Machine Learning Integration of Bulk and Single-Cell RNA-Seq Data Reveals Cathepsin B as a Central PANoptosis Regulator in Influenza
Abstract
1. Introduction
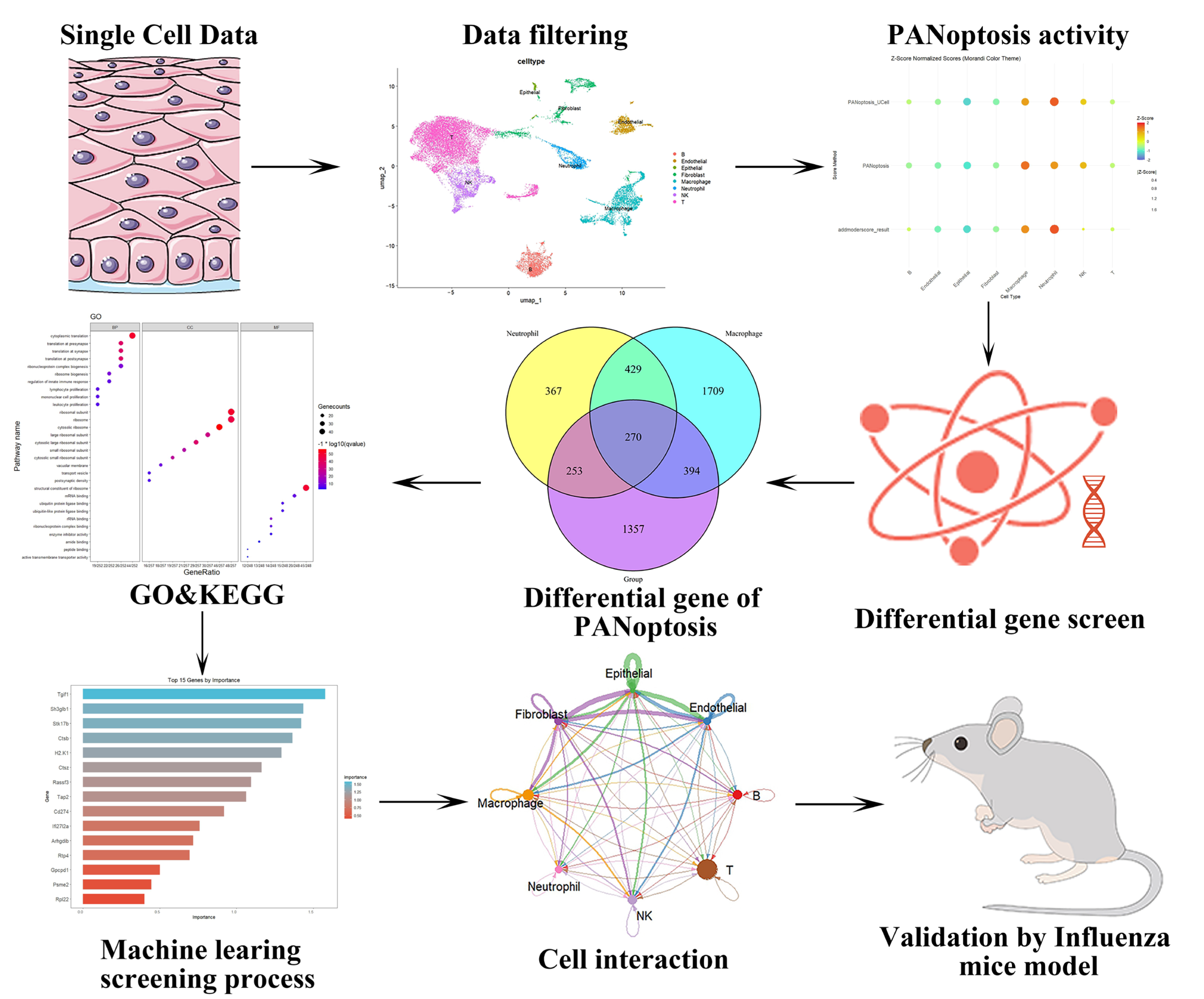
2. Results
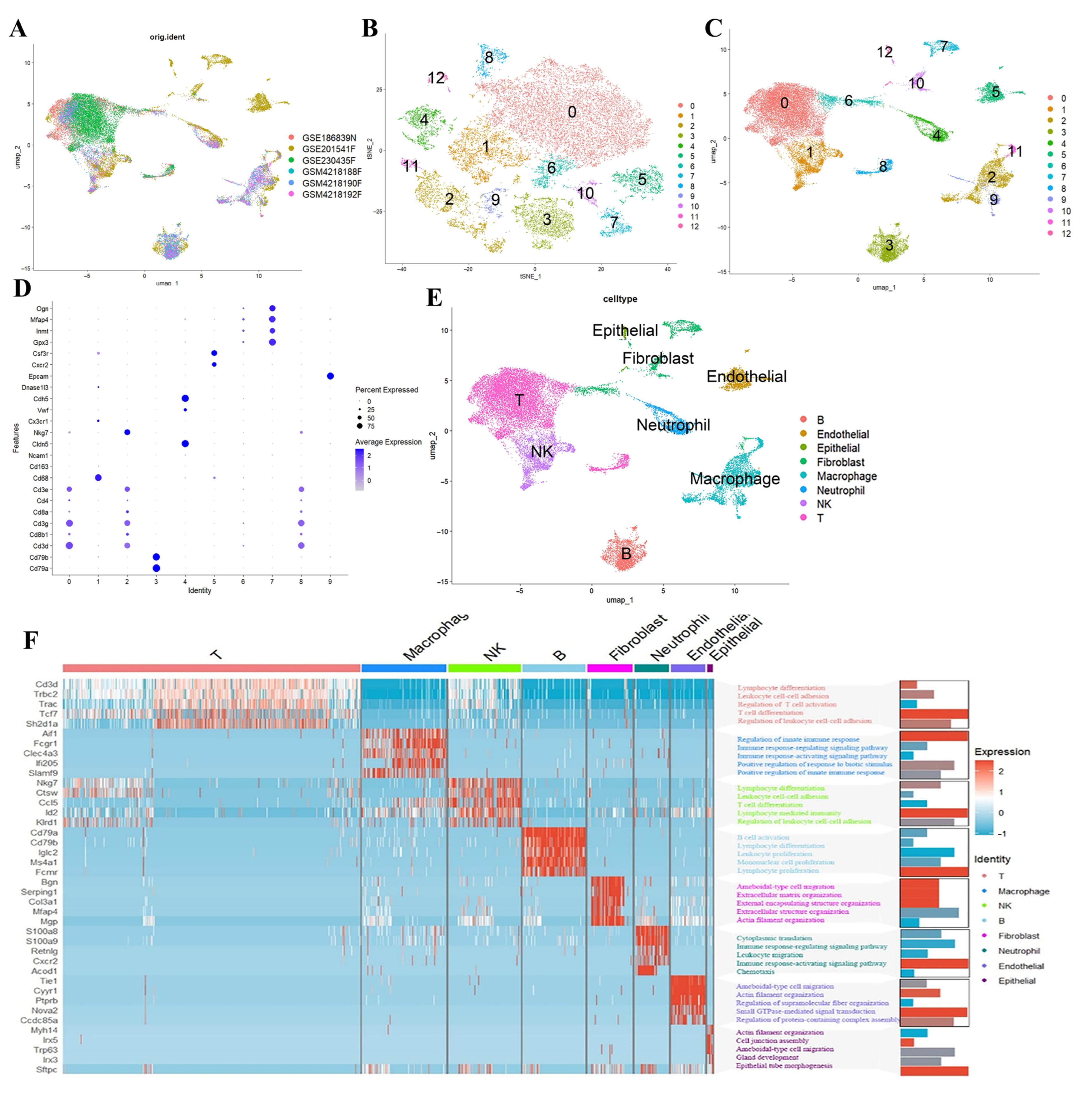
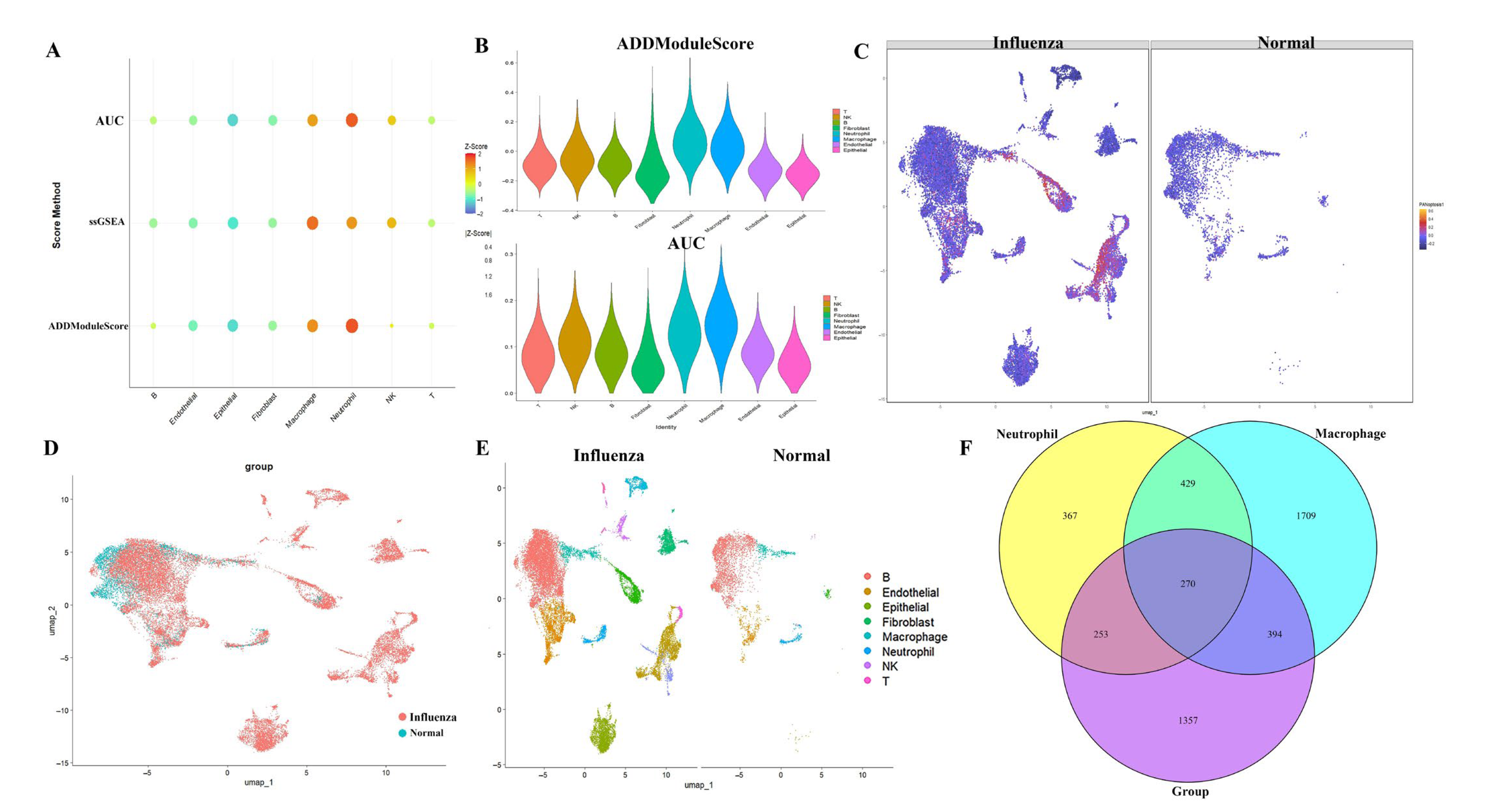
2.1. Analysis of PANoptosis-Related Features in Influenza Using scRNA-Seq Datasets
2.2. Results of GO Analysis and PPI Analysis
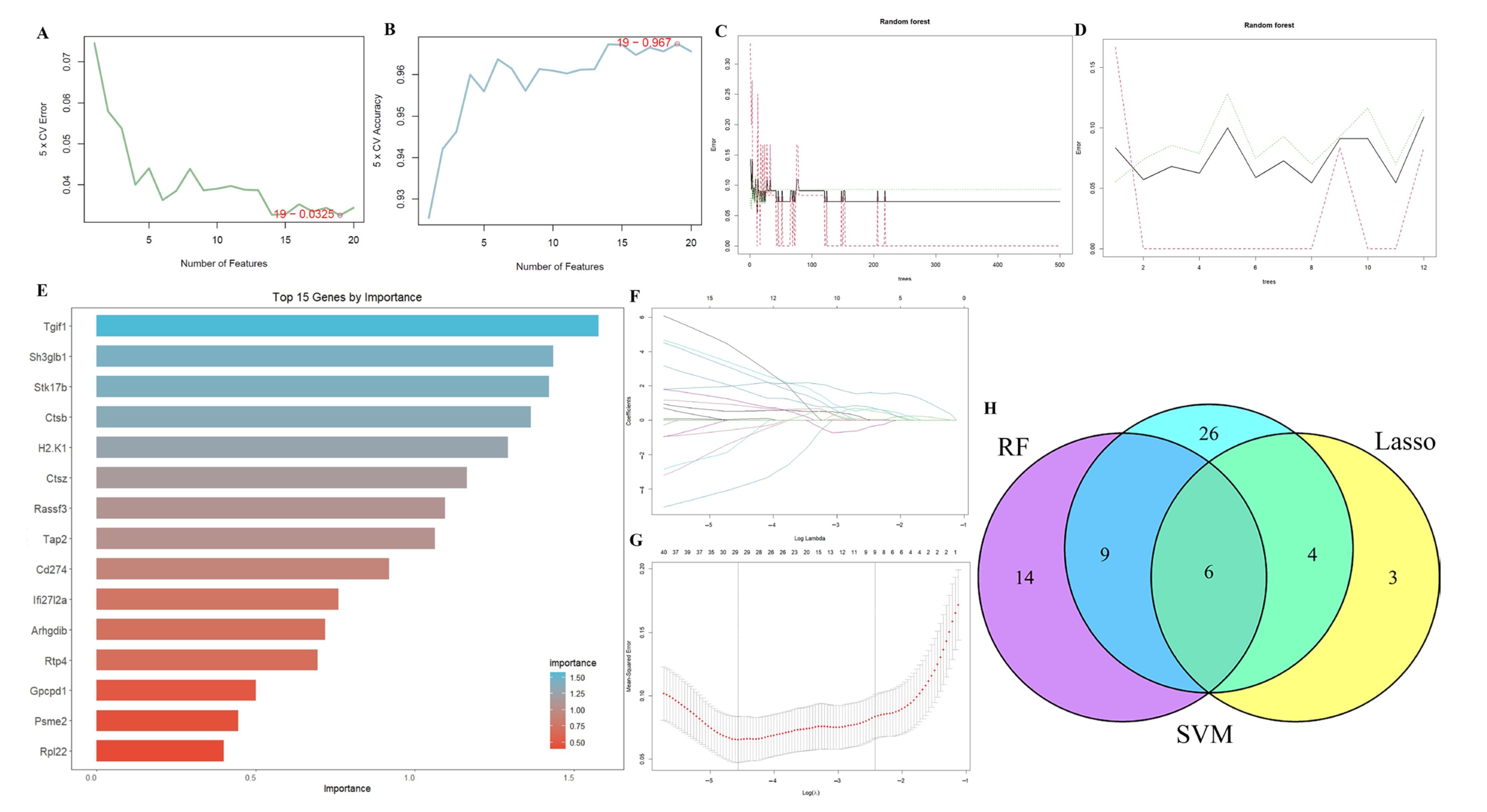
2.3. Results of Machine Learning Algorithms
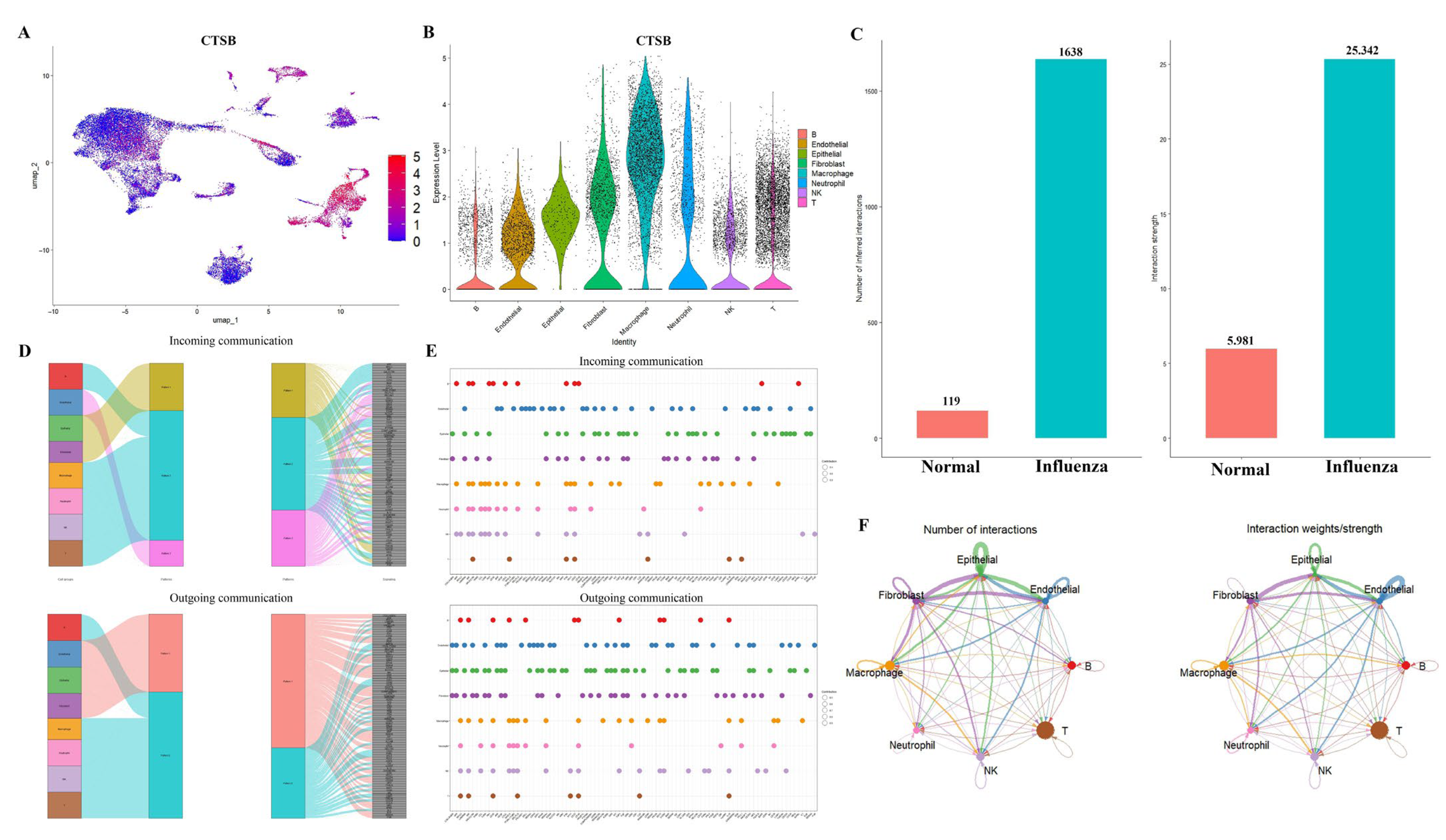
2.4. Comprehensive Validation of Core Feature Genes at Single-Cell Resolution and Analysis of Cellular Communication Networks
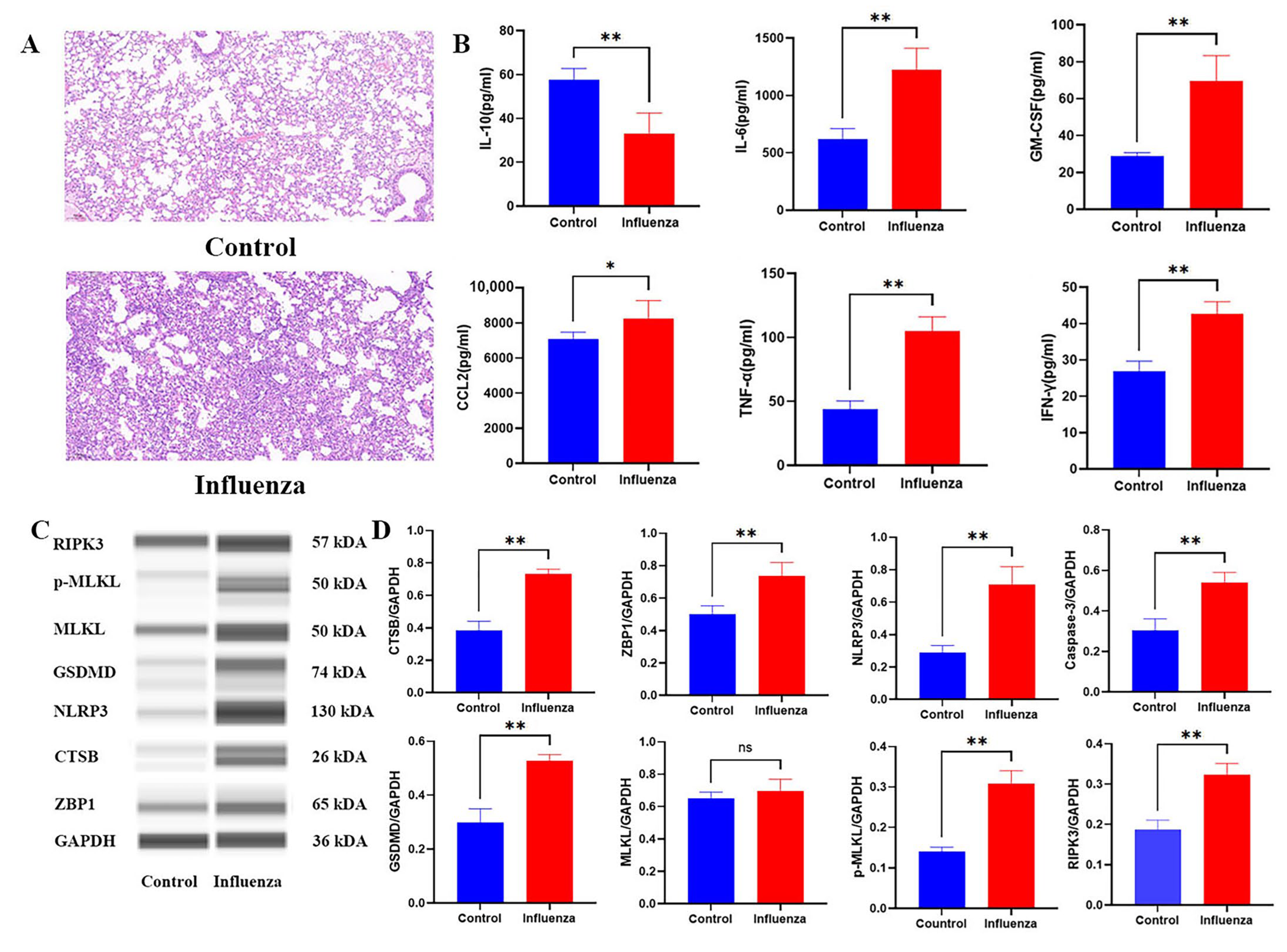
2.5. Histopathological and Molecular Characterization of Influenza-Induced PANoptosis
3. Discussion
4. Materials and Methods
4.1. Source of Data
4.2. scRNA-Seq Dataset Analysis
4.3. Evaluation of PANoptosis Activity
4.4. Enrichment Analysis
4.5. Machine Learning Algorithms for Identifying the Optimal Feature Genes
4.6. Interactions Between Intercellular Communication and Transcription Factors
4.7. Animals and Experimental Design
4.8. Evaluation of Influenza Model in Mice
4.9. Capillary Western Blotting
4.10. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUCell | Area Under the Recovery Curve algorithm |
| BP | Biological Process (in Gene Ontology analysis) |
| CC | Cellular Component (in Gene Ontology analysis) |
| CTSB | Cathepsin B |
| DEGs | Differentially Expressed Genes |
| GO | Gene Ontology |
| GSVA | Gene Set Variation Analysis |
| GSDMD | Gasdermin D |
| IAV | Influenza A Virus |
| IFN-γ | Interferon gamma |
| IL- | Interleukin (e.g., IL-1β, IL-6, IL-10) |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LMP | Lysosomal Membrane Permeabilization |
| MCODE | Molecular Complex Detection algorithm |
| MF | Molecular Function (in Gene Ontology analysis) |
| MLKL | Mixed Lineage Kinase Domain Like Pseudokinase |
