Analysis of Microarray and Single-Cell RNA-Seq Finds Gene Co-Expression and Tumor Environment Associated with Extracellular Matrix in Epithelial–Mesenchymal Transition in Prostate Cancer
Abstract
1. Introduction
2. Results
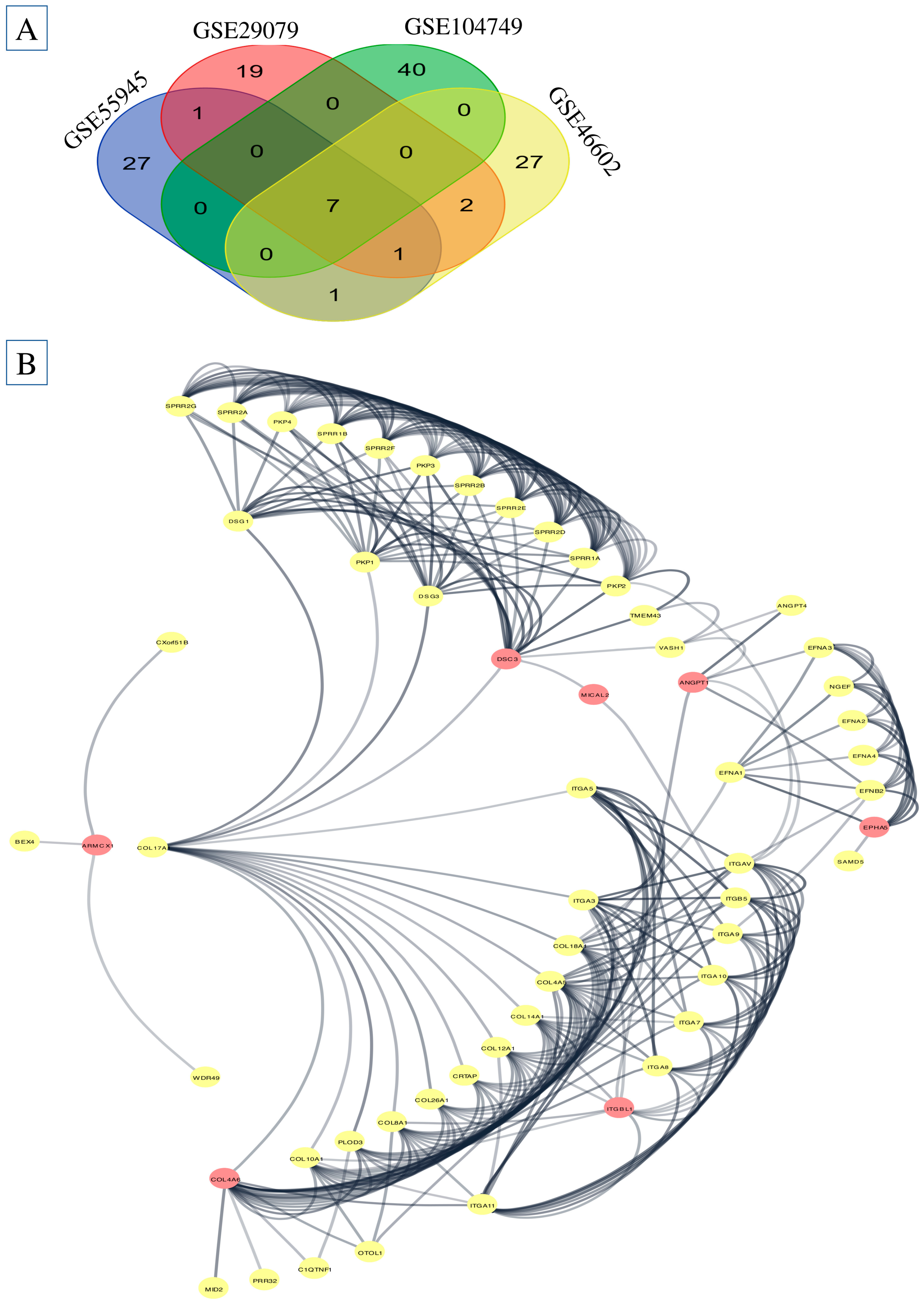
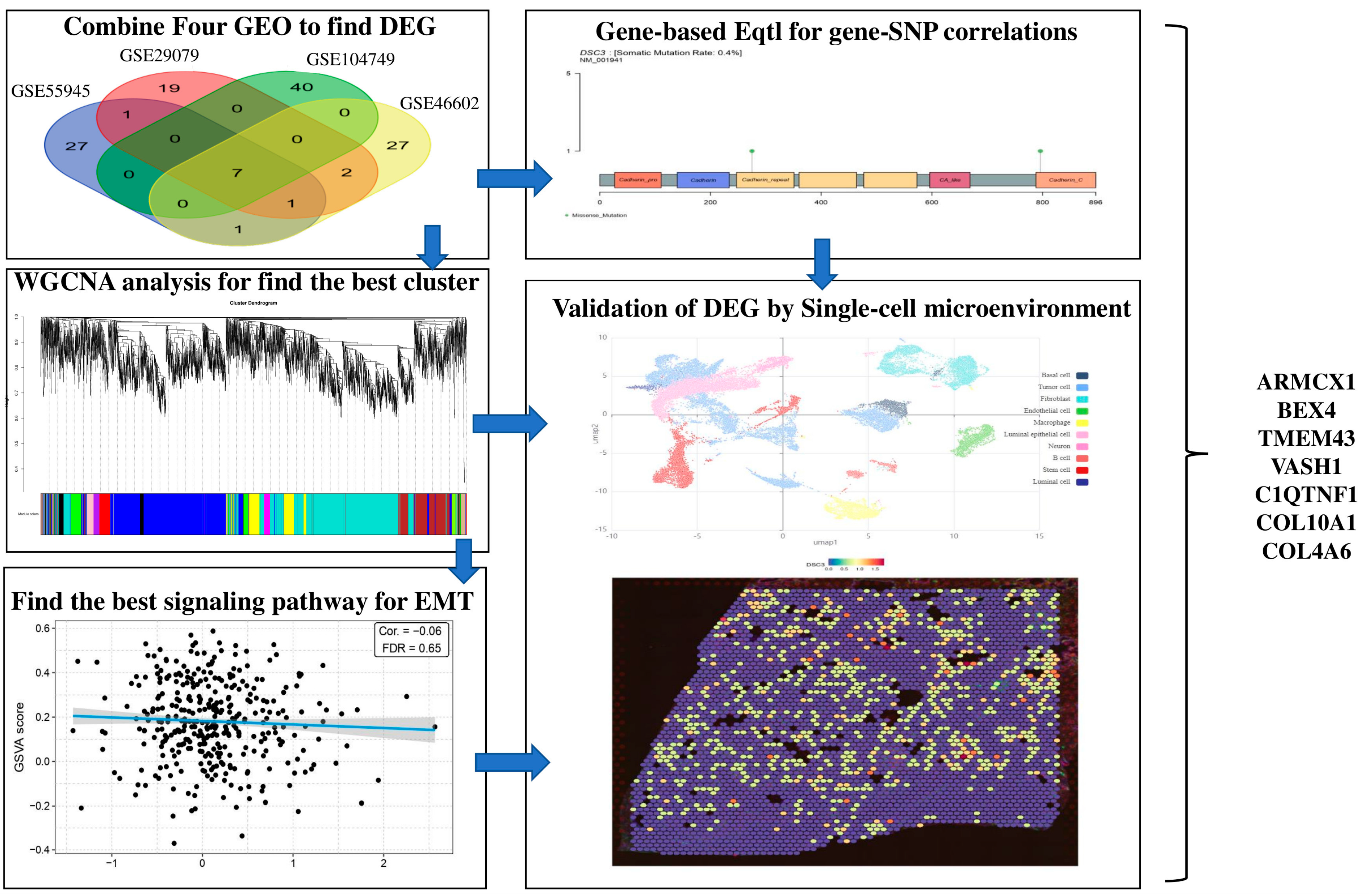
2.1. Differentially Expressed Genes (DEGs) Screening
2.2. Protein Class Sorting and Finding EMT Genes
2.3. PPI Analysis
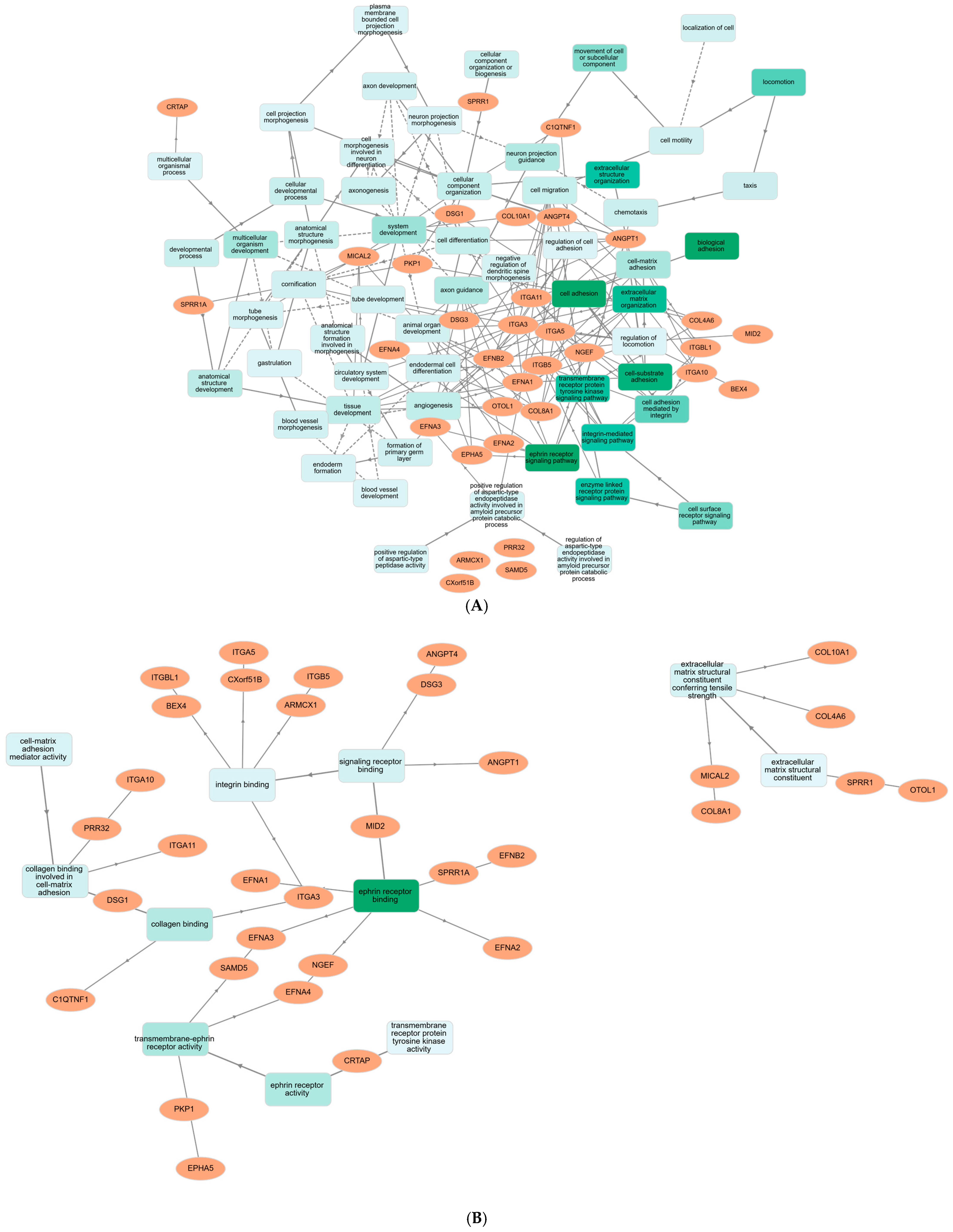
2.4. KEGG Pathway Enrichment
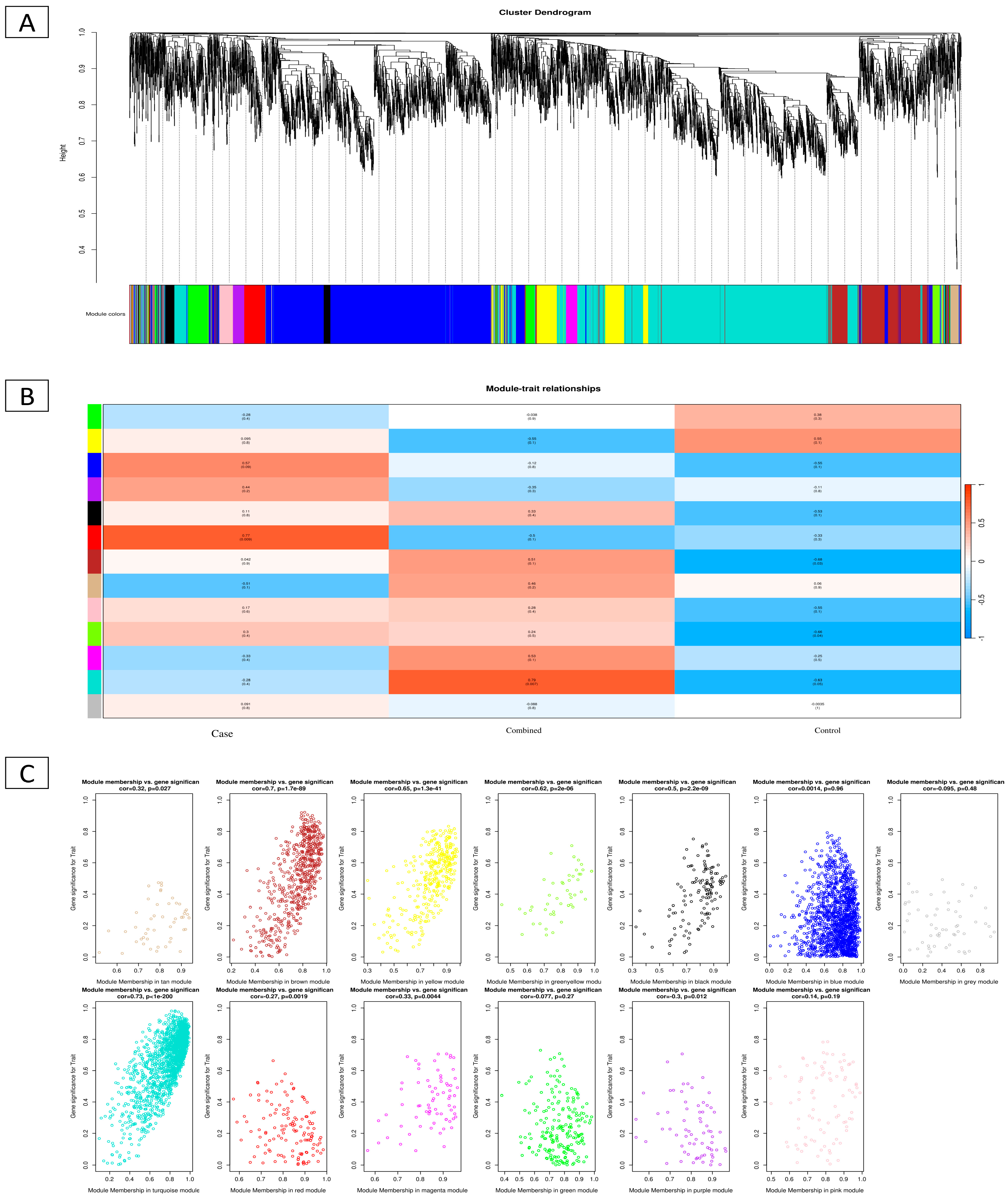
2.5. WGCNA of DEGs
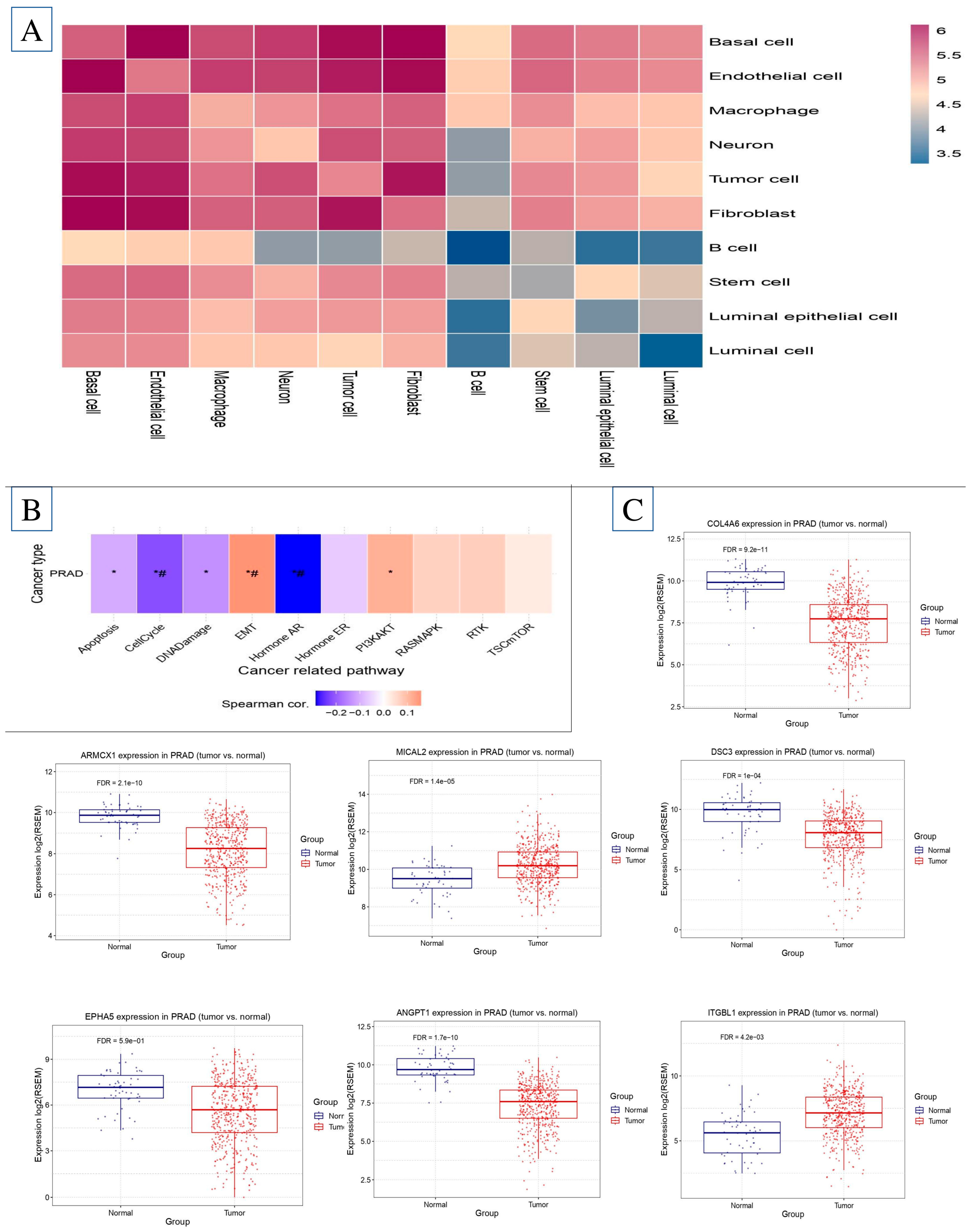
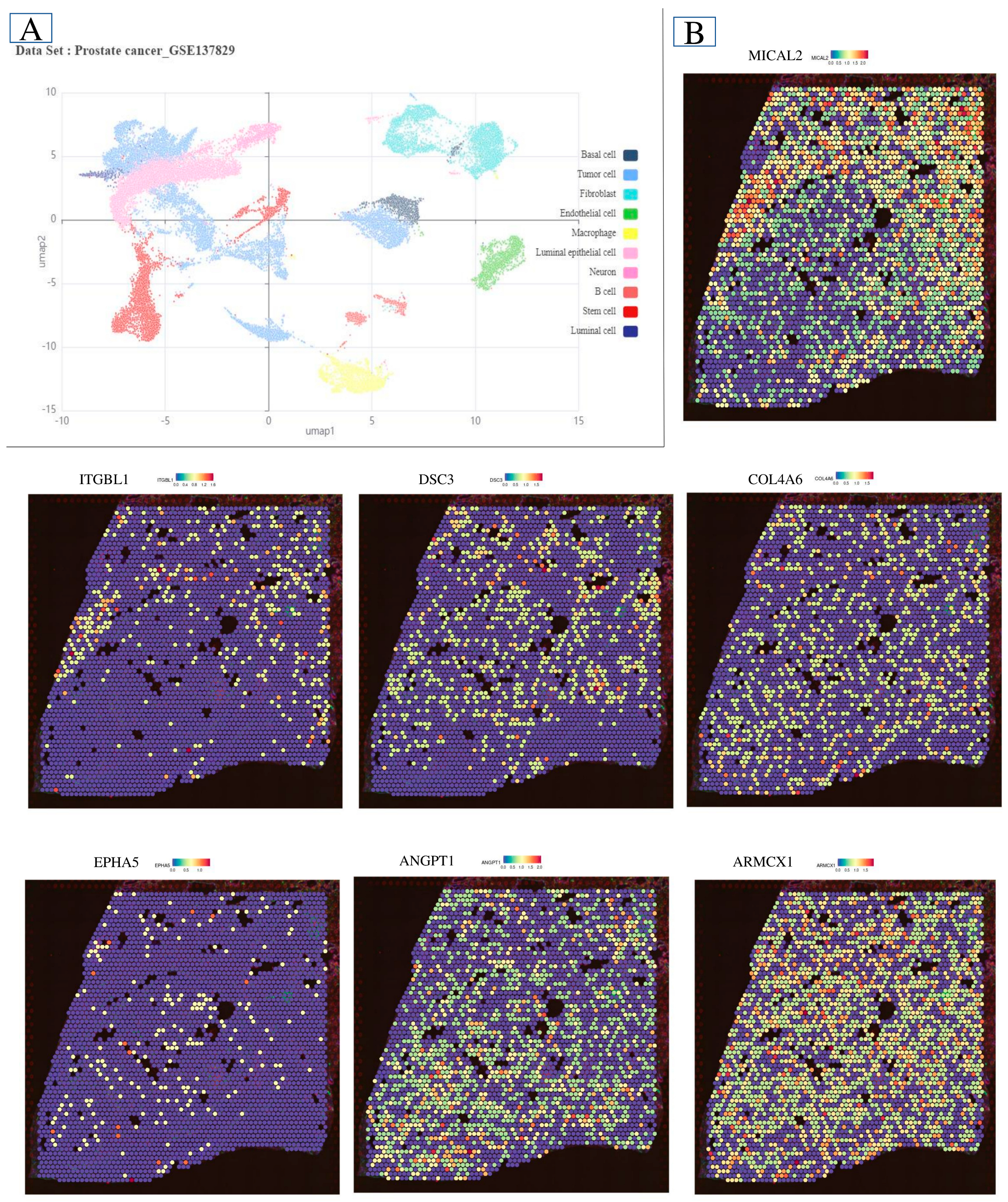
2.6. Single-Cell Microenvironment Gene Expression
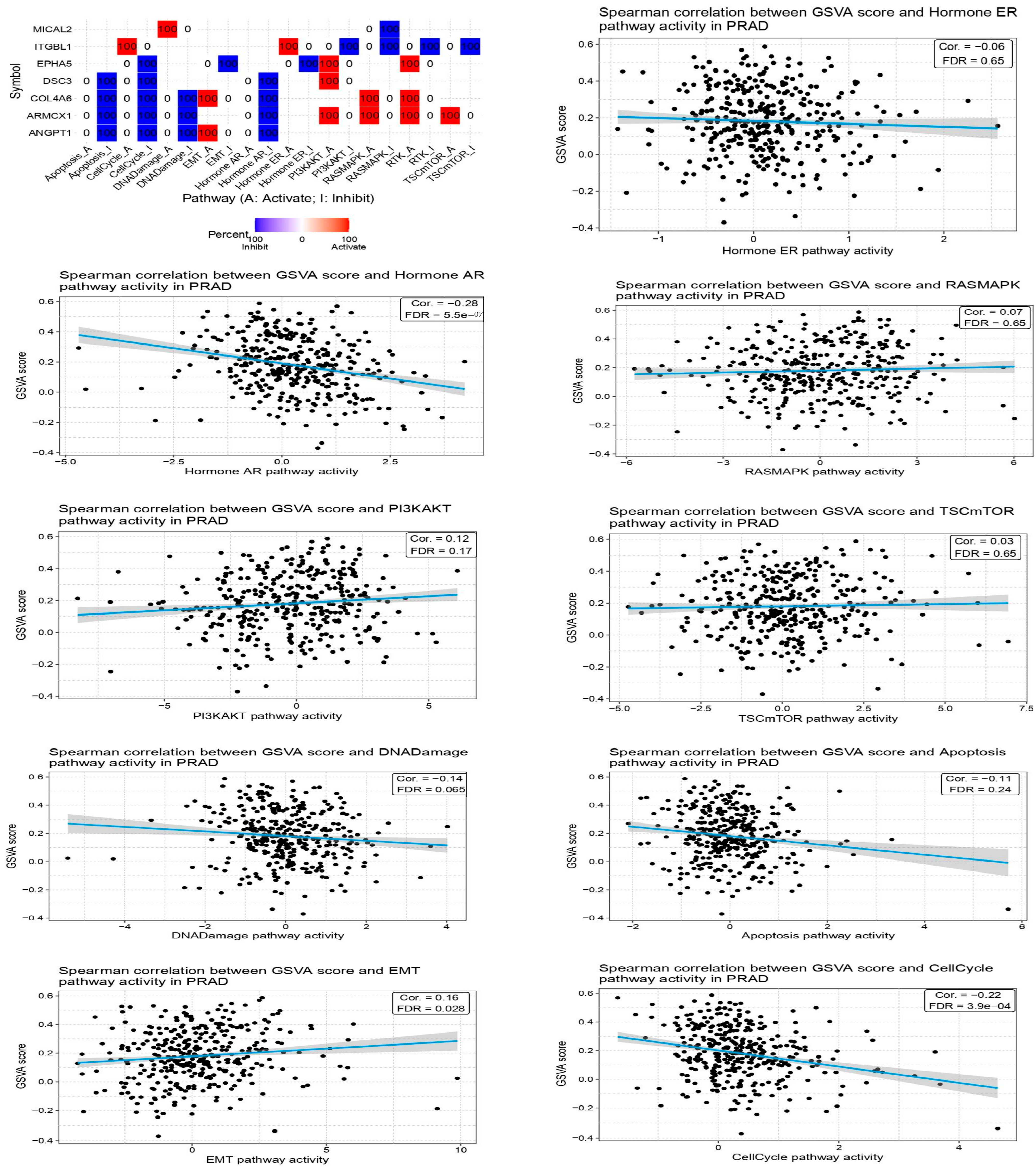
2.7. Investigation of Immune Infiltration and Prostate Cancer
2.8. Survival Analysis
2.9. Gene-Based eQTL
2.10. Characteristics of Significant Cis-eQTL Findings
3. Discussion
4. Materials and Methods
4.1. Microarray Data
4.2. Data Processing
4.3. Extracellular Matrix in Epithelial–Mesenchymal Transition Genes in PCa
4.4. Transcription Factors Regulating EMT Genes
4.5. EMT Co-Expressed Genes and PPI Network
4.6. EMT Genes Functional Enrichment Analysis
4.7. Exploring GO and KEGG Pathway
4.8. Regulatory Network Analysis of KGs
4.9. WGCNA
4.10. Data Collection and Preprocessing of Single Cells
4.11. EMT Genes Expression and Survival Analysis in PCa
4.12. Examining Gene Involved in EMT in Relation to Immunological Subtypes
4.13. Expression Analysis for KGs by GEPIA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, H.; Weinstein, H.N.W.; Allegakoen, P.; Wadsworth, M.H.; Xie, J.; Yang, H.; Castro, E.A.; Lu, K.L.; Stohr, B.A.; Feng, F.Y.; et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat. Commun. 2022, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Karoii, D.H.; Bavandi, S.; Djamali, M.; Abroudi, A.S. Exploring the interaction between immune cells in the prostate cancer microenvironment combining weighted correlation gene network analysis and single-cell sequencing: An integrated bioinformatics analysis. Discov. Oncol. 2024, 15, 513. [Google Scholar] [CrossRef] [PubMed]
- Amirian, M.; Azizi, H.; Hashemi Karoii, D.; Karoii, T. VASA protein and gene expression analysis of human non-obstructive azoospermia and normal by immunohistochemistry, immunocytochemistry, and bioinformatics analysis. Sci. Rep. 2022, 12, 17259. [Google Scholar] [CrossRef] [PubMed]
