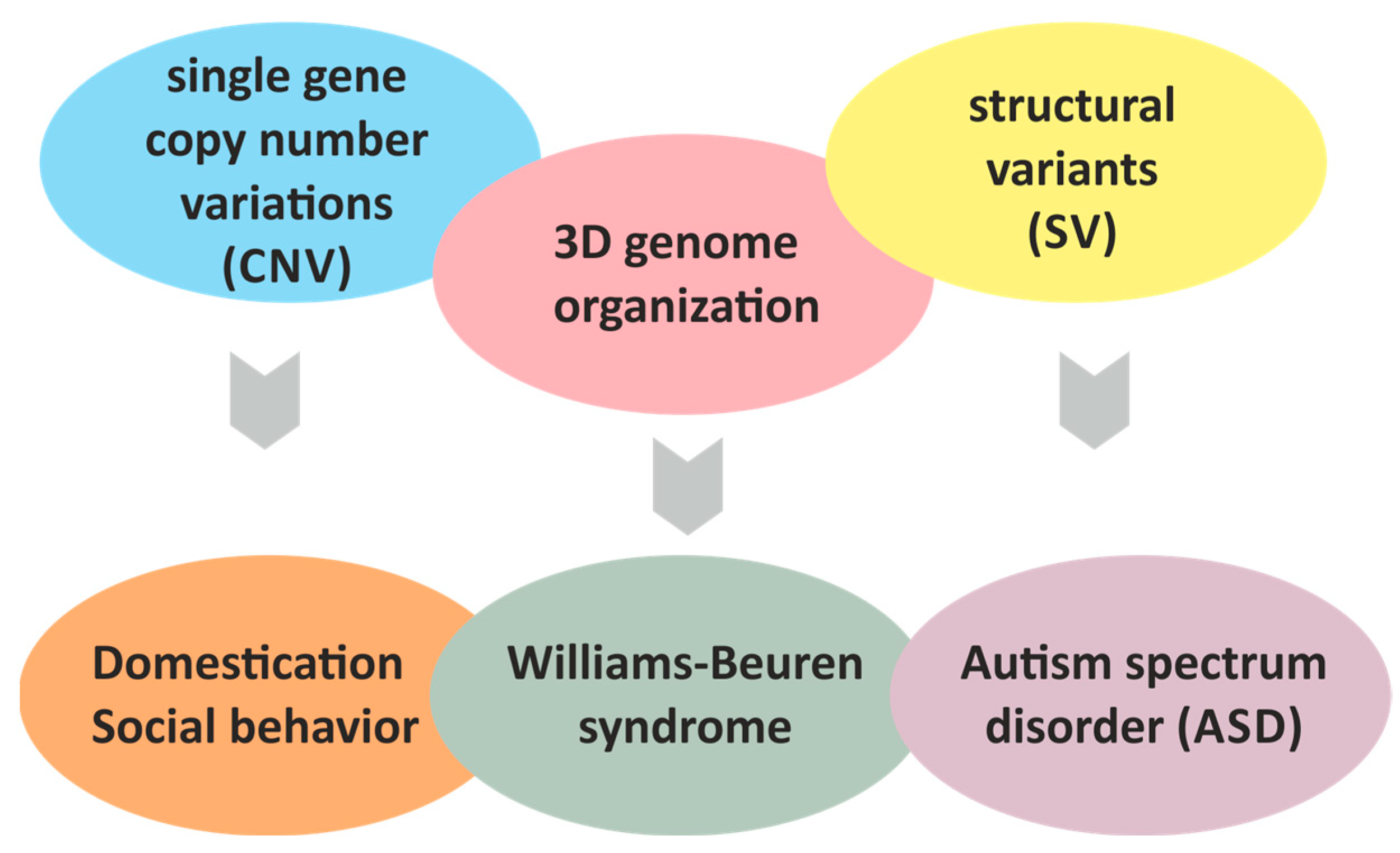
WBSCR Locus: At the Crossroads of Human Behavioral Disorders and Domestication of Animals
Abstract
1. Genetic Basis of Domestication
2. Genetic Basis of Behavioral Disorders in Human
2.1. The Genetics of Behavioral Disorders in Human
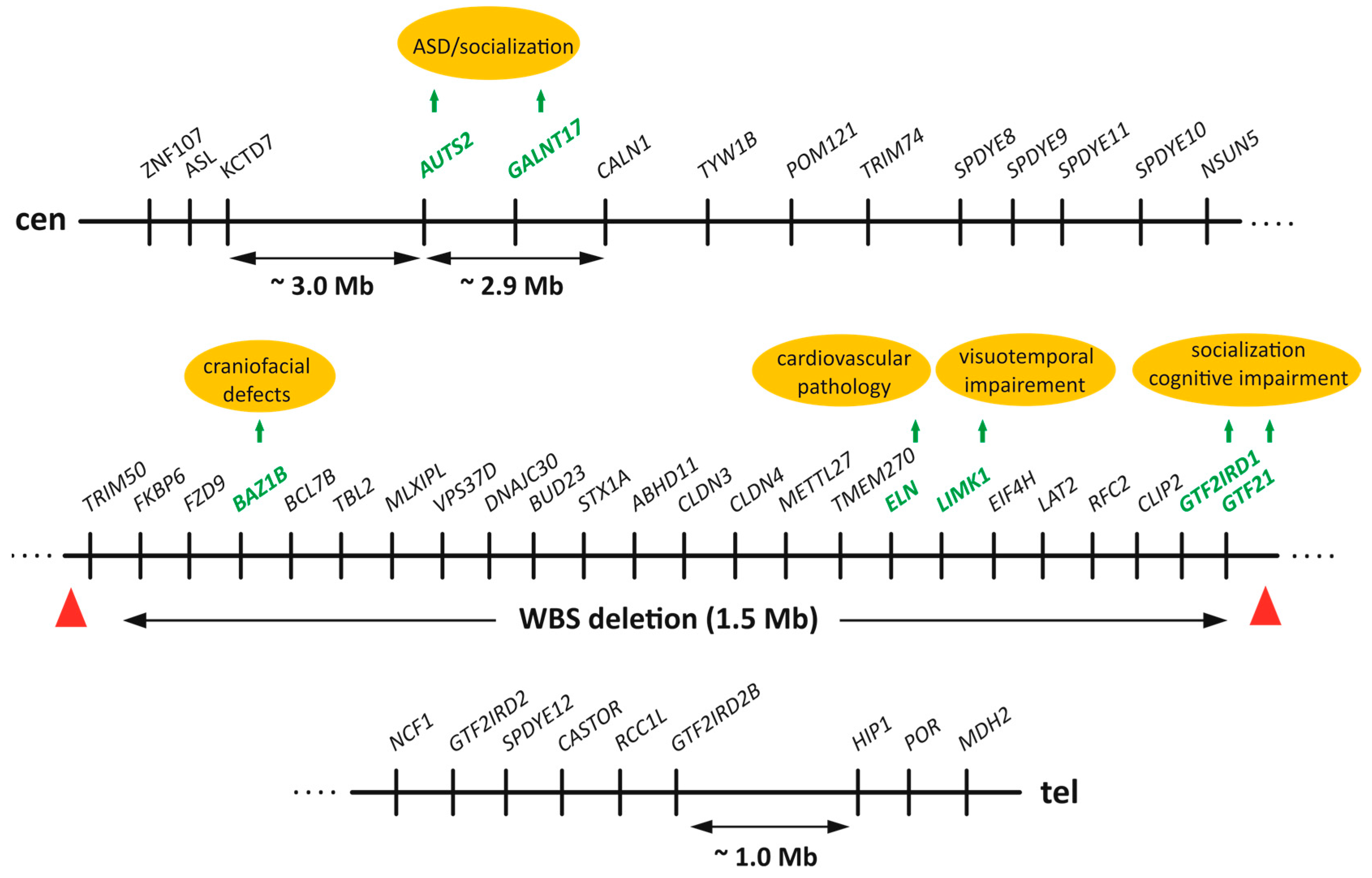
2.2. The Williams–Beuren Syndrome Critical Region (WBSCR) and Behavioral Disorders
3. WBSCR Locus and Dog Domestication
4. Three-Dimensional Genome Alterations in the WBSCR and Social Behavior
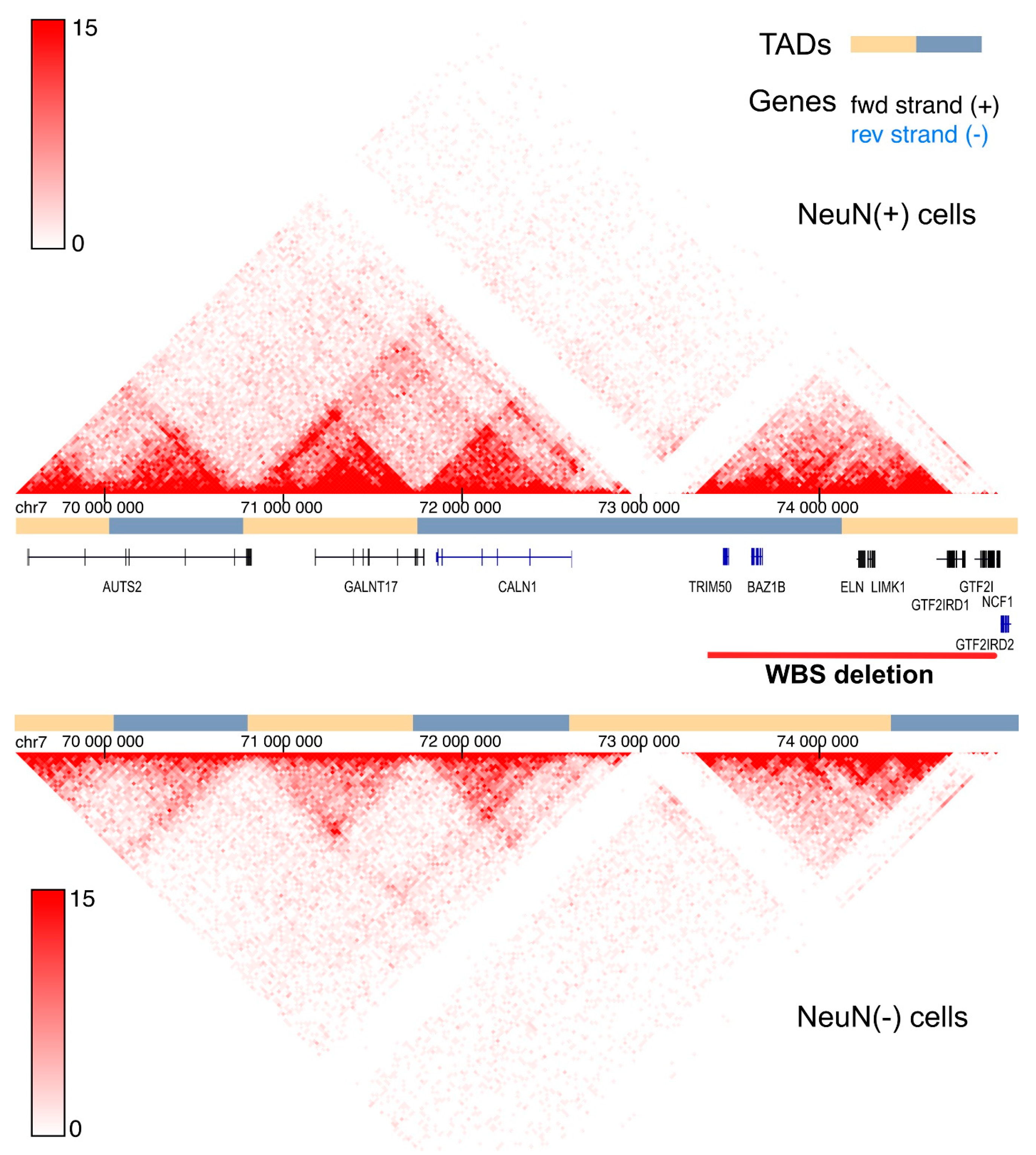
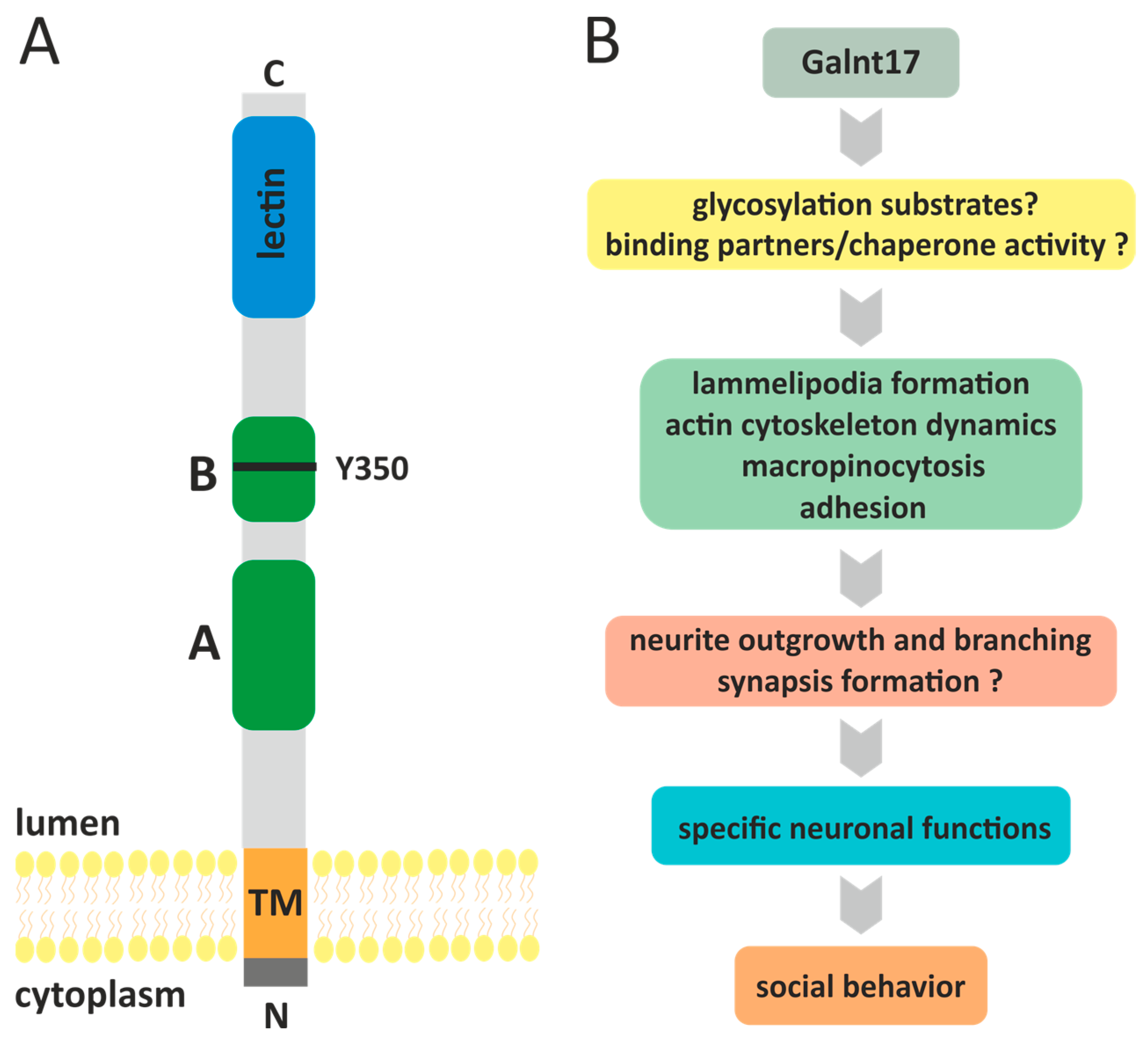
5. GALNT17 (WBSCR17) Gene
5.1. Cellular Functions of Galnt17
5.2. Galnt17 Knockout Mouse
5.3. Galnt17, Human Diseases and Phenotypic Traits in Animals
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum Disorder |
| CNS | Central nervous system |
| CNV | Copy Number Variant |
| GalNAc | N-acetylgalactosamine |
| GlcNAc | N-acetylglucoseamine |
| GWAS | Genome-wide association studies |
| iPSC | induced pluripotent stem cell |
| KO | knockout |
| LCR | low-copy repeat elements |
| NCC | neural crest cell |
| PD | Parkinson disease |
| SINE | Short-Interspersed Nucleotide Element |
| SNP | single-nucleotide polymorphism |
| SV | structural variant |
| TAD | topology associated domain |
| TE | transposon element |
| UDP-GalNAc | uridine-5′-diphospho-N-acetylgalactoseamine |
| WBDS | Williams–Beuren region duplication syndrome |
| WBS | Williams–Beuren syndrome |
| WBSCR | Williams–Beuren syndrome control region |
| WSTF | Williams syndrome transcription factor |
References
- Darwin, C. The Variation of Animals and Plants Under Domestication; John Murray: London, UK, 1868; Volume 2. [Google Scholar]
- Brown, T.A.; Jones, M.K.; Powell, W.; Allaby, R.G. The Complex Origins of Domesticated Crops in the Fertile Crescent. Trends Ecol. Evol. 2009, 24, 103–109. [Google Scholar] [CrossRef]
- Leach, H.M. Human Domestication Reconsidered. Curr. Anthropol. 2003, 44, 349–368. [Google Scholar] [CrossRef]
- Trut, L.N.; Plyusnina, I.Z.; Oskina, I.N. An Experiment on Fox Domestication and Debatable Issues of Evolution of the Dog. Russ. J. Genet. 2004, 40, 644–655. [Google Scholar] [CrossRef]
- Trut, L.; Oskina, I.; Kharlamova, A. Animal Evolution during Domestication: The Domesticated Fox as a Model. BioEssays News Rev. Mol. Cell. Dev. Biol. 2009, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F.; Opitz, J.M.; Raff, R.A. Resynthesizing Evolutionary and Developmental Biology. Dev. Biol. 1996, 173, 357–372. [Google Scholar] [CrossRef]
- Neural Crest Cells: Evolution, Development and Disease; Trainor, P., Ed.; Academic Press: Cambridge, MA, USA, 2013; ISBN 978-0-12-401730-6. [Google Scholar]
- Vega-Lopez, G.A.; Cerrizuela, S.; Aybar, M.J. Trunk Neural Crest Cells: Formation, Migration and Beyond. Int. J. Dev. Biol. 2017, 61, 5–15. [Google Scholar] [CrossRef]
- Wilkins, A.S.; Wrangham, R.W.; Fitch, W.T. The “Domestication Syndrome” in Mammals: A Unified Explanation Based on Neural Crest Cell Behavior and Genetics. Genetics 2014, 197, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.O.; Summers, K. Neural Crest Cell Genes and the Domestication Syndrome: A Comparative Analysis of Selection. PLoS ONE 2022, 17, e0263830. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, B.T.; Wilson, L.A.B. Shared Reproductive Disruption, Not Neural Crest or Tameness, Explains the Domestication Syndrome. Proc. R. Soc. B Biol. Sci. 2023, 290, 20222464. [Google Scholar] [CrossRef]
