Molecular and Cellular Effects of CT Scans in Human Adipose Mesenchymal Stem Cells
Abstract
1. Introduction
2. Results
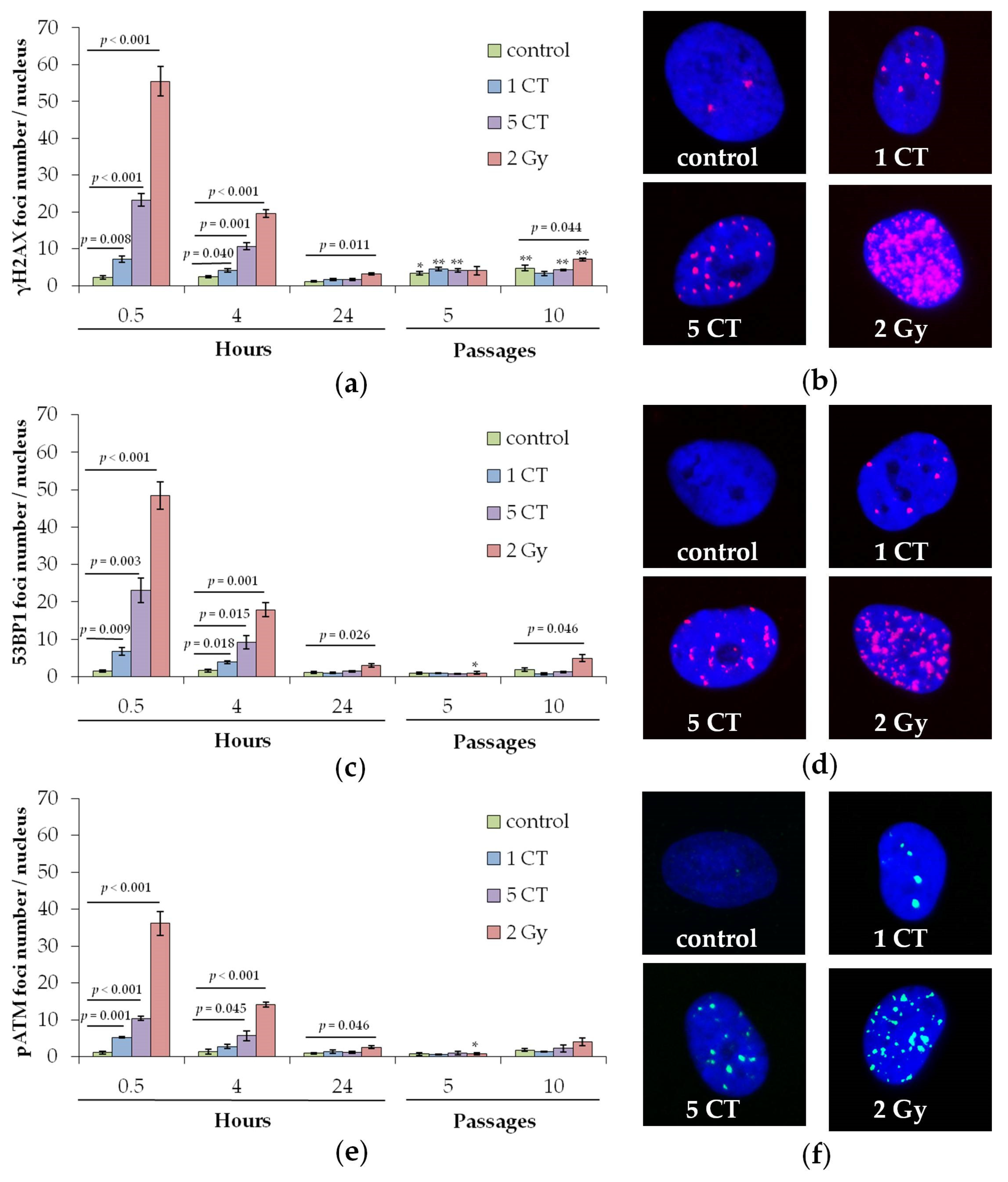
2.1. DNA Repair Foci
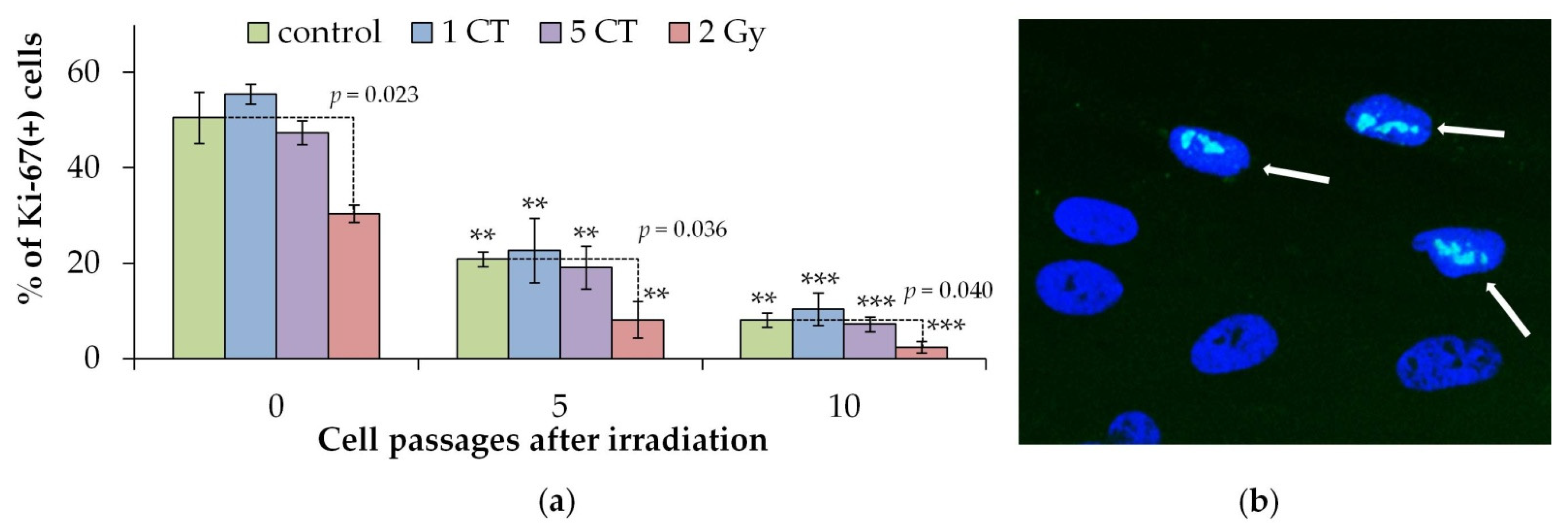
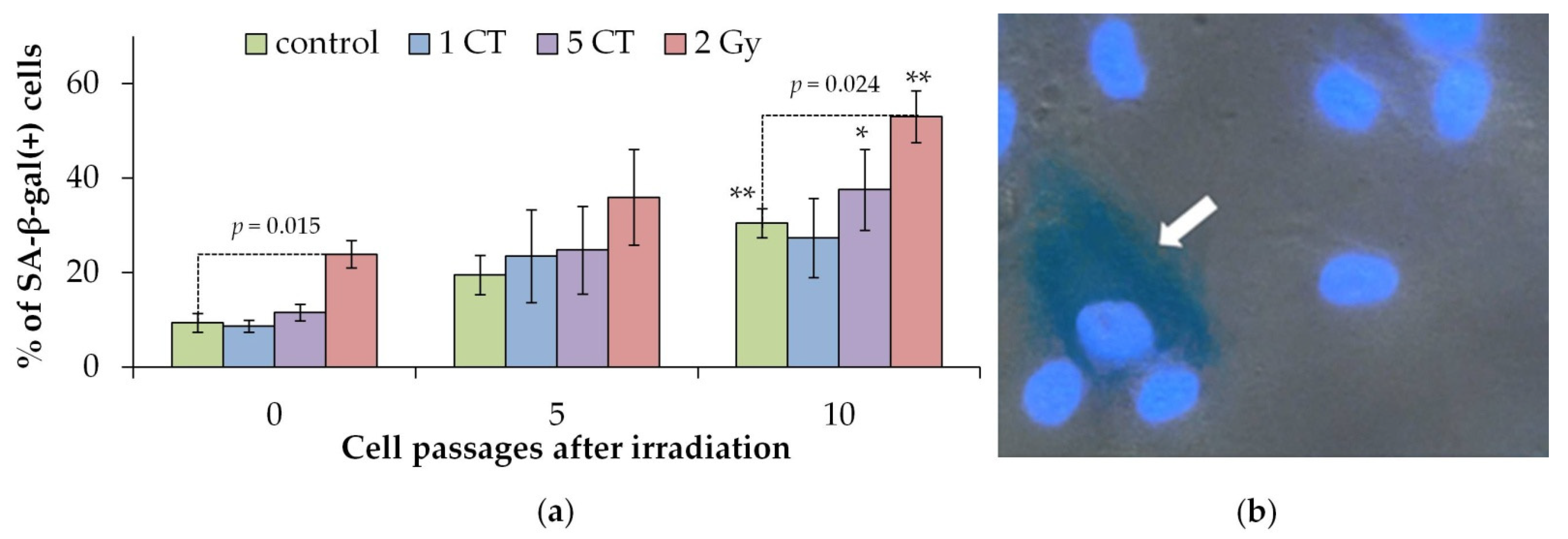
2.2. Cellular Proliferation and Senescence Associated β-Galactosidase Positive Cells
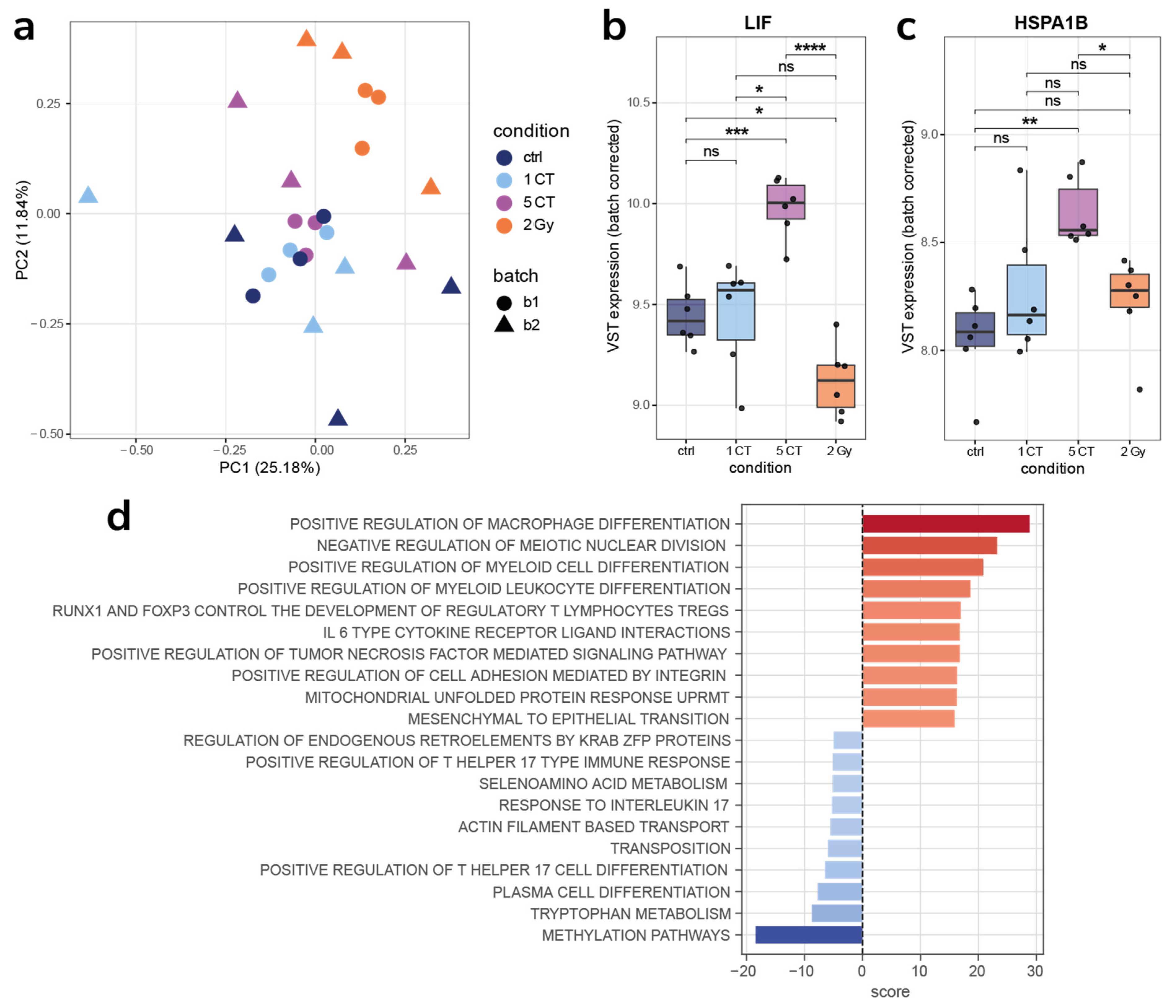
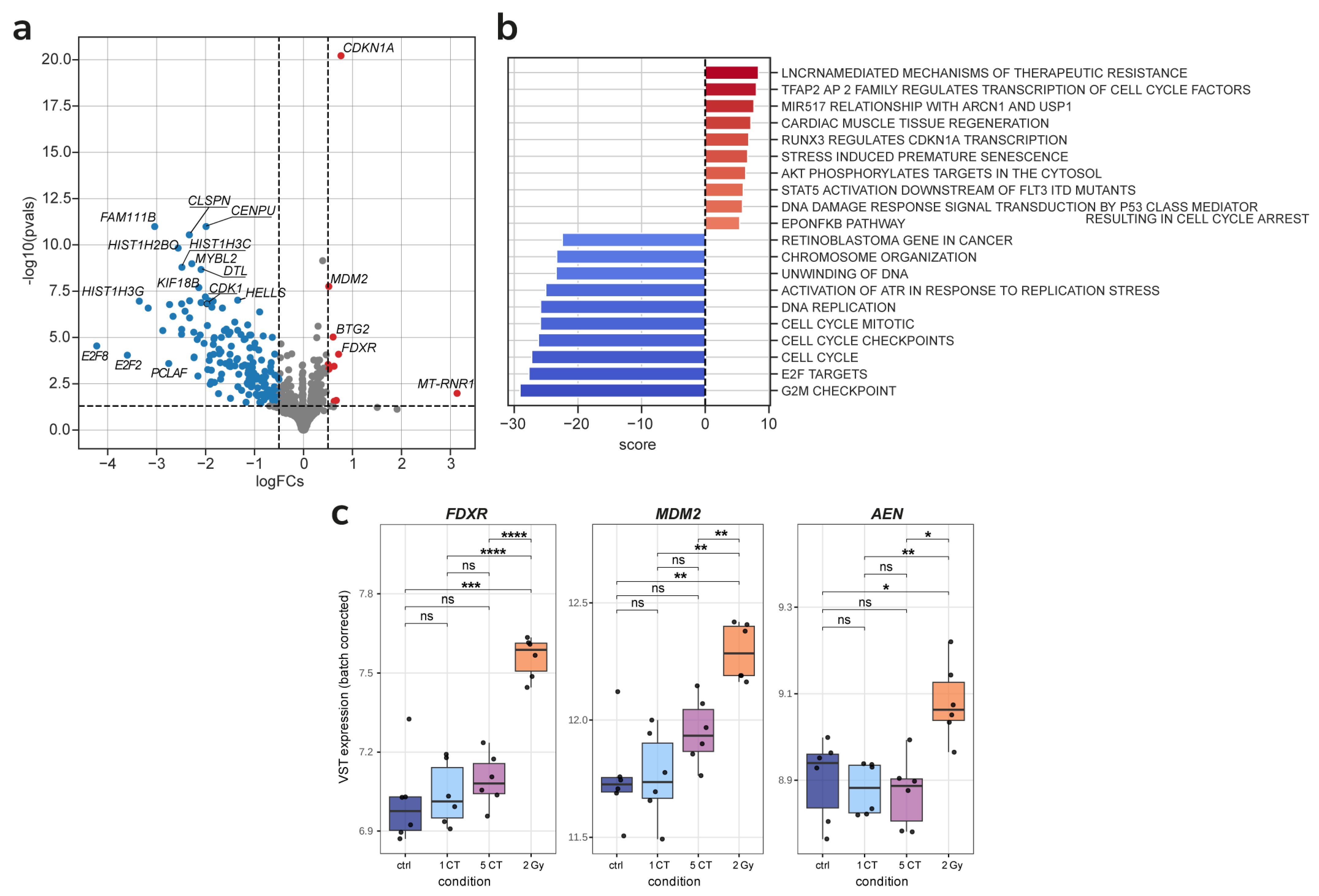
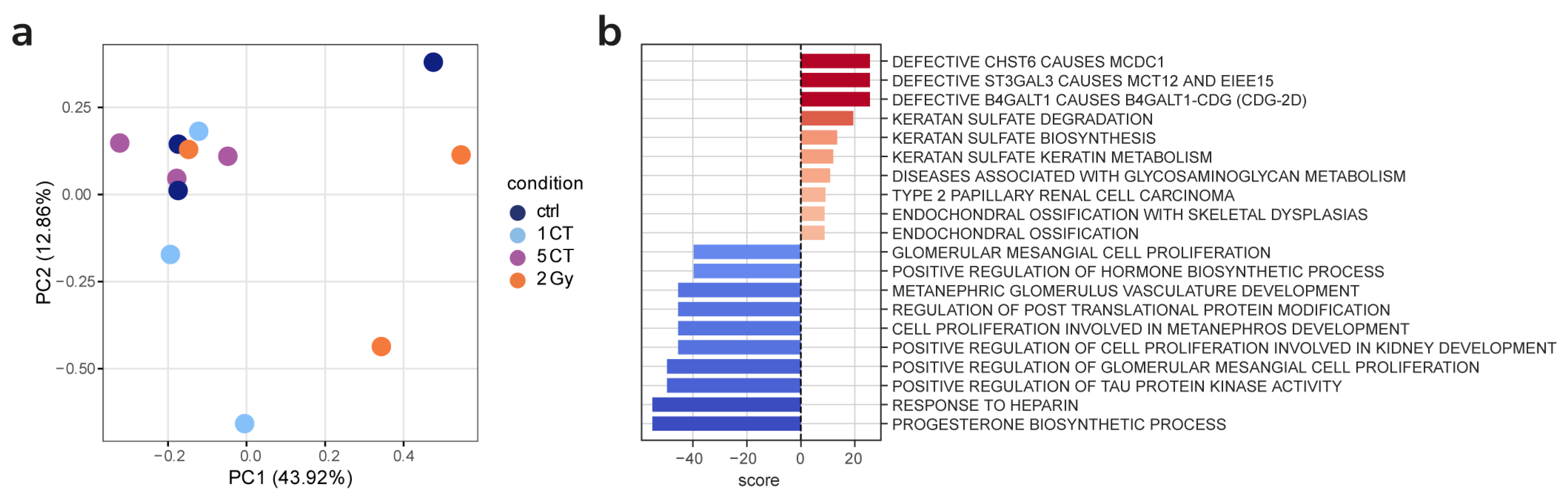
2.3. Gene Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Irradiation
4.3. Immunocytochemical Analysis
4.4. Assay of β-Galactosidase-Positive Cells
4.5. RNA Extraction and Sequencing
4.6. Read Mapping and Differential Expression
4.7. Pathway and Transcription Factor Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demissie, K.; Wu, Y.-J.; Kim, S.; Tsai, K.; Shao, Y.-H. Exposure to Tomographic Scans and Cancer Risks. JNCI Cancer Spectr. 2020, 4, pkz072. [Google Scholar] [CrossRef]
- Clement David-Olawade, A.; Olawade, D.B.; Vanderbloemen, L.; Rotifa, O.B.; Fidelis, S.C.; Egbon, E.; Akpan, A.O.; Adeleke, S.; Ghose, A.; Boussios, S. AI-Driven Advances in Low-Dose Imaging and Enhancement—A Review. Diagnostics 2025, 15, 689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhu, Z.; Yu, Z.; Sun, H.; Sun, Y.; Huang, H.; Xu, L.; Wan, J. Effectiveness of AI for Enhancing Computed Tomography Image Quality and Radiation Protection in Radiology: Systematic Review and Meta-Analysis. J. Med. Internet Res. 2025, 27, e66622. [Google Scholar] [CrossRef] [PubMed]
- Goldman, L.W. Principles of CT and CT Technology. J. Nucl. Med. Technol. 2007, 35, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Mettler, F.A.; Huda, W.; Yoshizumi, T.T.; Mahesh, M. Effective Doses in Radiology and Diagnostic Nuclear Medicine: A Catalog. Radiology 2008, 248, 254–263. [Google Scholar] [CrossRef]
- Berrington de González, A. Projected Cancer Risks From Computed Tomographic Scans Performed in the United States in 2007. Arch. Intern. Med. 2009, 169, 2071–2077. [Google Scholar] [CrossRef] [PubMed]
- Smith-Bindman, R.; Chu, P.W.; Azman Firdaus, H.; Stewart, C.; Malekhedayat, M.; Alber, S.; Bolch, W.E.; Mahendra, M.; Berrington de González, A.; Miglioretti, D.L. Projected Lifetime Cancer Risks From Current Computed Tomography Imaging. JAMA Intern. Med. 2025, 185, 710. [Google Scholar] [CrossRef]