- Azizi, H.; Karoii, D.H.; Skutella, T. Clinical management, differential diagnosis, follow-up and biomarkers of infertile men with nonobstructive azoospermia. Transl. Androl. Urol. 2024, 13, 359–362. [Google Scholar] [CrossRef]
- Nik, B.D.; Karoii, D.H.; Favaedi, R.; Ramazanali, F.; Jahangiri, M.; Movaghar, B.; Shahhoseini, M. Differential expression of ion channel coding genes in the endometrium of women experiencing recurrent implantation failures. Sci. Rep. 2024, 14, 19822. [Google Scholar] [CrossRef]
- Karoii, D.H.; Abroudi, A.S.; Darvar, M.; Djamali, M.; Azizi, H.; Skutella, T. Identification of novel long non-coding RNA involved in Sertoli cell of non-obstructive azoospermia based on microarray and bioinformatics analysis. Genomics 2025, 117, 111046. [Google Scholar] [CrossRef]
- Henry, G.H.; Malewska, A.; Joseph, D.B.; Malladi, V.S.; Lee, J.; Torrealba, J.; Mauck, R.J.; Gahan, J.C.; Raj, G.V.; Roehrborn, C.G.; et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep. 2018, 25, 3530–3542.e5. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H. A review of protein-protein interaction and signaling pathway of Vimentin in cell regulation, morphology and cell differentiation in normal cells. J. Recept. Signal Transduct. 2022, 42, 512–520. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H. OCT4 protein and gene expression analysis in the differentiation of spermatogonia stem cells into neurons by immunohistochemistry, immunocytochemistry, and bioinformatics analysis. Stem Cell Rev. Rep. 2023, 19, 1828–1844. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, J.K.; Phillips, J.W.; Huang, P.; Cheng, D.; Huang, J.; Witte, O.N. Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc. Natl. Acad. Sci. USA 2016, 113, 4482–4487. [Google Scholar] [CrossRef]
- Wei, Y.; Han, S.; Wen, J.; Liao, J.; Liang, J.; Yu, J.; Chen, X.; Xiang, S.; Huang, Z.; Zhang, B. E26 transformation-specific transcription variant 5 in development and cancer: Modification, regulation and function. J. Biomed. Sci. 2023, 30, 17. [Google Scholar] [CrossRef]
- Scaravilli, M.; Koivukoski, S.; Latonen, L. Androgen-Driven Fusion Genes and Chimeric Transcripts in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 623809. [Google Scholar] [CrossRef]