| NF-κB | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| NK cells | Natural Killer cells |
| NLRP3 | NLR Family Pyrin Domain Containing 3 |
| NASH | Non-Alcoholic Steatohepatitis |
| PAMP | Pathogen-Associated Molecular Pattern |
| PCA | Principal Component Analysis |
| PPI | Protein-Protein Interaction |
| RAGE | Receptor for Advanced Glycation Endproducts |
| RF | Random Forest |
| RIPK1/3 | Receptor-Interacting Serine/Threonine-Protein Kinase 1/3 |
| scRNA-seq | Single-Cell RNA Sequencing |
| SVM | Support Vector Machine |
| ssGSEA | Single-sample Gene Set Enrichment Analysis |
| SPF | Specific Pathogen-Free |
| TLR | Toll-Like Receptor |
| TNF-α | Tumor Necrosis Factor alpha |
| UMAP | Uniform Manifold Approximation and Projection |
| ZBP1 | Z-DNA Binding Protein 1 |
References
- Sharon, B.; Schleiss, M.R. Pandemic Influenza A (H1N1). Pediatr. Res. 2009, 66, 599. [Google Scholar] [CrossRef]
- Enserink, M.; Cohen, J. Virus of the Year. The Novel H1N1 Influenza. Science 2009, 326, 1607. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and Evolutionary Genomics of the 2009 Swine-Origin H1N1 Influenza A Epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The Mother of All Pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef]
- Yin, H.; Jiang, N.; Shi, W.; Chi, X.; Liu, S.; Chen, J.-L.; Wang, S. Development and Effects of Influenza Antiviral Drugs. Molecules 2021, 26, 810. [Google Scholar] [CrossRef]
- De Clercq, E. Antiviral Agents Active against Influenza A Viruses. Nat. Rev. Drug Discov. 2006, 5, 1015–1025. [Google Scholar] [CrossRef]
- O’Hanlon, R.; Shaw, M.L. Baloxavir Marboxil: The New Influenza Drug on the Market. Curr. Opin. Virol. 2019, 35, 14–18. [Google Scholar] [CrossRef]