- McCollough, C.H.; Primak, A.N.; Braun, N.; Kofler, J.; Yu, L.; Christner, J. Strategies for Reducing Radiation Dose in CT. Radiol. Clin. N. Am. 2009, 47, 27–40. [Google Scholar] [CrossRef]
- Cadavid, L.; Karout, L.; Kalra, M.K.; Morgado, F.; Londoño, M.A.; Pérez, L.; Galeano, M.; Montaño, M.; Wesley, L.; Almanza, J.; et al. Setting up regional diagnostic reference levels for pediatric computed tomography in Latin America: Preliminary results, challenges and the work ahead. Pediatr. Radiol. 2023, 54, 457–467. [Google Scholar] [CrossRef]
- Schauer, D.A.; Linton, O.W. National Council on Radiation Protection and Measurements Report Shows Substantial Medical Exposure Increase. Radiology 2009, 253, 293–296. [Google Scholar] [CrossRef]
- Hall, E.J.; Brenner, D.J. Cancer risks from diagnostic radiology. Br. J. Radiol. 2008, 81, 362–378. [Google Scholar] [CrossRef]
- Schultz, C.H.; Fairley, R.; Murphy, L.S.-L.; Doss, M. The Risk of Cancer from CT Scans and Other Sources of Low-Dose Radiation: A Critical Appraisal of Methodologic Quality. Prehospital Disaster Med. 2020, 35, 3–16. [Google Scholar] [CrossRef]
- Cao, C.-F.; Ma, K.-L.; Shan, H.; Liu, T.-F.; Zhao, S.-Q.; Wan, Y.; Jun, Z.; Wang, H.-Q. CT Scans and Cancer Risks: A Systematic Review and Dose-response Meta-analysis. BMC Cancer 2022, 22, 1238. [Google Scholar] [CrossRef] [PubMed]
- Rothkamm, K.; Lobrich, M. Misrepair of radiation-induced DNA double-strand breaks and its relevance for tumorigenesis and cancer treatment (review). Int. J. Oncol. 2002, 21, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Wu, J.; Ranjan, K.; Cui, X.; Wang, X.; Zhang, D.; Zhu, S. Key molecular DNA damage responses of human cells to radiation. Front. Cell Dev. Biol. 2024, 12, 1422520. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Mladenova, V.; Stuschke, M.; Iliakis, G. New Facets of DNA Double Strand Break Repair: Radiation Dose as Key Determinant of HR versus c-NHEJ Engagement. Int. J. Mol. Sci. 2023, 24, 14956. [Google Scholar] [CrossRef]
