Scaffold-Hopping Design and Synthesis of Thieno[3,2-d]pyrimidines: Anticancer Activity, Apoptosis Induction, and In Silico Inhibition of CDKs
Abstract
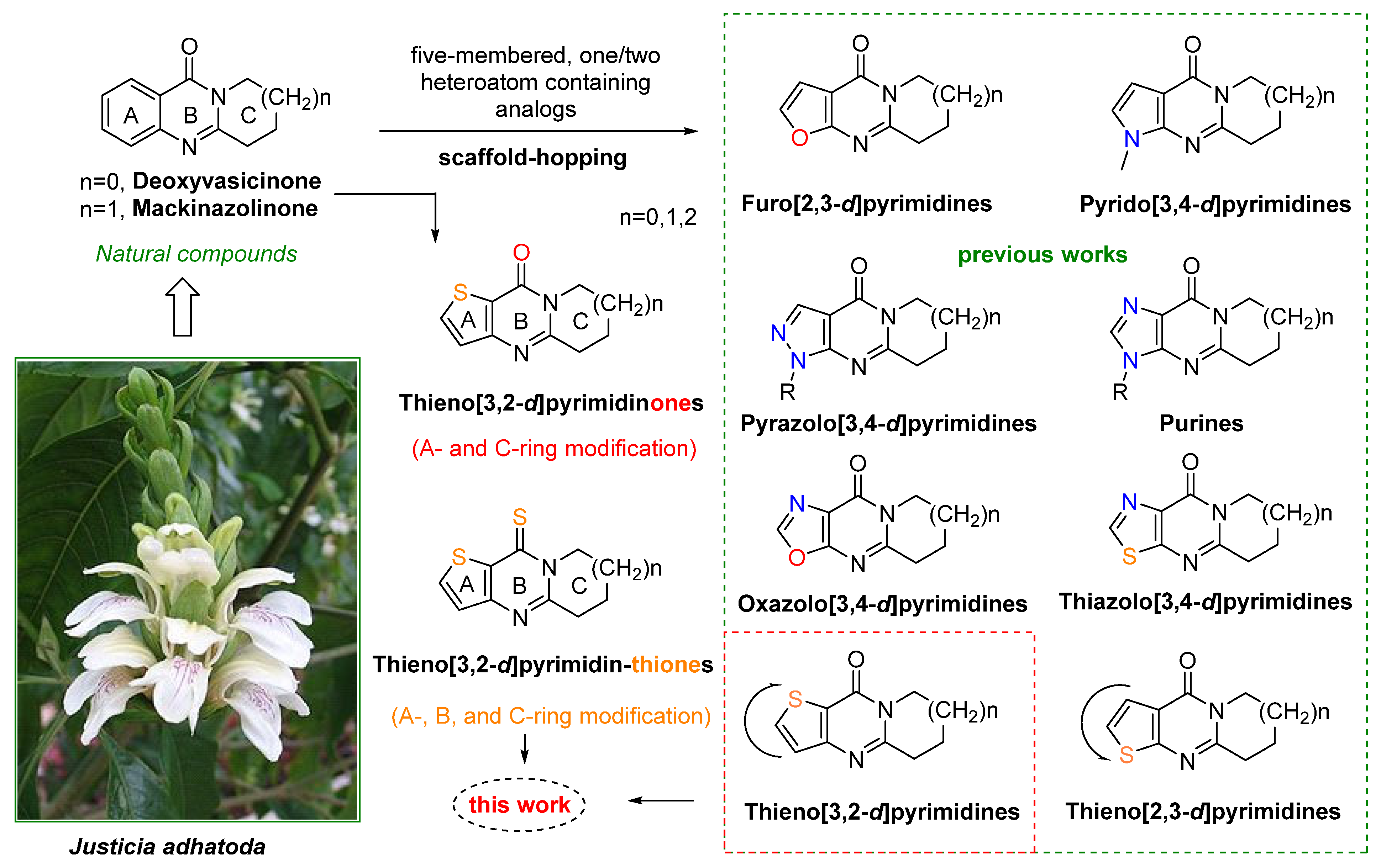