- Kuriakose, T.; Man, S.M.; Subbarao Malireddi, R.K.; Karki, R.; Kesavardhana, S.; Place, D.E.; Neale, G.; Vogel, P.; Kanneganti, T.D. ZBP1/DAI Is an Innate Sensor of Influenza Virus Triggering the NLRP3 Inflammasome and Programmed Cell Death Pathways. Sci. Immunol. 2016, 1, aag2045. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, R.; Ouyang, Y.; Gu, W.; Xiao, T.; Yang, H.; Tang, L.; Wang, H.; Xiang, B.; Chen, P. Pyroptosis in Health and Disease: Mechanisms, Regulation and Clinical Perspective. Signal Transduct. Target. Ther. 2024, 9, 245. [Google Scholar] [CrossRef]
- Sahoo, B.; Mou, Z.; Liu, W.; Dubyak, G.; Dai, X. How NINJ1 Mediates Plasma Membrane Rupture and Why NINJ2 Cannot. Cell 2025, 188, 292–302.e11. [Google Scholar] [CrossRef]
- Bai, Y.; Pan, Y.; Liu, X. Mechanistic Insights into Gasdermin-Mediated Pyroptosis. Nat. Rev. Mol. Cell Biol. 2025, 26, 501–521. [Google Scholar] [CrossRef]
- Yang, L.; Ren, Q.; Wang, Y.; Zheng, Y.; Du, F.; Wang, F.; Zhou, J.; Gui, L.; Chen, S.; Chen, X.; et al. Research Progress of Mitochondrial Dysfunction Induced Pyroptosis in Acute Lung Injury. Respir. Res. 2024, 25, 398. [Google Scholar] [CrossRef]
- Hedlund, E.; Deng, Q. Single-Cell RNA Sequencing: Technical Advancements and Biological Applications. Mol. Asp. Med. 2018, 59, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Duan, L.; Cui, D.; Xie, J. Exploration of Biomarkers for Systemic Lupus Erythematosus by Machine-Learning Analysis. BMC Immunol. 2023, 24, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; He, S.; Yu, X.; Lai, X.; Tang, S.; Mariya, M.E.A.; Wang, M.; Yan, H.; Huang, X.; Zeng, S.; et al. Analysis and Experimental Validation of Rheumatoid Arthritis Innate Immunity Gene CYFIP2 and Pan-Cancer. Front. Immunol. 2022, 13, 954848. [Google Scholar] [CrossRef] [PubMed]
- Messaoud-Nacer, Y.; Culerier, E.; Rose, S.; Maillet, I.; Rouxel, N.; Briault, S.; Ryffel, B.; Quesniaux, V.F.J.; Togbe, D. STING Agonist diABZI Induces PANoptosis and DNA Mediated Acute Respiratory Distress Syndrome (ARDS). Cell Death Dis. 2022, 13, 269. [Google Scholar] [CrossRef]