- Willers, H.; Dahm-Daphi, J.; Powell, S.N. Repair of radiation damage to DNA. Br. J. Cancer 2004, 90, 1297–1301. [Google Scholar] [CrossRef]
- Osipov, A.; Chigasova, A.; Yashkina, E.; Ignatov, M.; Vorobyeva, N.; Zyuzikov, N.; Osipov, A.N. Early and Late Effects of Low-Dose X-Ray Exposure in Human Fibroblasts: DNA Repair Foci, Proliferation, Autophagy, and Senescence. Int. J. Mol. Sci. 2024, 25, 8253. [Google Scholar] [CrossRef]
- Barbieri, S.; Babini, G.; Morini, J.; Friedland, W.; Buonanno, M.; Grilj, V.; Brenner, D.J.; Ottolenghi, A.; Baiocco, G. Predicting DNA damage foci and their experimental readout with 2D microscopy: A unified approach applied to photon and neutron exposures. Sci. Rep. 2019, 9, 14019. [Google Scholar] [CrossRef]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci: Meaning and significance. Env. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef]
- Penninckx, S.; Pariset, E.; Cekanaviciute, E.; Costes, S.V. Quantification of radiation-induced DNA double strand break repair foci to evaluate and predict biological responses to ionizing radiation. NAR Cancer 2021, 3, zcab046. [Google Scholar] [CrossRef]
- Belyaev, I.Y. Radiation-induced DNA repair foci: Spatio-temporal aspects of formation, application for assessment of radiosensitivity and biological dosimetry. Mutat. Res. 2010, 704, 132–141. [Google Scholar] [CrossRef]
- Wanotayan, R.; Chousangsuntorn, K.; Petisiwaveth, P.; Anuttra, T.; Lertchanyaphan, W.; Jaikuna, T.; Jangpatarapongsa, K.; Uttayarat, P.; Tongloy, T.; Chousangsuntorn, C.; et al. A deep learning model (FociRad) for automated detection of gamma-H2AX foci and radiation dose estimation. Sci. Rep. 2022, 12, 5527. [Google Scholar] [CrossRef]
- Belov, O.; Chigasova, A.; Pustovalova, M.; Osipov, A.; Eremin, P.; Vorobyeva, N.; Osipov, A.N. Dose-Dependent Shift in Relative Contribution of Homologous Recombination to DNA Repair after Low-LET Ionizing Radiation Exposure: Empirical Evidence and Numerical Simulation. Curr. Issues Mol. Biol. 2023, 45, 7352–7373. [Google Scholar] [CrossRef]
- Falaschi, A.; Chiaramonte, A.; Testi, S.; Scarpato, R. Dual Immunofluorescence of gammaH2AX and 53BP1 in Human Peripheral Lymphocytes. J. Vis. Exp. 2023, 197, e65472. [Google Scholar] [CrossRef]