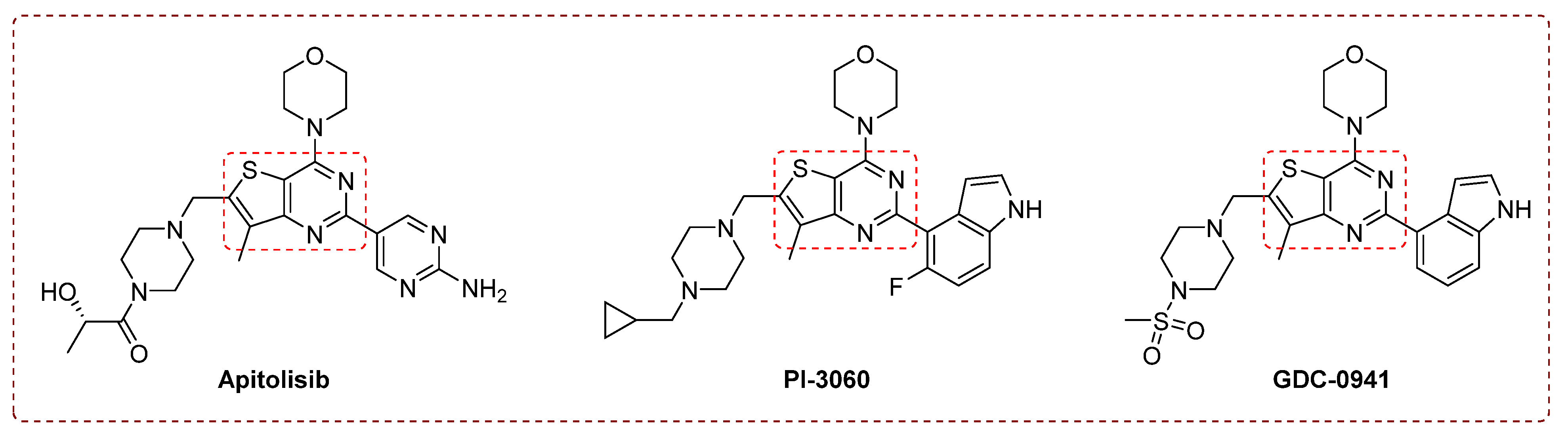
1. Introduction
- The total synthesis of other tricyclic analogs of deoxyvasicinone could provide novel tricyclic annulated heterocyclic systems for further research of