- Karki, R.; Lee, S.; Mall, R.; Pandian, N.; Wang, Y.; Sharma, B.R.; Malireddi, R.S.; Yang, D.; Trifkovic, S.; Steele, J.A.; et al. ZBP1-Dependent Inflammatory Cell Death, PANoptosis, and Cytokine Storm Disrupt IFN Therapeutic Efficacy during Coronavirus Infection. Sci. Immunol. 2022, 7, eabo6294. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ngunjiri, J.M.; Lee, C.-W. Influenza a Virus Subpopulations and Their Implication in Pathogenesis and Vaccine Development. Annu. Rev. Anim. Biosci. 2020, 8, 247–267. [Google Scholar] [CrossRef]
- Ampomah, P.B.; Lim, L.H.K. Influenza a Virus-Induced Apoptosis and Virus Propagation. Apoptosis Int. J. Program. Cell Death 2020, 25, 1–11. [Google Scholar] [CrossRef]
- Shirley, M. Baloxavir Marboxil: A Review in Acute Uncomplicated Influenza. Drugs 2020, 80, 1109–1118. [Google Scholar] [CrossRef]
- Zheng, M.; Kanneganti, T.-D. The Regulation of the ZBP1-NLRP3 Inflammasome and Its Implications in Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Immunol. Rev. 2020, 297, 26–38. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020, 180, 1115–1129.e13. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Lee, S. Recent Advances in ZBP1-Derived PANoptosis against Viral Infections. Front. Immunol. 2023, 14, 1148727. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Kanneganti, T.-D. PANoptosome Signaling and Therapeutic Implications in Infection: Central Role for ZBP1 to Activate the Inflammasome and PANoptosis. Curr. Opin. Immunol. 2023, 83, 102348. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Yang, H.-Y.; Li, Y.-P.; Shi, Z.-J.; Zhou, Z.-Y.; You, Y.-P.; Ke, H.-Y.; Yan, L.; Xu, L.-H.; Ouyang, D.-Y.; et al. Scutellarin Inhibits Inflammatory PANoptosis by Diminishing Mitochondrial ROS Generation and Blocking PANoptosome Formation. Int. Immunopharmacol. 2024, 139, 112710. [Google Scholar] [CrossRef]