- Graham, M.K.; Wang, R.; Chikarmane, R.; Abel, B.; Vaghasia, A.; Gupta, A.; Zheng, Q.; Hicks, J.; Sysa-Shah, P.; Pan, X.; et al. Convergent alterations in the tumor microenvironment of MYC-driven human and murine prostate cancer. Nat. Commun. 2024, 15, 7414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, S.; Liu, M.; Liu, Y.; Ye, Z.; Zeng, J.; Yao, M. Experimental models for cancer brain metastasis. Cancer Pathog. Ther. 2024, 2, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Jiang, J.; Lu, Y.; Nice, E.C.; Huang, C.; Zhang, J.; He, W. Emerging role of tumor cell plasticity in modifying therapeutic response. Signal Transduct. Target. Ther. 2020, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.D.; Gao, D.; Redfern, A.; Thompson, E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 2019, 19, 716–732. [Google Scholar] [CrossRef]
- Akhmetkaliyev, A.; Alibrahim, N.; Shafiee, D.; Tulchinsky, E. EMT/MET plasticity in cancer and Go-or-Grow decisions in quiescence: The two sides of the same coin? Mol. Cancer 2023, 22, 90. [Google Scholar] [CrossRef]
- Chaves, L.P.; Melo, C.M.; Saggioro, F.P.; Reis, R.B.D.; Squire, J.A. Epithelial-Mesenchymal Transition Signaling and Prostate Cancer Stem Cells: Emerging Biomarkers and Opportunities for Precision Therapeutics. Genes 2021, 12, 1900. [Google Scholar] [CrossRef]
- Chen, H.; Fang, S.; Zhu, X.; Liu, H. Cancer-associated fibroblasts and prostate cancer stem cells: Crosstalk mechanisms and implications for disease progression. Front. Cell Dev. Biol. 2024, 12, 1412337. [Google Scholar] [CrossRef]
- Zhang, H.; Yue, X.; Chen, Z.; Liu, C.; Wu, W.; Zhang, N.; Liu, Z.; Yang, L.; Jiang, Q.; Cheng, Q.; et al. Define cancer-associated fibroblasts (CAFs) in the tumor microenvironment: New opportunities in cancer immunotherapy and advances in clinical trials. Mol. Cancer 2023, 22, 159. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Liu, K.; Liu, Y.; Wang, S.; Jiang, W.; Lu, F.; Dang, Y. Targets of tumor microenvironment for potential drug development. MedComm Oncol. 2024, 3, e68. [Google Scholar] [CrossRef]
- Beltran, H.; Hruszkewycz, A.; Scher, H.I.; Hildesheim, J.; Isaacs, J.; Yu, E.Y.; Kelly, K.; Lin, D.; Dicker, A.; Arnold, J.; et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clin. Cancer Res. 2019, 25, 6916–6924. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.; Nouruzi, S.; Ganguli, D.; Namekawa, T.; Thaper, D.; Linder, S.; Karaoğlanoğlu, F.; Omur, M.E.; Kim, S.; Kobelev, M.; et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021, 23, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wang, C.; Wang, Y.; Xu, Y.; Li, X.; Johnson, N.A.; Mukherji, A.; Lo, U.G.; Xu, L.; Gonzalez, J.; et al. Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat. Cancer 2022, 3, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Li, F.; Zhang, Y.; Dai, P.; He, J.; Li, Y.; Zhu, Y.; Zheng, J.; Huang, H.; Bai, F.; et al. FOXA2 drives lineage plasticity and KIT pathway activation in neuroendocrine prostate cancer. Cancer Cell 2022, 40, 1306–1323.e8. [Google Scholar] [CrossRef]
- Chataigner, L.M.P.; Gogou, C.; den Boer, M.A.; Frias, C.P.; Thies-Weesie, D.M.E.; Granneman, J.C.M.; Heck, A.J.R.; Meijer, D.H.; Janssen, B.J.C. Structural insights into the contactin 1—Neurofascin 155 adhesion complex. Nat. Commun. 2022, 13, 6607. [Google Scholar] [CrossRef]
- Chen, D.H.; Yu, J.W.; Jiang, B.J. Contactin 1: A potential therapeutic target and biomarker in gastric cancer. World J. Gastroenterol. 2015, 21, 9707–9716. [Google Scholar] [CrossRef]
- Beier, A.K.; Puhr, M.; Stope, M.B.; Thomas, C.; Erb, H.H.H. Metabolic changes during prostate cancer development and progression. J. Cancer Res. Clin. Oncol. 2023, 149, 2259–2270. [Google Scholar] [CrossRef]
- Yu, W.; Wang, C.; Shang, Z.; Tian, J. Unveiling novel insights in prostate cancer through single-cell RNA sequencing. Front. Oncol. 2023, 13, 1224913. [Google Scholar] [CrossRef]
- Srinivasan, S.; Kryza, T.; Bock, N.; Tse, B.W.C.; Sokolowski, K.A.; Janaththani, P.; Fernando, A.; Moya, L.; Stephens, C.; Dong, Y.; et al. A PSA SNP associates with cellular function and clinical outcome in men with prostate cancer. Nat. Commun. 2024, 15, 9587. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H.; Darvari, M.; Qorbanee, A.; Hawezy, D.J. Identification of novel cytoskeleton protein involved in spermatogenic cells and sertoli cells of non-obstructive azoospermia based on microarray and bioinformatics analysis. BMC Med. Genom. 2025, 18, 19. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H.; Skutella, T. Altered G-Protein Transduction Protein Gene Expression in the Testis of Infertile Patients with Nonobstructive Azoospermia. DNA Cell Biol. 2023, 42, 617–637. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H.; Skutella, T. Microarray and in silico analysis of DNA repair genes between human testis of patients with nonobstructive azoospermia and normal cells. Cell Biochem. Funct. 