- Ignatov, M.A.; Chigasova, A.K.; Osipov, A.A.; Fedotov, Y.A.; Vorobyeva, N.Y.; Osipov, A.N. Effect of X-Ray Energy on the Quantitative Yield of γH2AX and 53BP1 Foci in Human Mesenchymal Stem Cells. Bull. Exp. Biol. Med. 2025, 178, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Slonina, D.; Kowalczyk, A.; Janecka-Widla, A.; Kabat, D.; Szatkowski, W.; Biesaga, B. Low-Dose Hypersensitive Response for Residual pATM and gammaH2AX Foci in Normal Fibroblasts of Cancer Patients. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Ulyanenko, S.; Pustovalova, M.; Koryakin, S.; Beketov, E.; Lychagin, A.; Ulyanenko, L.; Kaprin, A.; Grekhova, A.; Ozerova, A.M.; Ozerov, I.V.; et al. Formation of γH2AX and pATM Foci in Human Mesenchymal Stem Cells Exposed to Low Dose-Rate Gamma-Radiation. Int. J. Mol. Sci. 2019, 20, 2645. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Zhang, X.; Ye, C.; Sun, F.; Wei, W.; Hu, B.; Wang, J. Both Complexity and Location of DNA Damage Contribute to Cellular Senescence Induced by Ionizing Radiation. PLoS ONE 2016, 11, e0155725. [Google Scholar] [CrossRef]
- Pustovalova, M.; Astrelina, T.A.; Grekhova, A.; Vorobyeva, N.; Tsvetkova, A.; Blokhina, T.; Nikitina, V.; Suchkova, Y.; Usupzhanova, D.; Brunchukov, V.; et al. Residual γH2AX foci induced by low dose X-Ray radiation in bone marrow mesenchymal stem cells do not cause accelerated senescence in the progeny of irradiated cells. Aging 2017, 9, 2397–2410. [Google Scholar] [CrossRef][Green Version]
- Nicolay, N.H.; Perez, R.L.; Saffrich, R.; Huber, P.E. Radio-resistant mesenchymal stem cells: Mechanisms of resistance and potential implications for the clinic. Oncotarget 2015, 6, 19366–19380. [Google Scholar] [CrossRef]
- Cho, W.; Kim, E.S.; Kang, C.M.; Ji, Y.H.; Kim, J.I.; Park, S.J.; Son, Y.; Kim, C.H. Low-Dose Ionizing gamma-Radiation Promotes Proliferation of Human Mesenchymal Stem Cells and Maintains Their Stem Cell Characteristics. Tissue Eng. Regen. Med. 2017, 14, 421–432. [Google Scholar] [CrossRef]
- Tubiana, M.; Feinendegen, L.E.; Yang, C.; Kaminski, J.M. The Linear No-Threshold Relationship Is Inconsistent with Radiation Biologic and Experimental Data. Radiology 2009, 251, 13–22. [Google Scholar] [CrossRef]