- Guo, L.; Han, L.; Zhang, J.; Shen, M.; Li, J.; Zhang, K.; Chen, R.; Liu, H. HMGB1 Mediates Epithelial-Mesenchymal Transition and Fibrosis in Silicosis via RAGE/β-Catenin Signaling. Chem. Biol. Interact. 2025, 408, 111385. [Google Scholar] [CrossRef]
- Öz, H.H.; Cheng, E.-C.; Di Pietro, C.; Tebaldi, T.; Biancon, G.; Zeiss, C.; Zhang, P.-X.; Huang, P.H.; Esquibies, S.S.; Britto, C.J.; et al. Recruited Monocytes/Macrophages Drive Pulmonary Neutrophilic Inflammation and Irreversible Lung Tissue Remodeling in Cystic Fibrosis. Cell Rep. 2022, 41, 111797. [Google Scholar] [CrossRef]
- Duan, M.; Hibbs, M.L.; Chen, W. The Contributions of Lung Macrophage and Monocyte Heterogeneity to Influenza Pathogenesis. Immunol. Cell Biol. 2017, 95, 225–235. [Google Scholar] [CrossRef]
- Bi, X.; Li, M.; Guo, Y.; Hu, M.; Chen, Y.; Lian, N.; Chen, S.; Li, M.; Gu, H.; Chen, X. ZBP1-Mediated PANoptosis Is a Crucial Lethal Form in Diverse Keratinocyte Death Modalities in UVB-Induced Skin Injury. Cell Death Dis. 2025, 16, 44. [Google Scholar] [CrossRef]
- Batiha, G.E.-S.; Al-Gareeb, A.I.; Rotimi, D.; Adeyemi, O.S.; Al-Kuraishy, H.M. Common NLRP3 Inflammasome Inhibitors and COVID-19: Divide and Conquer. Sci. Afr. 2022, 18, e01407. [Google Scholar] [CrossRef] [PubMed]
- Adhikarla, S.V.; Jha, N.K.; Goswami, V.K.; Sharma, A.; Bhardwaj, A.; Dey, A.; Villa, C.; Kumar, Y.; Jha, S.K. TLR-Mediated Signal Transduction and Neurodegenerative Disorders. Brain Sci. 2021, 11, 1373. [Google Scholar] [CrossRef]
- Shi, L.; Chen, S.; Yang, L.; Li, Y. The Role of PD-1 and PD-L1 in T-Cell Immune Suppression in Patients with Hematological Malignancies. J. Hematol. Oncol. 2013, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.N.; Vanguri, V.K.; Ha, S.-J.; West, E.E.; Keir, M.E.; Glickman, J.N.; Sharpe, A.H.; Ahmed, R. PD-L1 Has Distinct Functions in Hematopoietic and Nonhematopoietic Cells in Regulating T Cell Responses during Chronic Infection in Mice. J. Clin. Investig. 2010, 120, 2508–2515. [Google Scholar] [CrossRef]