- Carneiro, M.; Piorno, V.; Rubin, C.-J.; Alves, J.M.; Ferrand, N.; Alves, P.C.; Andersson, L. Candidate Genes Underlying Heritable Differences in Reproductive Seasonality between Wild and Domestic Rabbits. Anim. Genet. 2015, 46, 418–425. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Zeder, M.A. Core Questions in Domestication Research. Proc. Natl. Acad. Sci. USA 2015, 112, 3191–3198. [Google Scholar] [CrossRef]
- Wang, G.; Zhai, W.; Yang, H.; Fan, R.; Cao, X.; Zhong, L.; Wang, L.; Liu, F.; Wu, H.; Cheng, L.; et al. The Genomics of Selection in Dogs and the Parallel Evolution between Dogs and Humans. Nat. Commun. 2013, 4, 1860. [Google Scholar] [CrossRef]
- Belyaev, D.K. Destabilizing Selection as a Factor in Domestication. J. Hered. 1979, 70, 301–308. [Google Scholar] [CrossRef] [PubMed]
- vonHoldt, B.M.; Ji, S.S.; Aardema, M.L.; Stahler, D.R.; Udell, M.A.R.; Sinsheimer, J.S. Activity of Genes with Functions in Human Williams-Beuren Syndrome Is Impacted by Mobile Element Insertions in the Gray Wolf Genome. Genome Biol. Evol. 2018, 10, 1546–1553. [Google Scholar] [CrossRef]
- Kozel, B.A.; Barak, B.; Kim, C.A.; Mervis, C.B.; Osborne, L.R.; Porter, M.; Pober, B.R. Williams Syndrome. Nat. Rev. Dis. Primer 2021, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Qi, F.; Bai, X.; Ren, T.; Shen, X.; Chu, Q.; Zhang, X.; Lu, X. Genome-Wide Analysis Reveals Molecular Convergence Underlying Domestication in 7 Bird and Mammals. BMC Genom. 2020, 21, 204. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Jones, B.M.; Traniello, I.M.; Bukhari, S.A.; Halfon, M.S.; Hofmann, H.A.; Huang, S.; Katz, P.S.; Keagy, J.; Lynch, V.J.; et al. Behavior-Related Gene Regulatory Networks: A New Level of Organization in the Brain. Proc. Natl. Acad. Sci. USA 2020, 117, 23270–23279. [Google Scholar] [CrossRef]
- López-Tobón, A.; Trattaro, S.; Testa, G. The Sociability Spectrum: Evidence from Reciprocal Genetic Copy Number Variations. Mol. Autism 2020, 11, 50. [Google Scholar] [CrossRef]
- Genovese, A.; Butler, M.G. The Autism Spectrum: Behavioral, Psychiatric and Genetic Associations. Genes 2023, 14, 677. [Google Scholar] [CrossRef]
- Madlon-Kay, S.; Montague, M.J.; Brent, L.J.N.; Ellis, S.; Zhong, B.; Snyder-Mackler, N.; Horvath, J.E.; Skene, J.H.P.; Platt, M.L. Weak Effects of Common Genetic Variation in Oxytocin and Vasopressin Receptor Genes on Rhesus Macaque Social Behavior. Am. J. Primatol. 2018, 80, e22873. [Google Scholar] [CrossRef] [PubMed]
- Ike, K.G.O.; Lamers, S.J.C.; Kaim, S.; de Boer, S.F.; Buwalda, B.; Billeter, J.-C.; Kas, M.J.H. The Human Neuropsychiatric Risk Gene Drd2 Is Necessary for Social Functioning across Evolutionary Distant Species. Mol. Psychiatry 2024, 29, 518–528. [Google Scholar] [CrossRef]
- Gao, W.-J.; Mack, N.R. From Hyposociability to Hypersociability—The Effects of PSD-95 Deficiency on the Dysfunctional Development of Social Behavior. Front. Behav. Neurosci. 2021, 15, 618397. [Google Scholar] [CrossRef]
- Feyder, M.; Karlsson, R.-M.; Mathur, P.; Lyman, M.; Bock, R.; Momenan, R.; Munasinghe, J.; Scattoni, M.L.; Ihne, J.; Camp, M.; et al. Association of Mouse Dlg4 (PSD-95) Gene Deletion and Human DLG4 Gene Variation With Phenotypes Relevant to Autism Spectrum Disorders and Williams’ Syndrome. Am. J. Psychiatry 2010, 167, 1508–1517. [Google Scholar] [CrossRef]
- Hörnberg, H.; Pérez-Garci, E.; Schreiner, D.; Hatstatt-Burklé, L.; Magara, F.; Baudouin, S.; Matter, A.; Nacro, K.; Pecho-Vrieseling, E.; Scheiffele, P. Rescue of Oxytocin Response and Social Behaviour in a Mouse Model of Autism. Nature 2020, 584, 252–256. [Google Scholar] [CrossRef]
- Leblond, C.S.; Nava, C.; Polge, A.; Gauthier, J.; Huguet, G.; Lumbroso, S.; Giuliano, F.; Stordeur, C.; Depienne, C.; Mouzat, K.; et al. Meta-Analysis of SHANK Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments. PLoS Genet. 2014, 10, e1004580. [Google Scholar] [CrossRef]