- Osipov, A.N.; Pustovalova, M.; Grekhova, A.; Eremin, P.; Vorobyova, N.; Pulin, A.; Zhavoronkov, A.; Roumiantsev, S.; Klokov, D.Y.; Eremin, I. Low doses of X-Rays induce prolonged and ATM-independent persistence of gammaH2AX foci in human gingival mesenchymal stem cells. Oncotarget 2015, 6, 27275–27287. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ingram, S.P.; Warmenhoven, J.W.; Henthorn, N.T.; Chadiwck, A.L.; Santina, E.E.; McMahon, S.J.; Schuemann, J.; Kirkby, N.F.; Mackay, R.I.; Kirkby, K.J.; et al. A computational approach to quantifying miscounting of radiation-induced double-strand break immunofluorescent foci. Commun. Biol. 2022, 5, 700. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Uxa, S.; Castillo-Binder, P.; Kohler, R.; Stangner, K.; Müller, G.A.; Engeland, K. Ki-67 gene expression. Cell Death Differ. 2021, 28, 3357–3370. [Google Scholar] [CrossRef] [PubMed]
- Valieva, Y.; Ivanova, E.; Fayzullin, A.; Kurkov, A.; Igrunkova, A. Senescence-Associated β-Galactosidase Detection in Pathology. Diagnostics 2022, 12, 2309. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef]
- Kaatsch, H.L.; Majewski, M.; Schrock, G.; Obermair, R.; Seidel, J.; Nestler, K.; Abend, M.; Waldeck, S.; Port, M.; Ullmann, R.; et al. CT Irradiation-induced Changes of Gene Expression within Peripheral Blood Cells. Health Phys. 2020, 119, 44–51. [Google Scholar] [CrossRef]
- Yachi, Y.; Yoshii, Y.; Matsuya, Y.; Mori, R.; Oikawa, J.; Date, H. Track Structure Study for Energy Dependency of Electrons and X-Rays on DNA Double-Strand Break Induction. Sci. Rep. 2019, 9, 17649. [Google Scholar] [CrossRef]
- Bellamy, M.; Puskin, J.; Hertel, N.; Eckerman, K. An empirical method for deriving RBE values associated with electrons, photons and radionuclides. Radiat. Prot. Dosim. 2015, 167, 664–670. [Google Scholar] [CrossRef]
- Noubissi, F.K.; McBride, A.A.; Leppert, H.G.; Millet, L.J.; Wang, X.; Davern, S.M. Detection and quantification of gamma-H2AX using a dissociation enhanced lanthanide fluorescence immunoassay. Sci. Rep. 2021, 11, 8945. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, K.S.; Kuttikrishnan, S.; Ahmad, N.; Habeeba, U.; Mariyam, Z.; Suleman, M.; Bhat, A.A.; Uddin, S. H2AX: A key player in DNA damage response and a promising target for cancer therapy. Biomed. Pharmacother. 2024, 175, 116663. [Google Scholar] [CrossRef] [PubMed]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008, 36, 5678–5694. [Google Scholar] [CrossRef]
