bacterial, viral, fungal, and parasitic) signatures associated with different breast cancer subtypes |
| Desalegn et al. (2023) [32] | 16S rRNA gene sequencing (V4)/ASVs using RDP’s Training Set 16 (11.5) database via the DADA2 pipeline | Ethiopia | Breast tissue (fresh frozen tissue) | 50 breast tumors and 50 paired normal adjacent tissues from breast cancer patients | Comparative analysis of breast tissue microbiota between tumor and normal adjacent tissues in Ethiopian women |
| Banerjee et al. (2021) [33] | PathoChip Array | USA | Breast tissue (FFPE) | 95–105 breast tissue samples each for the different breast cancer subtypes (ER+ or PgR+, HER2+, ER+ PgR+ HER2+, and triple-negative), 20 matched control samples, and 68 non-matched control samples | Study of microbial (bacterial, viral, fungal, and parasitic) signatures associated with different breast cancer subtypes, and association to disease outcome |
| Chan et al. (2016) [34] | 16S rRNA gene sequencing (V4)/OTUs using mothur pipeline RDP classifier training set v14 | USA | Breast (NAF) and skin control swabs | Nipple aspirate fluid from 25 breast cancer survivors and 23 healthy control women | Characterization of nipple aspirate fluid microbiome |
| Abstract from Jiwa et al. (2022) [35] | 16S rRNA gene sequencing (variable region unspecified)/ASVs (pipeline not specified) | UK | Breast (NAF), with nipple, breast and arm skin as controls | Both breasts of patients were sampled for nipple aspirate fluid, resulting in samples from 23 normal breasts and 22 breasts with tumor | Characterization of nipple aspirate fluid microbiota |
| Hieken et al. (2016) [36] | 16S rRNA gene sequencing (V3–V5)/OTUs to Greengenes 13.5 reference database using the IM-TORNADO pipeline | USA | Breast tissue (fresh frozen tissues) | Aseptically collected normal adjacent breast tissue, skin tissue, and skin swab from patients with benign and malignant breast disease | Comparative study of aseptically collected breast tissue, skin tissue and skin swabs in benign and malignant disease |
| Thyagarajan et al. (2020) [37] | 16S rRNA gene sequencing (V3–V4)/OTUs using the RDP classifier against the Greengenes database | USA | Breast tumor tissue (fresh frozen tissues) | Breast tumor tissue and normal adjacent tissue from a total of 23 white non-Hispanic (17 triple-positive, and 6 triple-negative breast cancer) and 10 black non-Hispanic (7 with triple-positive, 3 with triple-negative breast cancer) that were racial identity-confirmed through ancestry analysis | Comparative analysis of racial differences in breast tumor microbiome, and the differences between triple-positive and triple-negative breast cancer |
| Balmaganbetova et al. (2021) [38] | Femoflor reagent kit (qPCR) | Kazakhstan | Vagina | 278 women with breast cancer (147 luminal A, 57 luminal B, 26 HER2+, 48 triple negative) that comprised 174 patients that received combination therapy during the study and 104 patients that had breast cancer 2–4 years ago | Comparative analysis of vaginal microbiota in women with breast cancer |
| Peng et al. (2024) [39] | 16S rRNA gene sequencing (V3–V4)/ASVs against the SILVA 138 database using the QIIME2 pipeline | China | Blood | 107 breast cancer patients and 107 healthy controls | Comparison and correlation of blood microbiota and microbial metabolites between healthy controls and breast cancer patients |
| An et al. (2023) [40] | 16S rRNA gene sequencing (V3–V4)/OTUs using UCLUST against the SILVA 132 database via QIIME 1.9.1 pipeline | South Korea | Blood (isolated bacterial extracellular vesicles) | 96 patients with breast cancer and 192 healthy controls | Blood microbiota data for the development of a breast cancer diagnostic algorithm using blood microbiota patterns |
| Shi et al. (2019) [41] | 16S rRNA gene sequencing (V3–V4)/OTUs using RDP classifier v2.2 via the UPARSE pipeline | China | Gut (stool) | 80 breast cancer patients | Analysis of gut microbiota and its diversity in breast cancer in correlation to tumor infiltrating lymphocyte status |
| Klymiuk et al. (2022) [42] | 16S rRNA gene sequencing (V4–V5)/ASVs against the SILVA 138 database via the QIIME2 pipeline | Austria | Oral (saliva) | Breast cancer patients with non-metastatic breast cancer undergoing chemotherapy, samples were obtained over three timepoints | Analysis of chemotherapy-associated changes in oral microbiome |