- Miles, J.H. Autism Spectrum Disorders—A Genetics Review. Genet. Med. 2011, 13, 278–294. [Google Scholar] [CrossRef]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism Spectrum Disorder: Neuropathology and Animal Models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef] [PubMed]
- Peoples, R.; Franke, Y.; Wang, Y.-K.; Pérez-Jurado, L.; Paperna, T.; Cisco, M.; Francke, U. A Physical Map, Including a BAC/PAC Clone Contig, of the Williams-Beuren Syndrome–Deletion Region at 7q11.23. Am. J. Hum. Genet. 2000, 66, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Juárez, C.A.; Prieto-Corona, B.; Rodríguez-Camacho, M.; Sandoval-Lira, L.; Villalva-Sánchez, Á.F.; Yáñez-Téllez, M.G.; López, M.F.R. Neuropsychological Genotype-Phenotype in Patients with Williams Syndrome with Atypical Deletions: A Systematic Review. Neuropsychol. Rev. 2023, 33, 891–911. [Google Scholar] [CrossRef]
- Miezah, D.; Porter, M.; Rossi, A.; Kazzi, C.; Batchelor, J.; Reeve, J. Cognitive Profile of Young Children with Williams Syndrome. J. Intellect. Disabil. Res. JIDR 2021, 65, 784–794. [Google Scholar] [CrossRef]
- Bayés, M.; Magano, L.F.; Rivera, N.; Flores, R.; Pérez Jurado, L.A. Mutational Mechanisms of Williams-Beuren Syndrome Deletions. Am. J. Hum. Genet. 2003, 73, 131–151. [Google Scholar] [CrossRef]
- Etokebe, G.E.; Axelsson, S.; Svaerd, N.H.; Storhaug, K.; Dembić, Z. Detection of Hemizygous Chromosomal Copy Number Variants in Williams-Beuren Syndrome (WBS) by Duplex Quantitative PCR Array: An Unusual Type of WBS Genetic Defect. Int. J. Biomed. Sci. IJBS 2008, 4, 161–170. [Google Scholar]
- Ferrero, G.B.; Howald, C.; Micale, L.; Biamino, E.; Augello, B.; Fusco, C.; Turturo, M.G.; Forzano, S.; Reymond, A.; Merla, G. An Atypical 7q11.23 Deletion in a Normal IQ Williams–Beuren Syndrome Patient. Eur. J. Hum. Genet. 2010, 18, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Antonell, A.; Del Campo, M.; Magano, L.F.; Kaufmann, L.; de la Iglesia, J.M.; Gallastegui, F.; Flores, R.; Schweigmann, U.; Fauth, C.; Kotzot, D.; et al. Partial 7q11.23 Deletions Further Implicate GTF2I and GTF2IRD1 as the Main Genes Responsible for the Williams-Beuren Syndrome Neurocognitive Profile. J. Med. Genet. 2010, 47, 312–320. [Google Scholar] [CrossRef]
- Campbell, L.E.; Daly, E.; Toal, F.; Stevens, A.; Azuma, R.; Karmiloff-Smith, A.; Murphy, D.G.M.; Murphy, K.C. Brain Structural Differences Associated with the Behavioural Phenotype in Children with Williams Syndrome. Brain Res. 2009, 1258, 96–107. [Google Scholar] [CrossRef]
- Sanders, S.J.; Ercan-Sencicek, A.G.; Hus, V.; Luo, R.; Murtha, M.T.; Moreno-De-Luca, D.; Chu, S.H.; Moreau, M.P.; Gupta, A.R.; Thomson, S.A.; et al. Multiple Recurrent de Novo Copy Number Variations (CNVs), Including Duplications of the 7q11.23 Williams-Beuren Syndrome Region, Are Strongly Associated with Autism. Neuron 2011, 70, 863–885. [Google Scholar] [CrossRef]
- Van der Aa, N.; Rooms, L.; Vandeweyer, G.; van den Ende, J.; Reyniers, E.; Fichera, M.; Romano, C.; Delle Chiaie, B.; Mortier, G.; Menten, B.; et al. Fourteen New Cases Contribute to the Characterization of the 7q11.23 Microduplication Syndrome. Eur. J. Med. Genet. 2009, 52, 94–100. [Google Scholar] [CrossRef]
- López-Tobón, A.; Shyti, R.; Villa, C.E.; Cheroni, C.; Fuentes-Bravo, P.; Trattaro, S.; Caporale, N.; Troglio, F.; Tenderini, E.; Mihailovich, M.; et al. GTF2I Dosage Regulates Neuronal Differentiation and Social Behavior in 7q11.23 Neurodevelopmental Disorders. Sci. Adv. 2023, 9, eadh2726. [Google Scholar] [CrossRef] [PubMed]