- Kiang, J.G.; Garrison, B.R.; Smith, J.T.; Fukumoto, R. Ciprofloxacin as a potential radio-sensitizer to tumor cells and a radio-protectant for normal cells: Differential effects on γ-H2AX formation, p53 phosphorylation, Bcl-2 production, and cell death. Mol. Cell. Biochem. 2014, 393, 133–143. [Google Scholar] [CrossRef]
- Rass, E.; Willaume, S.; Bertrand, P. 53BP1: Keeping It under Control, Even at a Distance from DNA Damage. Genes 2022, 13, 2390. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Du, S.; Peng, Z.; Chen, L. Multifaceted regulation and functions of 53BP1 in NHEJ-mediated DSB repair (Review). Int. J. Mol. Med. 2022, 50, 90. [Google Scholar] [CrossRef]
- Bartova, E.; Legartova, S.; Dundr, M.; Suchankova, J. A role of the 53BP1 protein in genome protection: Structural and functional characteristics of 53BP1-dependent DNA repair. Aging 2019, 11, 2488–2511. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. ATM’s Role in the Repair of DNA Double-Strand Breaks. Genes 2021, 12, 1370. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Phan, L.M.; Rezaeian, A.H. ATM: Main Features, Signaling Pathways, and Its Diverse Roles in DNA Damage Response, Tumor Suppression, and Cancer Development. Genes 2021, 12, 845. [Google Scholar] [CrossRef]
- Zyuzikov, N.A.; Coates, P.J.; Parry, J.M.; Lorimore, S.A.; Wright, E.G. Lack of Nontargeted Effects in Murine Bone Marrow after Low-Dose In Vivo X Irradiation. Radiat. Res. 2011, 175, 322–327. [Google Scholar] [CrossRef]
- Nicola, N.A.; Babon, J.J. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 2015, 26, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.C.; Silva, D.N.; Fortuna, V.; Silveira, B.M.; Orge, I.D.; de Santana, T.A.; Sampaio, G.L.; Paredes, B.D.; Ribeiro-dos-Santos, R.; Soares, M.B.P. Leukemia Inhibitory Factor (LIF) Overexpression Increases the Angiogenic Potential of Bone Marrow Mesenchymal Stem/Stromal Cells. Front. Cell Dev. Biol. 2020, 8, 778. [Google Scholar] [CrossRef]