| Study (Year) | Niche | Breast Cancer Microbiota Signature Utilized | AUC |
|---|---|---|---|
| Zhu et al. (2018) [10] | Gut (stool) | Fusobacterium varium, Shigella_sp_D9, Desulfovibrio piger, Escherichia_sp_1_1_43, Shigella sonnei, Eubacterium eligens, Escherichia_sp_3_2_53FAA, Vibrio cholerae, Acinetobacter baumannii, Proteus mirabilis, Fusobacterium nucleatum, Campylobacter concisus, Escherichia coli, and Porphyromonas uenonis | 87.25% (95% CI 77.57–93.47%) on the training sample cohort of postmenopausal patients, 72% (95% CI 56.01–88.44%) on the test sample cohort consisting of both pre- and postmenopausal patients |
| Hou et al. (2021) [11] | Gut (stool) | Premenopausal: Bacteroides fragilis, Anaerostipes, Haemophilus parainfluenzae, Sutterella, Faecalibacterium prausnitzii, Bifidobacterium adolescentis, Bifidobacterium longum, Bifidobacterium bifidum, Ruminococcus gnavus, Rothia mucilaginosa Postmenopausal: Klebsiella pneumoniae, Haemophilus parainfluenzae, Sutterella, Akkermansia muciniphila, Phascolarctobacterium, Ruminococcus gnavus, Rothia mucilaginosa Both pre- and postmenopausal: Haemophilus parainfluenzae, Sutterella, Faecalibacterium prausnitzii, Ruminococcus gnavus, Rothia mucilaginosa | 0.826 for premenopausal women, 0.887 for postmenopausal women, 0.791 for both pre- and postmenopausal women |
| Zeber-Lubecka et al. (2024) [13] | Gut (stool) | Premenopausal: Bacteroides vulgatus, Eubacterium eligens, Bifidobacterium adolescentis, Parabacteroides distasonis, Instestinimonas butyriciproducens, Alistipes finegoldii, Gordonibacter pamelaeae, Ruthenibacterium lactatiformans, Gemmiger formicilis, Alistipes shahii, Roseburia intestinalis, Collinsella intestinalis, Pseudoflavonifractor sp. An 194, Enterorhabdus caecimuris, Faecalibacterium prausnitzii Postmenopausal: Alistipes finegoldii, Faecalibacterium prausnitzii, Barnesiella intestinihominis, Parabacteroides distasonis, Dorea longicatena, Alistipes putredinis, Eubacterium ramulus, Alistipes indistinctus, Coprobacter fastidious, Eubacterium ventriosum, Eubacterium sp. CAG 38, Agathobaculum butyriciproducens, Ruminococcus bromii, Enterorhabdus caecimuris, Asacharobacter celatus | 0.866 (95% CI 0.717–1.000) in premenopausal women, 0.810 (95% CI 0.579–1.000) in postmenopausal women |
| Esposito et al. (2022) [30] | Breast tissue | Propinionibacterium acnes, Acinetobacter johnsonii, Bacillus sp. YDWLR1, Pseudomonas putida, Actinetobacter junii, Xanthomonas citri, Diaphorobacter, Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas stutzeri, and Enterobacter aerogenes | 89% in their patient cohort (reported as diagnosis accuracy) |
| An et al. (2023) [40] | Blood | Enterobacter, Bacteroides, Kluyvera, Pseudomonas, Parabacteroides, Enterobacter, Pseudomonas, Bacteroides, Staphylococcus, Acinetobacter, and Corynebacterium 1 | 0.978–0.996 in their cohort of training and test set at an 80:20 ratio |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, A.Y.W.; Bicchieraro, G.; Palumbo, I.; Ciabattoni, A.; Aristei, C.; Spaccapelo, R. Microbial Signatures in Breast Cancer: Exploring New Potentials Across Body Niches. Int. J. Mol. Sci. 2025, 26, 8654. https://doi.org/10.3390/ijms26178654
Wong AYW, Bicchieraro G, Palumbo I, Ciabattoni A, Aristei C, Spaccapelo R. Microbial Signatures in Breast Cancer: Exploring New Potentials Across Body Niches. International Journal of Molecular Sciences. 2025; 26(17):8654. https://doi.org/10.3390/ijms26178654
Chicago/Turabian StyleWong, Alicia Yoke Wei, Giulia Bicchieraro, Isabella Palumbo, Antonella Ciabattoni, Cynthia Aristei, and Roberta Spaccapelo. 2025. "Microbial Signatures in Breast Cancer: Exploring New Potentials Across Body Niches" International Journal of Molecular Sciences 26, no. 17: 8654. https://doi.org/10.3390/ijms26178654
APA StyleWong, A. Y. W., Bicchieraro, G., Palumbo, I., Ciabattoni, A., Aristei, C., & Spaccapelo, R. (2025). Microbial Signatures in Breast Cancer: Exploring New Potentials Across Body Niches. International Journal of Molecular Sciences, 26(17), 8654. https://doi.org/10.3390/ijms26178654