2022, 40, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Steinbuch, S.C.; Lüß, A.-M.; Eltrop, S.; Götte, M.; Kiesel, L. Endometriosis-Associated Ovarian Cancer: From Molecular Pathologies to Clinical Relevance. Int. J. Mol. Sci. 2024, 25, 4306. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.P.; Green, K.J. Structure, function, and regulation of desmosomes. Prog. Mol. Biol. Transl. Sci. 2013, 116, 95–118. [Google Scholar] [PubMed]
- Song, K.; Yu, Z.; Zu, X.; Li, G.; Hu, Z.; Xue, Y. Collagen Remodeling along Cancer Progression Providing a Novel Opportunity for Cancer Diagnosis and Treatment. Int. J. Mol. Sci. 2022, 23, 10509. [Google Scholar] [CrossRef]
- Nong, B.; Su, T.; Jin, M.; Huang, J.; Huang, A.; Fang, D.; Wei, J. Immune-related gene ANGPT1 is an adverse biomarker for endometrial carcinoma. Transl. Cancer Res. 2021, 10, 2962–2976. [Google Scholar] [CrossRef]
- Hu, Z.; Wu, Y.; Sun, X.; Tong, Y.; Qiu, H.; Zhuo, E. ARMCX1 inhibits lung adenocarcinoma progression by recruiting FBXW7 for c-Myc degradation. Biol. Direct 2024, 19, 82. [Google Scholar] [CrossRef]
- Mariotti, S.; Barravecchia, I.; Vindigni, C.; Pucci, A.; Balsamo, M.; Libro, R.; Senchenko, V.; Dmitriev, A.; Jacchetti, E.; Cecchini, M.; et al. MICAL2 is a novel human cancer gene controlling mesenchymal to epithelial transition involved in cancer growth and invasion. Oncotarget 2016, 7, 1808–1825. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, D.; Guan, X.; Ma, S.; Zhou, W.; Peng, J.; Yuan, L.; Wang, Y.; Jin, S.; Xu, Q.; et al. Identification of distinct tumor cell patterns with single-cell RNA sequencing integrating primary lung adenocarcinoma and brain metastasis tumor. Transl. Lung Cancer Res. 2023, 12, 547–565. [Google Scholar] [CrossRef]
- Zhong, Z.; Hou, J.; Yao, Z.; Dong, L.; Liu, F.; Yue, J.; Wu, T.; Zheng, J.; Ouyang, G.; Yang, C.; et al. Domain generalization enables general cancer cell annotation in single-cell and spatial transcriptomics. Nat. Commun. 2024, 15, 1929. [Google Scholar] [CrossRef]
- Arredouani, M.S.; Lu, B.; Bhasin, M.; Eljanne, M.; Yue, W.; Mosquera, J.M.; Bubley, G.J.; Li, V.; Rubin, M.A.; Libermann, T.A.; et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin. Cancer Res. 2009, 15, 5794–5802. [Google Scholar] [CrossRef]