pharmacological interest in related fields.
- Convenient synthetic methods for these heterocycles could be used to modify quinazolinone scaffolds via a scaffold-hopping strategy.
2. Results and Discussion
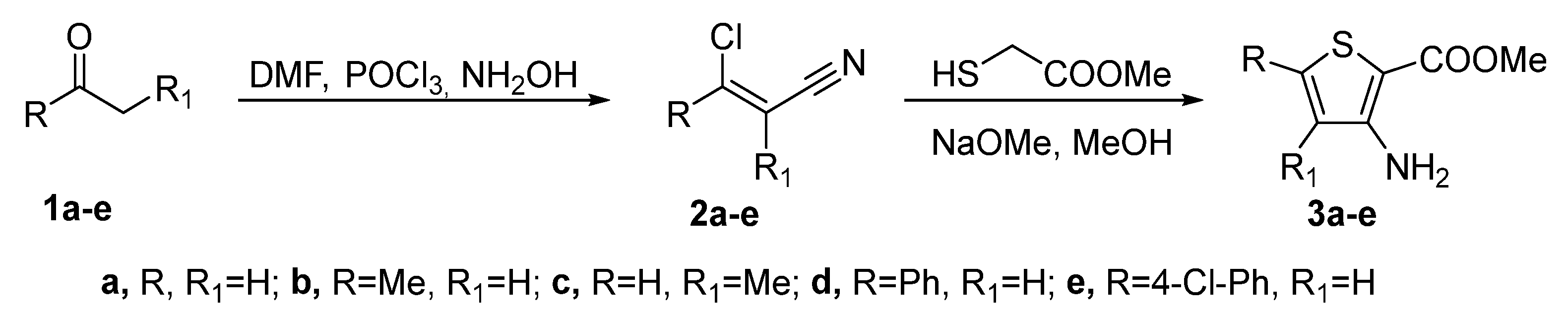
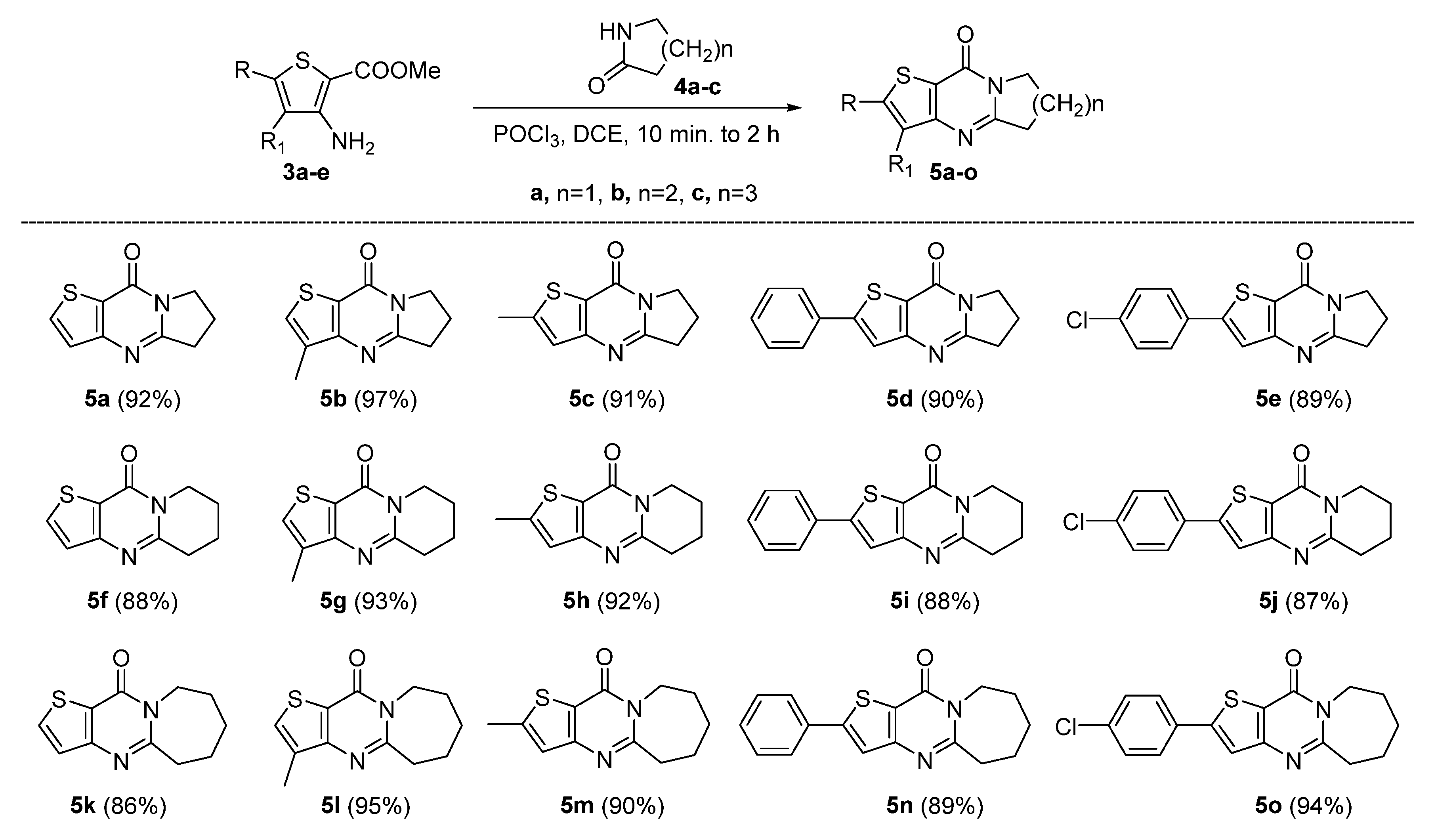
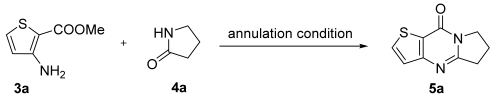
2.1. Synthesis
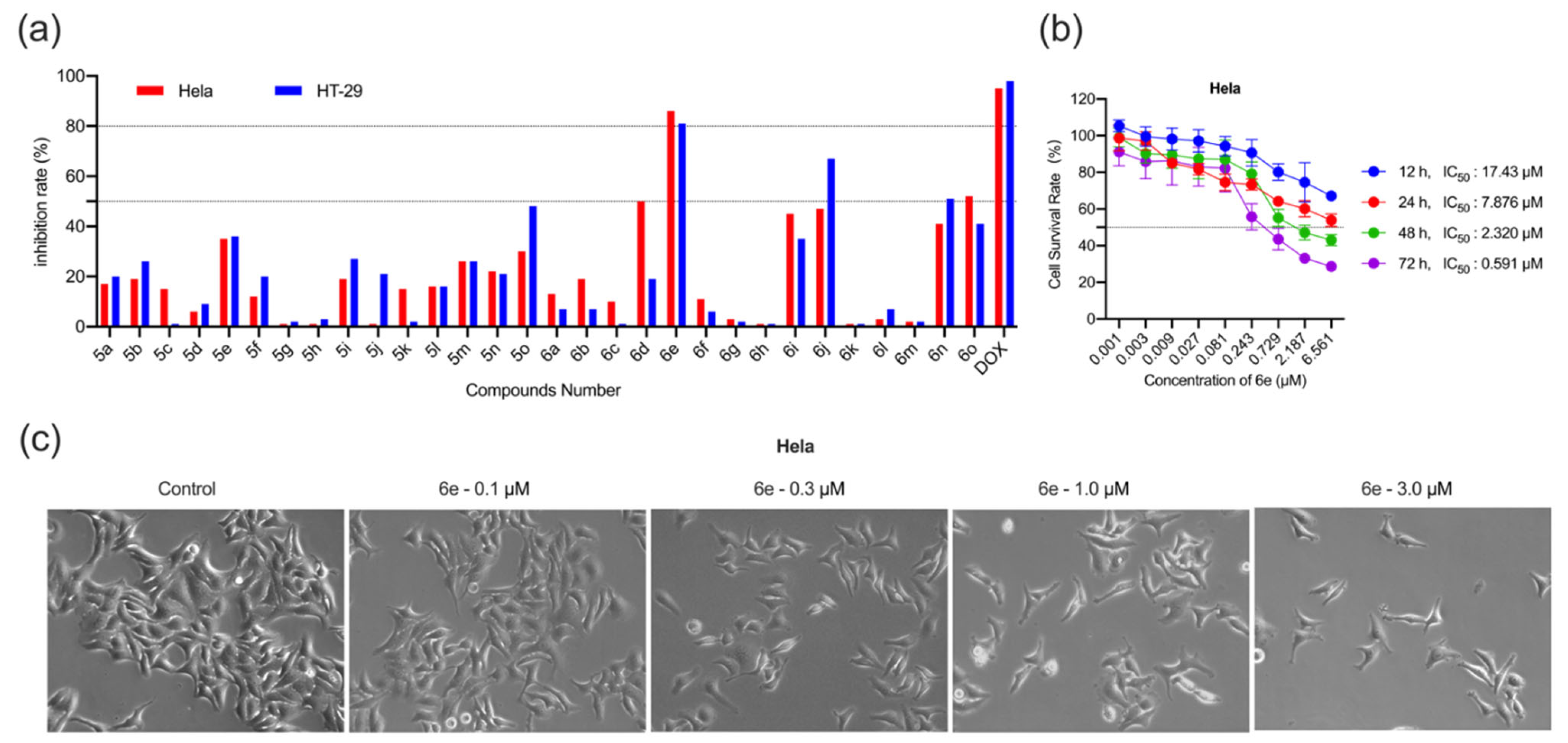
2.2. Antiproliferative Activity