- Mikhailova, E.; Sokolenko, A.; Combs, S.E.; Shevtsov, M. Modulation of Heat Shock Proteins Levels in Health and Disease: An Integrated Perspective in Diagnostics and Therapy. Cells 2025, 14, 979. [Google Scholar] [CrossRef]
- Albakova, Z.; Mangasarova, Y.; Albakov, A.; Gorenkova, L. HSP70 and HSP90 in Cancer: Cytosolic, Endoplasmic Reticulum and Mitochondrial Chaperones of Tumorigenesis. Front. Oncol. 2022, 12, 829520. [Google Scholar] [CrossRef] [PubMed]
- Hunziker, A.; Jensen, M.H.; Krishna, S. Stress-specific response of the p53-Mdm2 feedback loop. BMC Syst. Biol. 2010, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zheng, S.; Liu, M.; Wang, G. Novel Anti-Cancer Stem Cell Compounds: A Comprehensive Review. Pharmaceutics 2024, 16, 1024. [Google Scholar] [CrossRef]
- Koo, N.; Sharma, A.K.; Narayan, S. Therapeutics Targeting p53-MDM2 Interaction to Induce Cancer Cell Death. Int. J. Mol. Sci. 2022, 23, 5005. [Google Scholar] [CrossRef]
- Imamichi, Y.; Mizutani, T.; Ju, Y.; Matsumura, T.; Kawabe, S.; Kanno, M.; Yazawa, T.; Miyamoto, K. Transcriptional regulation of human ferredoxin reductase through an intronic enhancer in steroidogenic cells. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 33–42. [Google Scholar] [CrossRef]
- O’Brien, G.; Cruz-Garcia, L.; Majewski, M.; Grepl, J.; Abend, M.; Port, M.; Tichý, A.; Sirak, I.; Malkova, A.; Donovan, E.; et al. FDXR is a biomarker of radiation exposure in vivo. Sci. Rep. 2018, 8, 684. [Google Scholar] [CrossRef]
- Brzóska, K.; Abend, M.; O’Brien, G.; Gregoire, E.; Port, M.; Badie, C. Calibration curve for radiation dose estimation using FDXR gene expression biodosimetry—Premises and pitfalls. Int. J. Radiat. Biol. 2024, 100, 1202–1212. [Google Scholar] [CrossRef]
- Kawase, T.; Ichikawa, H.; Ohta, T.; Nozaki, N.; Tashiro, F.; Ohki, R.; Taya, Y. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene 2008, 27, 3797–3810. [Google Scholar] [CrossRef]
- Chuang, L.S.H.; Matsuo, J.; Douchi, D.; Bte Mawan, N.A.; Ito, Y. RUNX3 in Stem Cell and Cancer Biology. Cells 2023, 12, 408. [Google Scholar] [CrossRef]
- Ge, X.; Shen, Z.; Yin, Y. Comprehensive review of LncRNA-mediated therapeutic resistance in non-small cell lung cancer. Cancer Cell Int. 2024, 24, 369. [Google Scholar] [CrossRef]