- Beunders, G.; van de Kamp, J.M.; Veenhoven, R.H.; van Hagen, J.M.; Nieuwint, A.W.M.; Sistermans, E.A. A Triplication of the Williams-Beuren Syndrome Region in a Patient with Mental Retardation, a Severe Expressive Language Delay, Behavioural Problems and Dysmorphisms. J. Med. Genet. 2010, 47, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Segura-Puimedon, M.; Sahún, I.; Velot, E.; Dubus, P.; Borralleras, C.; Rodrigues, A.J.; Valero, M.C.; Valverde, O.; Sousa, N.; Herault, Y.; et al. Heterozygous Deletion of the Williams–Beuren Syndrome Critical Interval in Mice Recapitulates Most Features of the Human Disorder. Hum. Mol. Genet. 2014, 23, 6481–6494. [Google Scholar] [CrossRef]
- Morris, C.A.; Mervis, C.B.; Hobart, H.H.; Gregg, R.G.; Bertrand, J.; Ensing, G.J.; Sommer, A.; Moore, C.A.; Hopkin, R.J.; Spallone, P.A.; et al. GTF2I Hemizygosity Implicated in Mental Retardation in Williams Syndrome: Genotype–Phenotype Analysis of Five Families with Deletions in the Williams Syndrome Region. Am. J. Med. Genet. A. 2003, 123A, 45–59. [Google Scholar] [CrossRef]
- van Hagen, J.M.; van der Geest, J.N.; van der Giessen, R.S.; Lagers-van Haselen, G.C.; Eussen, H.J.F.M.M.; Gille, J.J.P.; Govaerts, L.C.P.; Wouters, C.H.; de Coo, I.F.M.; Hoogenraad, C.C.; et al. Contribution of CYLN2 and GTF2IRD1 to Neurological and Cognitive Symptoms in Williams Syndrome. Neurobiol. Dis. 2007, 26, 112–124. [Google Scholar] [CrossRef]
- Makeyev, A.V.; Bayarsaihan, D. Molecular Basis of Williams-Beuren Syndrome: TFII-I Regulated Targets Involved in Craniofacial Development. Cleft Palate-Craniofacial J. Off. Publ. Am. Cleft Palate-Craniofacial Assoc. 2011, 48, 109–116. [Google Scholar] [CrossRef]
- Lopatina, O.L.; Komleva, Y.K.; Gorina, Y.V.; Olovyannikova, R.Y.; Trufanova, L.V.; Hashimoto, T.; Takahashi, T.; Kikuchi, M.; Minabe, Y.; Higashida, H.; et al. Oxytocin and Excitation/Inhibition Balance in Social Recognition. Neuropeptides 2018, 72, 1–11. [Google Scholar] [CrossRef]
- Sohal, V.S.; Rubenstein, J.L.R. Excitation-Inhibition Balance as a Framework for Investigating Mechanisms in Neuropsychiatric Disorders. Mol. Psychiatry 2019, 24, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Enkhmandakh, B.; Bitchevaia, N.; Ruddle, F.; Bayarsaihan, D. The Early Embryonic Expression of TFII-I during Mouse Preimplantation Development. Gene Expr. Patterns 2004, 4, 25–28. [Google Scholar] [CrossRef]
- Roy, A.L. Biochemistry and Biology of the Inducible Multifunctional Transcription Factor TFII-I: 10years Later. Gene 2012, 492, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, A.V.; Bayarsaihan, D. New TFII-I Family Target Genes Involved in Embryonic Development. Biochem. Biophys. Res. Commun. 2009, 386, 554–558. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chimge, N.-O.; Makeyev, A.V.; Ruddle, F.H.; Bayarsaihan, D. Identification of the TFII-I Family Target Genes in the Vertebrate Genome. Proc. Natl. Acad. Sci. USA 2008, 105, 9006–9010. [Google Scholar] [CrossRef]
- Nir Sade, A.; Levy, G.; Schokoroy Trangle, S.; Elad Sfadia, G.; Bar, E.; Ophir, O.; Fischer, I.; Rokach, M.; Atzmon, A.; Parnas, H.; et al. Neuronal Gtf2i Deletion Alters Mitochondrial and Autophagic Properties. Commun. Biol. 2023, 6, 1269. [Google Scholar] [CrossRef] [PubMed]
- Mervis, C.B.; Dida, J.; Lam, E.; Crawford-Zelli, N.A.; Young, E.J.; Henderson, D.R.; Onay, T.; Morris, C.A.; Woodruff-Borden, J.; Yeomans, J.; et al. Duplication of GTF2I Results in Separation Anxiety in Mice and Humans. Am. J. Hum. Genet. 2012, 90, 1064–1070. [Google Scholar] [CrossRef]