- Shan, M.; Xia, Q.; Yan, D.; Zhu, Y.; Zhang, X.; Zhang, G.; Guo, J.; Hou, J.; Chen, W.; Zhu, T.; et al. Molecular analyses of prostate tumors for diagnosis of malignancy on fine-needle aspiration biopsies. Oncotarget 2017, 8, 104761–104771. [Google Scholar] [CrossRef]
- Mortensen, M.M.; Høyer, S.; Lynnerup, A.S.; Ørntoft, T.F.; Sørensen, K.D.; Borre, M.; Dyrskjøt, L. Expression profiling of prostate cancer tissue delineates genes associated with recurrence after prostatectomy. Sci. Rep. 2015, 5, 16018. [Google Scholar] [CrossRef]
- Kuner, R.; Fälth, M.; Pressinotti, N.C.; Brase, J.C.; Puig, S.B.; Metzger, J.; Gade, S.; Schäfer, G.; Bartsch, G.; Steiner, E.; et al. The maternal embryonic leucine zipper kinase (MELK) is upregulated in high-grade prostate cancer. J. Mol. Med. 2013, 91, 237–248. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Karoii, D.H.; Azizi, H.; Amirian, M. Signaling pathways and protein–protein interaction of vimentin in invasive and migration cells: A review. Cell. Reprogram. 2022, 24, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Karoii, D.H.; Azizi, H.; Skutella, T. Whole transcriptome analysis to identify non-coding RNA regulators and hub genes in sperm of non-obstructive azoospermia by microarray, single-cell RNA sequencing, weighted gene co-expression network analysis, and mRNA-miRNA-lncRNA interaction analysis. BMC Genom. 2024, 25, 583. [Google Scholar] [CrossRef] [PubMed]
- Niazi Tabar, A.; Azizi, H.; Hashemi Karoii, D.; Skutella, T. Testicular Localization and Potential Function of Vimentin Positive Cells during Spermatogonial Differentiation Stages. Animals 2022, 12, 268. [Google Scholar] [CrossRef]
- Shams, A.A.; Vesal, S.; Karoii, D.H.; Vesali, S.; Alizadeh, A.; Shahhoseini, M. Paternal trans fatty acid and vitamin E diet affect the expression pattern of androgen signaling pathway genes in the testis of rat offspring. Theriogenology 2025, 231, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hashemi Karoii, D.; Azizi, H.; Skutella, T. Integrating Microarray Data and Single-Cell RNA-Seq Reveals Key Gene Involved in Spermatogonia Stem Cell Aging. Int. J. Mol. Sci. 2024, 25, 11653. [Google Scholar] [CrossRef] [PubMed]
- Hashemi Karoii, D.; Baghaei, H.; Abroudi, A.S.; Djamali, M.; Hasani Mahforoozmahalleh, Z.; Azizi, H.; Skutella, T. Alteration of the metabolite interconversion enzyme in sperm and Sertoli cell of non-obstructive azoospermia: A microarray data and in-silico analysis. Sci. Rep. 2024, 14, 25965. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, J.E.; Xie, Z.; Marino, G.B.; Nguyen, N.; Clarke, D.J.; Ma’ayan, A. Enrichr-KG: Bridging enrichment analysis across multiple libraries. Nucleic Acids Res. 2023, 51, W168–W179. [Google Scholar] [CrossRef]