- Zheng, M.; Kanneganti, T.-D. Newly Identified Function of Caspase-6 in ZBP1-Mediated Innate Immune Responses, NLRP3 Inflammasome Activation, PANoptosis, and Host Defense. J. Cell. Immunol. 2020, 2, 341. [Google Scholar] [CrossRef]
- Choudhury, S.M.; Sarkar, R.; Karki, R.; Kanneganti, T.-D. A Comparative Study of Apoptosis, Pyroptosis, Necroptosis, and PANoptosis Components in Mouse and Human Cells. PLoS ONE 2024, 19, e0299577. [Google Scholar] [CrossRef]
- Man, S.M.; Kanneganti, T.-D. Regulation of Lysosomal Dynamics and Autophagy by CTSB/Cathepsin, B. Autophagy 2016, 12, 2504–2505. [Google Scholar] [CrossRef]
- Gaire, B.P.; Subedi, L.; Teramoto, H.; Hu, B. The Role of Cathepsin B in Ischemia-Reperfusion Injury after Stroke. In Cerebral Ischemia; Exon Publications: Brisbane, QLD, Australia, 2021. [Google Scholar]
- Norton, E.S.; Whaley, L.A.; Jones, V.K.; Brooks, M.M.; Russo, M.N.; Morderer, D.; Jessen, E.; Schiapparelli, P.; Ramos-Fresnedo, A.; Zarco, N.; et al. Cell-Specific Cross-Talk Proteomics Reveals Cathepsin B Signaling as a Driver of Glioblastoma Malignancy near the Subventricular Zone. Sci. Adv. 2024, 10, eadn1607. [Google Scholar] [CrossRef]
- Xu, L.-B.; Qin, Y.-F.; Su, L.; Huang, C.; Xu, Q.; Zhang, R.; Shi, X.-D.; Sun, R.; Chen, J.; Song, Z.; et al. Cathepsin-Facilitated Invasion of BMI1-High Hepatocellular Carcinoma Cells Drives Bile Duct Tumor Thrombi Formation. Nat. Commun. 2023, 14, 7033. [Google Scholar] [CrossRef]
- Liu, M.; Lu, J.; Hu, J.; Chen, Y.; Deng, X.; Wang, J.; Zhang, S.; Guo, J.; Li, W.; Guan, S. Sodium Sulfite Triggered Hepatic Apoptosis, Necroptosis, and Pyroptosis by Inducing Mitochondrial Damage in Mice and AML-12 Cells. J. Hazard. Mater. 2024, 467, 133719. [Google Scholar] [CrossRef] [PubMed]
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza a Virus-Infected Human Primary Macrophages-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/21589892/ (accessed on 23 July 2025).