- Twite, M.D.; Stenquist, S.; Ing, R.J. Williams Syndrome. Pediatr. Anesth. 2019, 29, 483–490. [Google Scholar] [CrossRef]
- Li, D.Y.; Faury, G.; Taylor, D.G.; Davis, E.C.; Boyle, W.A.; Mecham, R.P.; Stenzel, P.; Boak, B.; Keating, M.T. Novel Arterial Pathology in Mice and Humans Hemizygous for Elastin. J. Clin. Invest. 1998, 102, 1783–1787. [Google Scholar] [CrossRef]
- Zanella, M.; Vitriolo, A.; Andirko, A.; Martins, P.T.; Sturm, S.; O’Rourke, T.; Laugsch, M.; Malerba, N.; Skaros, A.; Trattaro, S.; et al. Dosage Analysis of the 7q11.23 Williams Region Identifies BAZ1B as a Major Human Gene Patterning the Modern Human Face and Underlying Self-Domestication. Sci. Adv. 2019, 5, eaaw7908. [Google Scholar] [CrossRef]
- Barnett, C.; Yazgan, O.; Kuo, H.-C.; Malakar, S.; Thomas, T.; Fitzgerald, A.; Harbour, W.; Henry, J.J.; Krebs, J.E. Williams Syndrome Transcription Factor Is Critical for Neural Crest Cell Function in Xenopus Laevis. Mech. Dev. 2012, 129, 324–338. [Google Scholar] [CrossRef]
- Gregory, M.D.; Mervis, C.B.; Elliott, M.L.; Kippenhan, J.S.; Nash, T.; Czarapata, J.B.; Prabhakaran, R.; Roe, K.; Eisenberg, D.P.; Kohn, P.D.; et al. Williams Syndrome Hemideletion and LIMK1 Variation Both Affect Dorsal Stream Functional Connectivity. Brain 2019, 142, 3963–3974. [Google Scholar] [CrossRef]
- Todorovski, Z.; Asrar, S.; Liu, J.; Saw, N.M.N.; Joshi, K.; Cortez, M.A.; Snead, O.C.; Xie, W.; Jia, Z. LIMK1 Regulates Long-Term Memory and Synaptic Plasticity via the Transcriptional Factor CREB. Mol. Cell. Biol. 2015, 35, 1316–1328. [Google Scholar] [CrossRef]
- Biel, A.; Castanza, A.S.; Rutherford, R.; Fair, S.R.; Chifamba, L.; Wester, J.C.; Hester, M.E.; Hevner, R.F. AUTS2 Syndrome: Molecular Mechanisms and Model Systems. Front. Mol. Neurosci. 2022, 15, 858582. [Google Scholar] [CrossRef] [PubMed]
- Weisner, P.A.; Chen, C.-Y.; Sun, Y.; Yoo, J.; Kao, W.-C.; Zhang, H.; Baltz, E.T.; Troy, J.M.; Stubbs, L. A Mouse Mutation That Dysregulates Neighboring Galnt17 and Auts2 Genes Is Associated with Phenotypes Related to the Human AUTS2 Syndrome. G3 Genes Genomes Genet. 2019, 9, 3891–3906. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Abe, M.; Yamazaki, M.; Sakimura, K.; Yamada, K.; Hoshino, M. Heterozygous Disruption of Autism Susceptibility Candidate 2 Causes Impaired Emotional Control and Cognitive Memory. PLoS ONE 2015, 10, e0145979. [Google Scholar] [CrossRef]
- Foo, J.N.; Chew, E.G.Y.; Chung, S.J.; Peng, R.; Blauwendraat, C.; Nalls, M.A.; Mok, K.Y.; Satake, W.; Toda, T.; Chao, Y.; et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol. 2020, 77, 746–754. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Seward, C.H.; Song, Y.; Inamdar, M.; Leddy, A.M.; Zhang, H.; Yoo, J.; Kao, W.-C.; Pawlowski, H.; Stubbs, L.J. Galnt17 Loss-of-Function Leads to Developmental Delay and Abnormal Coordination, Activity, and Social Interactions with Cerebellar Vermis Pathology. Dev. Biol. 2022, 490, 155–171. [Google Scholar] [CrossRef]
- Liu, M.; Chen, Y.; Sun, M.; Du, Y.; Bai, Y.; Lei, G.; Zhang, C.; Zhang, M.; Zhang, Y.; Xi, C.; et al. Auts2 Regulated Autism-like Behavior, Glucose Metabolism and Oxidative Stress in Mice. Exp. Neurol. 2023, 361, 114298. [Google Scholar] [CrossRef]