- Pei, G.; Chen, L.; Zhang, W. WGCNA application to proteomic and metabolomic data analysis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 585, pp. 135–158. [Google Scholar]
- Pereira, W.J.; Almeida, F.M.; Conde, D.; Balmant, K.; Triozzi, P.; Schmidt, H.; Dervinis, C.; Pappas, G.; Kirst, M. Asc-Seurat: Analytical single-cell Seurat-based web application. BMC Bioinform. 2021, 22, 556. [Google Scholar] [CrossRef]
- Dong, B.; Miao, J.; Wang, Y.; Luo, W.; Ji, Z.; Lai, H.; Zhang, M.; Cheng, X.; Wang, J.; Fang, Y.; et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun. Biol. 2020, 3, 778. [Google Scholar] [CrossRef]
- Azizi, H.; Hashemi Karoii, D.; Skutella, T. Whole Exome Sequencing and In Silico Analysis of Human Sertoli in Patients with Non-Obstructive Azoospermia. Int. J. Mol. Sci. 2022, 23, 12570. [Google Scholar] [CrossRef]
- Hashemi Karoii, D.; Azizi, H. Functions and mechanism of noncoding RNA in regulation and differentiation of male mammalian reproduction. Cell Biochem. Funct. 2023, 41, 767–778. [Google Scholar] [CrossRef]


| Dataset | GEO Accession | Platform | Total Samples | Cancer Samples | Normal Samples | Assigned Subtypes | Clinical Metadata |
|---|---|---|---|---|---|---|---|
| GSE55945 | GPL570 | Affymetrix Human Genome U133 Plus 2.0 Array | 72 | 36 | 36 | Luminal A (12), Luminal B (14), Basal (10) | Limited (age, diagnosis) |
| GSE104749 | GPL10558 | Illumina HumanHT-12 V4.0 Expression BeadChip | 20 | 16 | 4 | Luminal B (6), Basal (5), Neuroendocrine (5) | Gleason score provided |
| GSE46602 | GPL570 | Affymetrix Human Genome U133 Plus 2.0 Array | 50 | 36 | 14 | Luminal A (10), Basal (15), Neuroendocrine (11) | Gleason score, TNM stage |
| GSE32571 | GPL6244 | Affymetrix Human Gene 1.0 ST Array | 22 | 13 | 9 | Luminal B (4), Basal (5), Neuroendocrine (4) | Very limited |
| Gene | log2FC (GSE55945) | log2FC (GSE104749) | log2FC (GSE46602) | log2FC (GSE32571) | FDR (Avg) | Subtype(s) Highly Expressed |
|---|---|---|---|---|---|---|
| ARMCX1 | +2.4 | +1.9 | +2.7 | +2.1 | <0.001 | Luminal A, Basal |
| BEX4 | +1.8 | +2.0 | +2.5 | +2.2 | <0.001 | Basal |