- Qi, X.; Man, S.M.; Malireddi, R.K.S.; Karki, R.; Lupfer, C.; Gurung, P.; Neale, G.; Guy, C.S.; Lamkanfi, M.; Kanneganti, T.-D. Cathepsin B Modulates Lysosomal Biogenesis and Host Defense against Francisella Novicida Infection. J. Exp. Med. 2016, 213, 2081–2097. [Google Scholar] [CrossRef]
- Man, Y.; Zhai, Y.; Jiang, A.; Bai, H.; Gulati, A.; Plebani, R.; Mannix, R.J.; Merry, G.E.; Goyal, G.; Belgur, C.; et al. Exacerbation of Influenza Virus Induced Lung Injury by Alveolar Macrophages and Its Suppression by Pyroptosis Blockade in a Human Lung Alveolus Chip. bioRxiv Preprint 2024. [Google Scholar] [CrossRef]
- Cui, Z.; Li, Y.; Bi, Y.; Li, W.; Piao, J.; Ren, X. PANoptosis: A New Era for Anti-Cancer Strategies. Life Sci. 2024, 359, 123241. [Google Scholar] [CrossRef]
- Lu, D.; Zhang, W.; Li, R.; Tan, S.; Zhang, Y. Targeting Necroptosis in Alzheimer’s Disease: Can Exercise Modulate Neuronal Death? Front. Aging Neurosci. 2025, 17, 1499871. [Google Scholar] [CrossRef]
- Blevins, H.M.; Xu, Y.; Biby, S.; Zhang, S. The NLRP3 Inflammasome Pathway: A Review of Mechanisms and Inhibitors for the Treatment of Inflammatory Diseases. Front. Aging Neurosci. 2022, 14, 879021. [Google Scholar] [CrossRef] [PubMed]
- Paniri, A.; Akhavan-Niaki, H. Emerging Role of IL-6 and NLRP3 Inflammasome as Potential Therapeutic Targets to Combat COVID-19: Role of lncRNAs in Cytokine Storm Modulation. Life Sci. 2020, 257, 118114. [Google Scholar] [CrossRef] [PubMed]
- Baral, H.; Kaundal, R.K. Novel Insights into Neuroinflammatory Mechanisms in Traumatic Brain Injury: Focus on Pattern Recognition Receptors as Therapeutic Targets. Cytokine Growth Factor Rev. 2025, 83, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, J.; Zhang, Y.; Wang, C.; Zhang, L. IL-10: The Master Immunomodulatory Cytokine in Allergen Immunotherapy. Expert Rev. Clin. Immunol. 2025, 21, 17–28. [Google Scholar] [CrossRef]
- Pang, J.; Vince, J.E. The Role of Caspase-8 in Inflammatory Signalling and Pyroptotic Cell Death. Semin. Immunol. 2023, 70, 101832. [Google Scholar] [CrossRef]
- Carlini, V.; Noonan, D.M.; Abdalalem, E.; Goletti, D.; Sansone, C.; Calabrone, L.; Albini, A. The Multifaceted Nature of IL-10: Regulation, Role in Immunological Homeostasis and Its Relevance to Cancer, COVID-19 and Post-COVID Conditions. Front. Immunol. 2023, 14, 1161067. [Google Scholar] [CrossRef]
- Jürgensen, H.J.; Silva, L.M.; Krigslund, O.; van Putten, S.; Madsen, D.H.; Behrendt, N.; Engelholm, L.H.; Bugge, T.H. CCL2/MCP-1 Signaling Drives Extracellular Matrix Turnover by Diverse Macrophage Subsets. Matrix Biol. Plus 2019, 1, 100003. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Wang, C.; Liu, Z.; Xin, W.; Zhang, Q.; Ali, A.; Zeng, X.; Li, Z.; Ma, C. Machine Learning Algorithms Integrate Bulk and Single-Cell RNA Data to Unveil Oxidative Stress Following Intracerebral Hemorrhage. Int. Immunopharmacol. 2024, 137, 112449. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Zhang, X.; Xu, Y.; Chen, L.; Zhang, W.; Liu, E.; Xiao, C.; Kou, Q. Cathepsin B Aggravates Acute Pancreatitis by Activating the NLRP3 Inflammasome and Promoting the Caspase-1-Induced Pyroptosis. Int. Immunopharmacol. 2021, 94, 107496. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Sharkey, D.; Cantley, L.G. Tubular GM-CSF Promotes Late MCP-1/CCR2-Mediated Fibrosis and Inflammation after Ischemia/Reperfusion Injury. J. Am. Soc. Nephrol. JASN 2019, 30, 1825–1840. [Google Scholar] [CrossRef]
- Lukic, A.; Larssen, P.; Fauland, A.; Samuelsson, B.; Wheelock, C.E.; Gabrielsson, S.; Radmark, O. GM-CSF- and M-CSF-Primed Macrophages Present Similar Resolving but Distinct Inflammatory Lipid Mediator Signatures. Fed. Am. Soc. Exp. Biol. 2017, 31, 4370–4381. [Google Scholar] [CrossRef]
- Siwetz, M.; Blaschitz, A.; El-Heliebi, A.; Hiden, U.; Desoye, G.; Huppertz, B.; Gauster, M. TNF-α Alters the Inflammatory Secretion Profile of Human First Trimester Placenta. Lab. Investig. J. Tech. Methods Pathol 2016, 96, 428–438. [Google Scholar] [CrossRef]
- Kandhaya-Pillai, R.; Yang, X.; Tchkonia, T.; Martin, G.M.; Kirkland, J.L.; Oshima, J. TNF-α/IFN-γ Synergy Amplifies Senescence-associated Inflammation and SARS-CoV-2 Receptor Expression via Hyper-activated JAK/STAT1. Aging Cell 2022, 21, e13646. [Google Scholar] [CrossRef]