- Sanchez-Jimeno, C.; Blanco-Kelly, F.; López-Grondona, F.; Losada-Del Pozo, R.; Moreno, B.; Rodrigo-Moreno, M.; Martinez-Cayuelas, E.; Riveiro-Alvarez, R.; Fenollar-Cortés, M.; Ayuso, C.; et al. Attention Deficit Hyperactivity and Autism Spectrum Disorders as the Core Symptoms of AUTS2 Syndrome: Description of Five New Patients and Update of the Frequency of Manifestations and Genotype-Phenotype Correlation. Genes 2021, 12, 1360. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Pollinger, J.P.; Lohmueller, K.E.; Han, E.; Parker, H.G.; Quignon, P.; Degenhardt, J.D.; Boyko, A.R.; Earl, D.A.; Auton, A.; et al. Genome-Wide SNP and Haplotype Analyses Reveal a Rich History Underlying Dog Domestication. Nature 2010, 464, 898–902. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Shuldiner, E.; Koch, I.J.; Kartzinel, R.Y.; Hogan, A.; Brubaker, L.; Wanser, S.; Stahler, D.; Wynne, C.D.L.; Ostrander, E.A.; et al. Structural Variants in Genes Associated with Human Williams-Beuren Syndrome Underlie Stereotypical Hypersociability in Domestic Dogs. Sci. Adv. 2017, 3, e1700398. [Google Scholar] [CrossRef] [PubMed]
- Tandon, D.; Ressler, K.; Petticord, D.; Papa, A.; Jiranek, J.; Wilkinson, R.; Kartzinel, R.Y.; Ostrander, E.A.; Burney, N.; Borden, C.; et al. Homozygosity for Mobile Element Insertions Associated with WBSCR17 Could Predict Success in Assistance Dog Training Programs. Genes 2019, 10, 439. [Google Scholar] [CrossRef]
- Glazko, V.I.; Kosovsky, G.Y.; Blokhina, T.V.; Zhirkova, A.A.; Glazko, T.T. Socialization and Genetic Variability as a Driver of Domestication (by the Example of Dog Breeds). Selskokhozyaistvennaya Biol. Agric. Biol. 2021, 56, 292–303. [Google Scholar] [CrossRef]
- Gnanadesikan, G.E.; Tandon, D.; Bray, E.E.; Kennedy, B.S.; Tennenbaum, S.R.; MacLean, E.L.; vonHoldt, B.M. Transposons in the Williams–Beuren Syndrome Critical Region Are Associated with Social Behavior in Assistance Dogs. Behav. Genet. 2023, 54, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Tiukacheva, E.A.; Ulianov, S.V.; Karpukhina, A.; Razin, S.V.; Vassetzky, Y. 3D Genome Alterations and Editing in Pathology. Mol. Ther. J. Am. Soc. Gene Ther. 2023, 31, 924–933. [Google Scholar] [CrossRef]
- Okhovat, M.; VanCampen, J.; Nevonen, K.A.; Harshman, L.; Li, W.; Layman, C.E.; Ward, S.; Herrera, J.; Wells, J.; Sheng, R.R.; et al. TAD Evolutionary and Functional Characterization Reveals Diversity in Mammalian TAD Boundary Properties and Function. Nat. Commun. 2023, 14, 8111. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.N.K.; Quaid, K.; Xing, X.; Schmidt, H.; Wang, T. Widespread Contribution of Transposable Elements to the Rewiring of Mammalian 3D Genomes. Nat. Commun. 2023, 14, 634. [Google Scholar] [CrossRef]
- Merla, G.; Howald, C.; Henrichsen, C.N.; Lyle, R.; Wyss, C.; Zabot, M.-T.; Antonarakis, S.E.; Reymond, A. Submicroscopic Deletion in Patients with Williams-Beuren Syndrome Influences Expression Levels of the Nonhemizygous Flanking Genes. Am. J. Hum. Genet. 2006, 79, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, N.; Witwicki, R.M.; Migliavacca, E.; Leleu, M.; Didelot, G.; Harewood, L.; Rougemont, J.; Reymond, A. Structural Variation-Associated Expression Changes Are Paralleled by Chromatin Architecture Modifications. PLoS ONE 2013, 8, e79973. [Google Scholar] [CrossRef]
- Wang, Y.; Song, F.; Zhang, B.; Zhang, L.; Xu, J.; Kuang, D.; Li, D.; Choudhary, M.N.K.; Li, Y.; Hu, M.; et al. The 3D Genome Browser: A Web-Based Browser for Visualizing 3D Genome Organization and Long-Range Chromatin Interactions. Genome Biol. 2018, 19, 151. [Google Scholar] [CrossRef]
- Pletenev, I.A.; Bazarevich, M.; Zagirova, D.R.; Kononkova, A.D.; Cherkasov, A.V.; Efimova, O.I.; Tiukacheva, E.A.; Morozov, K.V.; Ulianov, K.A.; Komkov, D.; et al. Extensive Long-Range Polycomb Interactions and Weak Compartmentalization Are Hallmarks of Human Neuronal 3D Genome. Nucleic Acids Res. 2024, 52, 6234–6252. [Google Scholar] [CrossRef]