| TMEM43 | −2.1 | −2.3 | −1.9 | −2.0 | <0.001 | Downregulated in all |
| VASH1 | +1.6 | +1.7 | +2.1 | +1.9 | 0.002 | Luminal B |
| C1QTNF1 | +2.2 | +2.5 | +2.8 | +2.6 | <0.001 | Neuroendocrine |
| COL10A1 | +2.9 | +3.0 | +3.4 | +3.1 | <0.001 | All subtypes |
| COL4A6 | −2.5 | −2.1 | −2.7 | −2.3 | <0.001 | Downregulated in all |
| Sample/Accession No. | Prostate Cancer | Benign Prostate Hyperplasia | Sample Type | Platform | RNA Type |
|---|---|---|---|---|---|
| GSE55945 | 8 | 8 | tissue | Affymetrix GPL570 | mRNA |
| GSE104749 | 4 | 4 | tissue | Affymetrix GPL570 | mRNA |
| GSE46602 | 36 | 14 | tissue | Affymetrix GPL570 | mRNA |
| GSE32571 | 59 | 39 | tissue | Affymetrix GPL6947 | mRNA |
| GSE137829 | 6 | 6 | Tissue | GPL24676 Illumina NovaSeq 6000 | mRNA/Single cell |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shakeri Abroudi, A.; Mashhouri Moghaddam, M.; Hashemi Karoii, D.; Djamali, M.; Azizi, H.; Skutella, T. Analysis of Microarray and Single-Cell RNA-Seq Finds Gene Co-Expression and Tumor Environment Associated with Extracellular Matrix in Epithelial–Mesenchymal Transition in Prostate Cancer. Int. J. Mol. Sci. 2025, 26, 8575. https://doi.org/10.3390/ijms26178575
Shakeri Abroudi A, Mashhouri Moghaddam M, Hashemi Karoii D, Djamali M, Azizi H, Skutella T. Analysis of Microarray and Single-Cell RNA-Seq Finds Gene Co-Expression and Tumor Environment Associated with Extracellular Matrix in Epithelial–Mesenchymal Transition in Prostate Cancer. International Journal of Molecular Sciences. 2025; 26(17):8575. https://doi.org/10.3390/ijms26178575
Chicago/Turabian StyleShakeri Abroudi, Ali, Mahtab Mashhouri Moghaddam, Danial Hashemi Karoii, Melika Djamali, Hossein Azizi, and Thomas Skutella. 2025. "Analysis of Microarray and Single-Cell RNA-Seq Finds Gene Co-Expression and Tumor Environment Associated with Extracellular Matrix in Epithelial–Mesenchymal Transition in Prostate Cancer" International Journal of Molecular Sciences 26, no. 17: 8575. https://doi.org/10.3390/ijms26178575
APA StyleShakeri Abroudi, A., Mashhouri Moghaddam, M., Hashemi Karoii, D., Djamali, M., Azizi, H., & Skutella, T. (2025). Analysis of Microarray and Single-Cell RNA-Seq Finds Gene Co-Expression and Tumor Environment Associated with Extracellular Matrix in Epithelial–Mesenchymal Transition in Prostate Cancer. International Journal of Molecular Sciences, 26(17), 8575. https://doi.org/10.3390/ijms26178575