- Li, X.; Zhu, L.; Wang, B.; Yuan, M.; Zhu, R. Drugs and Targets in Fibrosis. Front. Pharmacol. 2017, 8, 855. [Google Scholar] [CrossRef]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.-J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine Inhibits Autophagic Flux by Decreasing Autophagosome-Lysosome Fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef]
- Tang, T.-T.; Lv, L.-L.; Pan, M.-M.; Wen, Y.; Wang, B.; Li, Z.-L.; Wu, M.; Wang, F.-M.; Crowley, S.D.; Liu, B.-C. Hydroxychloroquine Attenuates Renal Ischemia/Reperfusion Injury by Inhibiting Cathepsin Mediated NLRP3 Inflammasome Activation. Cell Death Dis. 2018, 9, 351. [Google Scholar] [CrossRef]
- McAfee, Q.; Zhang, Z.; Samanta, A.; Levi, S.M.; Ma, X.-H.; Piao, S.; Lynch, J.P.; Uehara, T.; Sepulveda, A.R.; Davis, L.E.; et al. Autophagy Inhibitor Lys05 Has Single-Agent Antitumor Activity and Reproduces the Phenotype of a Genetic Autophagy Deficiency. Proc. Natl. Acad. Sci. USA 2012, 109, 8253–8258. [Google Scholar] [CrossRef] [PubMed]
- Gies, V.; Bekaddour, N.; Dieudonné, Y.; Guffroy, A.; Frenger, Q.; Gros, F.; Rodero, M.P.; Herbeuval, J.-P.; Korganow, A.-S. Beyond Anti-Viral Effects of Chloroquine/Hydroxychloroquine. Front. Immunol. 2020, 11, 1409. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.K. Repurposing New Use for Old Drug Chloroquine against Metabolic Syndrome: A Review on Animal and Human Evidence. Int. J. Med. Sci. 2021, 18, 2673–2688. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Zeng, T.; Wang, Y.; Zhang, H.; Wan, Y.; Shi, Z.; Cao, R.; Tang, H. Integrated Bulk and Single-Cell Transcriptomes Reveal Pyroptotic Signature in Prognosis and Therapeutic Options of Hepatocellular Carcinoma by Combining Deep Learning. Briefings Bioinf. 2023, 25, bbad487. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, Z.; He, D.; Zhu, Y.; Chen, X.; Cao, K. Identification of Novel Subtypes Based on ssGSEA in Immune-Related Prognostic Signature for Tongue Squamous Cell Carcinoma. Cancer Med. 2021, 10, 8693–8707. [Google Scholar] [CrossRef]
- Mei, Y.; Li, M.; Wen, J.; Kong, X.; Li, J. Single-Cell Characteristics and Malignancy Regulation of Alpha-Fetoprotein-Producing Gastric Cancer. Cancer Med. 2023, 12, 12018–12033. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar] [CrossRef]
- Luo, G.; Zhu, Y.; Wang, R.; Tong, Y.; Lu, W.; Wang, H. Random Forest-Based Classsification and Analysis of Hemiplegia Gait Using Low-Cost Depth Cameras. Med. Biol. Eng. Comput. 2020, 58, 373–382. [Google Scholar] [CrossRef]
- Kang, J.; Choi, Y.J.; Kim, I.-K.; Lee, H.S.; Kim, H.; Baik, S.H.; Kim, N.K.; Lee, K.Y. LASSO-Based Machine Learning Algorithm for Prediction of Lymph Node Metastasis in T1 Colorectal Cancer. Cancer Res. Treat. 2021, 53, 773–783. [Google Scholar] [CrossRef]
- de Abreu, R.C.; Ramos, C.V.; Becher, C.; Lino, M.; Jesus, C.; da Costa Martins, P.A.; Martins, P.A.T.; Moreno, M.J.; Fernandes, H.; Ferreira, L. Exogenous Loading of miRNAs into Small Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12111, Erratum in J. Extracell. Vesicles 2021, 10, e12149. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Zhu, L.; Zhang, C.; Wang, D.; Liu, H.; Liu, J.; Sun, J.; Feng, X.; Yang, W. Machine Learning Integration of Bulk and Single-Cell RNA-Seq Data Reveals Cathepsin B as a Central PANoptosis Regulator in Influenza. Int. J. Mol. Sci. 2025, 26, 8533. https://doi.org/10.3390/ijms26178533
Liu B, Zhu L, Zhang C, Wang D, Liu H, Liu J, Sun J, Feng X, Yang W. Machine Learning Integration of Bulk and Single-Cell RNA-Seq Data Reveals Cathepsin B as a Central PANoptosis Regulator in Influenza. International Journal of Molecular Sciences. 2025; 26(17):8533. https://doi.org/10.3390/ijms26178533
Chicago/Turabian StyleLiu, Bin, Lin Zhu, Caijuan Zhang, Dunfang Wang, Haifan Liu, Jianyao Liu, Jingwei Sun, Xue Feng, and Weipeng Yang. 2025. "Machine Learning Integration of Bulk and Single-Cell RNA-Seq Data Reveals Cathepsin B as a Central PANoptosis Regulator in Influenza" International Journal of Molecular Sciences 26, no. 17: 8533. https://doi.org/10.3390/ijms26178533
APA StyleLiu, B., Zhu, L., Zhang, C., Wang, D., Liu, H., Liu, J., Sun, J., Feng, X., & Yang, W. (2025). Machine Learning Integration of Bulk and Single-Cell RNA-Seq Data Reveals Cathepsin B as a Central PANoptosis Regulator in Influenza. International Journal of Molecular Sciences, 26(17), 8533. https://doi.org/10.3390/ijms26178533