- Engmann, O.; Labonté, B.; Mitchell, A.; Bashtrykov, P.; Calipari, E.S.; Rosenbluh, C.; Loh, Y.-H.E.; Walker, D.M.; Burek, D.; Hamilton, P.J.; et al. Cocaine-Induced Chromatin Modifications Associate With Increased Expression and Three-Dimensional Looping of Auts2. Biol. Psychiatry 2017, 82, 794–805. [Google Scholar] [CrossRef]
- Tandon, D.; Kubinyi, E.; Sándor, S.; Faughnan, H.; Miklósi, Á.; vonHoldt, B.M. Canine Hyper-Sociability Structural Variants Associated with Altered Three-Dimensional Chromatin State. BMC Genom. 2024, 25, 767. [Google Scholar] [CrossRef] [PubMed]
- Merla, G.; Ucla, C.; Guipponi, M.; Reymond, A. Identification of Additional Transcripts in the Williams-Beuren Syndrome Critical Region. Hum. Genet. 2002, 110, 429–438. [Google Scholar] [CrossRef]
- Nakamura, N.; Toba, S.; Hirai, M.; Morishita, S.; Mikami, T.; Konishi, M.; Itoh, N.; Kurosaka, A. Cloning and Expression of a Brain-Specific Putative UDP-GalNAc: Polypeptide N-Acetylgalactosaminyltransferase Gene. Biol. Pharm. Bull. 2005, 28, 429–433. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Hussain, M.R.M.; Hoessli, D.C.; Fang, M. N-Acetylgalactosaminyltransferases in Cancer. Oncotarget 2016, 7, 54067–54081. [Google Scholar] [CrossRef]
- Nakayama, Y.; Nakamura, N.; Oki, S.; Wakabayashi, M.; Ishihama, Y.; Miyake, A.; Itoh, N.; Kurosaka, A. A Putative Polypeptide N-Acetylgalactosaminyltransferase/Williams-Beuren Syndrome Chromosome Region 17 (WBSCR17) Regulates Lamellipodium Formation and Macropinocytosis. J. Biol. Chem. 2012, 287, 32222–32235. [Google Scholar] [CrossRef]
- Tenno, M.; Saeki, A.; Elhammer, Å.P.; Kurosaka, A. Function of Conserved Aromatic Residues in the Gal/GalNAc-glycosyltransferase Motif of UDP-GalNAc:Polypeptide N-acetylgalactosaminyltransferase 1. FEBS J. 2007, 274, 6037–6045. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Li, W.; Xu, Y.; Shao, D.; Xie, Y.; Xie, W.; Kubota, T.; Narimatsu, H.; Zhang, Y. Characterization of ppGalNAc-T18, a Member of the Vertebrate-Specific Y Subfamily of UDP-N-Acetyl-α-D-Galactosamine:Polypeptide N-Acetylgalactosaminyltransferases. Glycobiology 2012, 22, 602–615. [Google Scholar] [CrossRef]
- Shan, A.; Lu, J.; Xu, Z.; Li, X.; Xu, Y.; Li, W.; Liu, F.; Yang, F.; Sato, T.; Narimatsu, H.; et al. Polypeptide N-Acetylgalactosaminyltransferase 18 Non-Catalytically Regulates the ER Homeostasis and O-Glycosylation. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 870–882. [Google Scholar] [CrossRef]
- Krause, M.; Gautreau, A. Steering Cell Migration: Lamellipodium Dynamics and the Regulation of Directional Persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Dent, E.W.; Gupton, S.L.; Gertler, F.B. The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harb. Perspect. Biol. 2011, 3, a001800. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Metz, I.; Rust, M.B. Regulation of Actin Filament Assembly and Disassembly in Growth Cone Motility and Axon Guidance. Brain Res. Bull. 2023, 192, 21–35. [Google Scholar] [CrossRef]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Taya, S.; Hashimoto, R.; Hayashi, T.; Abe, M.; Yamazaki, M.; Nakao, K.; et al. Cytoskeletal Regulation by AUTS2 in Neuronal Migration and Neuritogenesis. Cell Rep. 2014, 9, 2166–2179. [Google Scholar] [CrossRef]
- Nakayama, Y.; Nakamura, N.; Kawai, T.; Kaneda, E.; Takahashi, Y.; Miyake, A.; Itoh, N.; Kurosaka, A. Identification and Expression Analysis of Zebrafish Polypeptide α-N-Acetylgalactosaminyltransferase Y-Subfamily Genes during Embryonic Development. Gene Expr. Patterns GEP 2014, 16, 1–7. [Google Scholar] [CrossRef]
- Lau, K.S.; Khan, S.; Dennis, J.W. Genome-Scale Identification of UDP-GlcNAc-Dependent Pathways. Proteomics 2008, 8, 3294–3302. [Google Scholar] [CrossRef]