- Nakayama, F.; Umeda, S.; Ichimiya, T.; Kamiyama, S.; Hazawa, M.; Yasuda, T.; Nishihara, S.; Imai, T. Sulfation of keratan sulfate proteoglycan reduces radiation-induced apoptosis in human Burkitt’s lymphoma cell lines. FEBS Lett. 2012, 587, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Osipov, A.; Chigasova, A.; Yashkina, E.; Ignatov, M.; Fedotov, Y.; Molodtsova, D.; Vorobyeva, N.; Osipov, A.N. Residual Foci of DNA Damage Response Proteins in Relation to Cellular Senescence and Autophagy in X-Ray Irradiated Fibroblasts. Cells 2023, 12, 1209. [Google Scholar] [CrossRef] [PubMed]
- Zorin, V.; Zorina, A.; Smetanina, N.; Kopnin, P.; Ozerov, I.V.; Leonov, S.; Isaev, A.; Klokov, D.; Osipov, A.N. Diffuse colonies of human skin fibroblasts in relation to cellular senescence and proliferation. Aging 2017, 9, 1404–1413. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ignatov, M.; Markelova, E.E.; Chigasova, A.; Osipov, A.; Buianov, I.; Fedotov, Y.; Eremin, P.; Vorobyeva, N.; Zyuzikov, N.; Osipov, A.N. Molecular and Cellular Effects of CT Scans in Human Adipose Mesenchymal Stem Cells. Int. J. Mol. Sci. 2025, 26, 8584. https://doi.org/10.3390/ijms26178584
Ignatov M, Markelova EE, Chigasova A, Osipov A, Buianov I, Fedotov Y, Eremin P, Vorobyeva N, Zyuzikov N, Osipov AN. Molecular and Cellular Effects of CT Scans in Human Adipose Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2025; 26(17):8584. https://doi.org/10.3390/ijms26178584
Chicago/Turabian StyleIgnatov, Maxim, Ekaterina E. Markelova, Anna Chigasova, Andrey Osipov, Ilia Buianov, Yuriy Fedotov, Petr Eremin, Natalia Vorobyeva, Nikolay Zyuzikov, and Andreyan N. Osipov. 2025. "Molecular and Cellular Effects of CT Scans in Human Adipose Mesenchymal Stem Cells" International Journal of Molecular Sciences 26, no. 17: 8584. https://doi.org/10.3390/ijms26178584
APA StyleIgnatov, M., Markelova, E. E., Chigasova, A., Osipov, A., Buianov, I., Fedotov, Y., Eremin, P., Vorobyeva, N., Zyuzikov, N., & Osipov, A. N. (2025). Molecular and Cellular Effects of CT Scans in Human Adipose Mesenchymal Stem Cells. International Journal of Molecular Sciences, 26(17), 8584. https://doi.org/10.3390/ijms26178584