- Grover, S.; Kumar-Sreelatha, A.A.; Bobbili, D.R.; May, P.; Domenighetti, C.; Sugier, P.-E.; Schulte, C.; COURAGE-PD Consortium; Elbaz, A.; Krüger, R.; et al. Replication of a Novel Parkinson’s Locus in a European Ancestry Population. Mov. Disord. Off. J. Mov. Disord. Soc. 2021, 36, 1689–1695. [Google Scholar] [CrossRef]
- Welton, T.; Teo, T.W.J.; Chan, L.L.; Tan, E.-K.; Tan, L.C.S. Parkinson’s Disease Risk Variant Rs9638616 Is Non-Specifically Associated with Altered Brain Structure and Function. J. Park. Dis. 2024, 14, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Jee, D.; Kang, S.; Park, S. Association of Age-Related Cataract Risk with High Polygenetic Risk Scores Involved in Galactose-Related Metabolism and Dietary Interactions. Lifestyle Genom. 2022, 15, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, H.; Li, J.; Liu, H.; Wang, F.; Wei, Y.; Su, J.; Zhang, D.; Liu, T.; Zhang, Y. The Identification of Specific Methylation Patterns across Different Cancers. PLoS ONE 2015, 10, e0120361. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, N.A.; Robinson, R.C.; Barile, D.; Larsen, L.B.; Buitenhuis, B. A Genome-Wide Association Study Reveals Specific Transferases as Candidate Loci for Bovine Milk Oligosaccharides Synthesis. BMC Genom. 2019, 20, 404. [Google Scholar] [CrossRef]
- Forde, N.; Duffy, G.B.; McGettigan, P.A.; Browne, J.A.; Mehta, J.P.; Kelly, A.K.; Mansouri-Attia, N.; Sandra, O.; Loftus, B.J.; Crowe, M.A.; et al. Evidence for an Early Endometrial Response to Pregnancy in Cattle: Both Dependent upon and Independent of Interferon Tau. Physiol. Genom. 2012, 44, 799–810. [Google Scholar] [CrossRef]
- Thelin, E.P.; Just, D.; Frostell, A.; Häggmark-Månberg, A.; Risling, M.; Svensson, M.; Nilsson, P.; Bellander, B.-M. Protein Profiling in Serum after Traumatic Brain Injury in Rats Reveals Potential Injury Markers. Behav. Brain Res. 2018, 340, 71–80. [Google Scholar] [CrossRef]
- Li, G.; Qin, Y.; Qin, S.; Zhou, X.; Zhao, W.; Zhang, D. Circ_WBSCR17 Aggravates Inflammatory Responses and Fibrosis by Targeting miR-185-5p/SOX6 Regulatory Axis in High Glucose-Induced Human Kidney Tubular Cells. Life Sci. 2020, 259, 118269. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shepelev, M.V.; Skobel, O.I.; Glazko, T.T.; Popov, D.V.; Vysotskii, D.E.; Georgiev, P.G.; Maksimenko, O.G.; Kosovsky, G.Y.; Silaeva, Y.Y. WBSCR Locus: At the Crossroads of Human Behavioral Disorders and Domestication of Animals. Int. J. Mol. Sci. 2025, 26, 8549. https://doi.org/10.3390/ijms26178549
Shepelev MV, Skobel OI, Glazko TT, Popov DV, Vysotskii DE, Georgiev PG, Maksimenko OG, Kosovsky GY, Silaeva YY. WBSCR Locus: At the Crossroads of Human Behavioral Disorders and Domestication of Animals. International Journal of Molecular Sciences. 2025; 26(17):8549. https://doi.org/10.3390/ijms26178549
Chicago/Turabian StyleShepelev, Mikhail V., Olga I. Skobel, Tatiana T. Glazko, Dmitry V. Popov, Denis E. Vysotskii, Pavel G. Georgiev, Oksana G. Maksimenko, Gleb Y. Kosovsky, and Yuliya Y. Silaeva. 2025. "WBSCR Locus: At the Crossroads of Human Behavioral Disorders and Domestication of Animals" International Journal of Molecular Sciences 26, no. 17: 8549. https://doi.org/10.3390/ijms26178549
APA StyleShepelev, M. V., Skobel, O. I., Glazko, T. T., Popov, D. V., Vysotskii, D. E., Georgiev, P. G., Maksimenko, O. G., Kosovsky, G. Y., & Silaeva, Y. Y. (2025). WBSCR Locus: At the Crossroads of Human Behavioral Disorders and Domestication of Animals. International Journal of Molecular Sciences, 26(17), 8549. https://doi.org/10.3390/ijms26178549