2.2.1. Effect of 6e on Morphological Changes in HeLa Cells
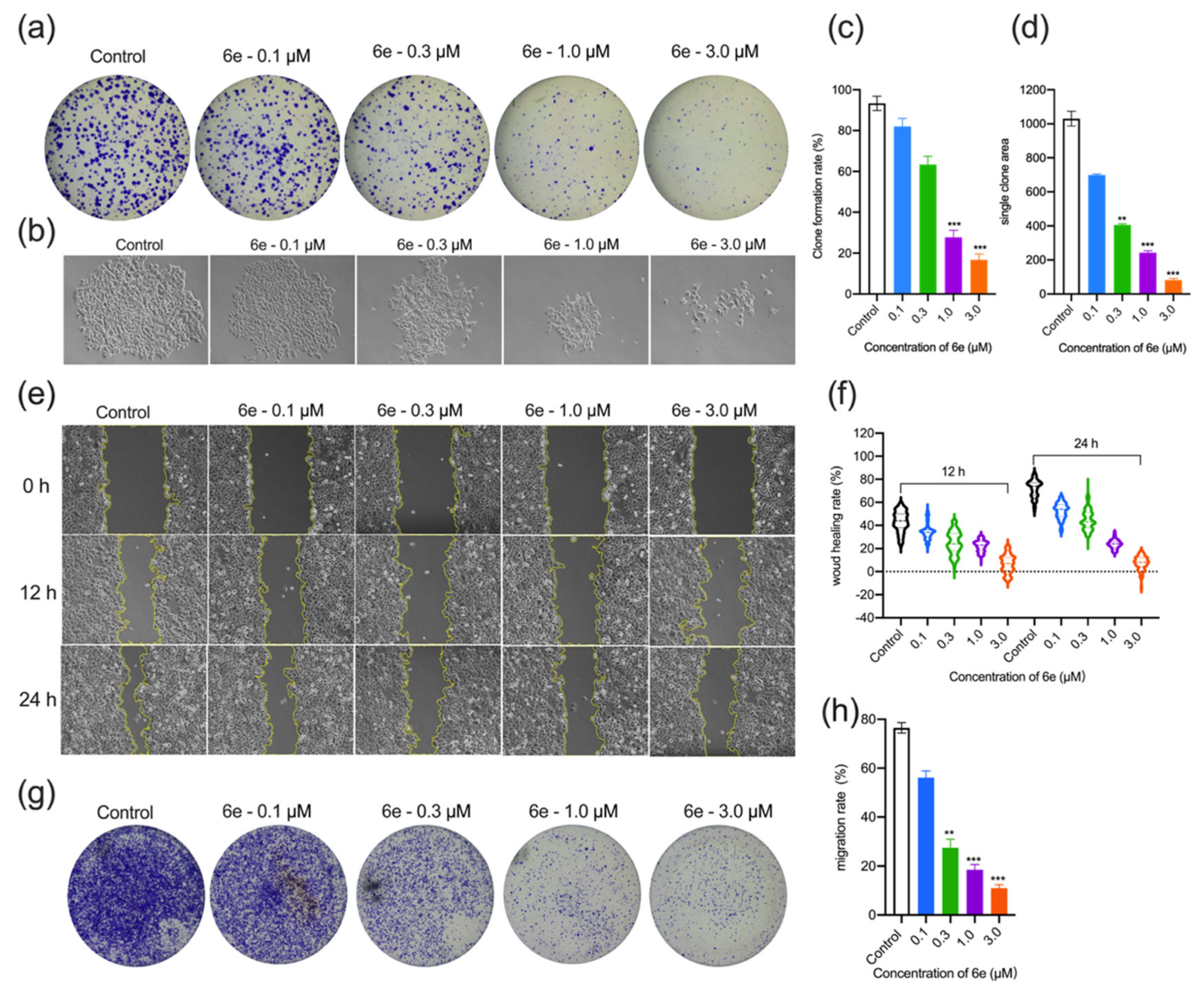
2.2.2. Effect of 6e on Colony Formation, Wound Healing, and Migration Ability of HeLa Cells
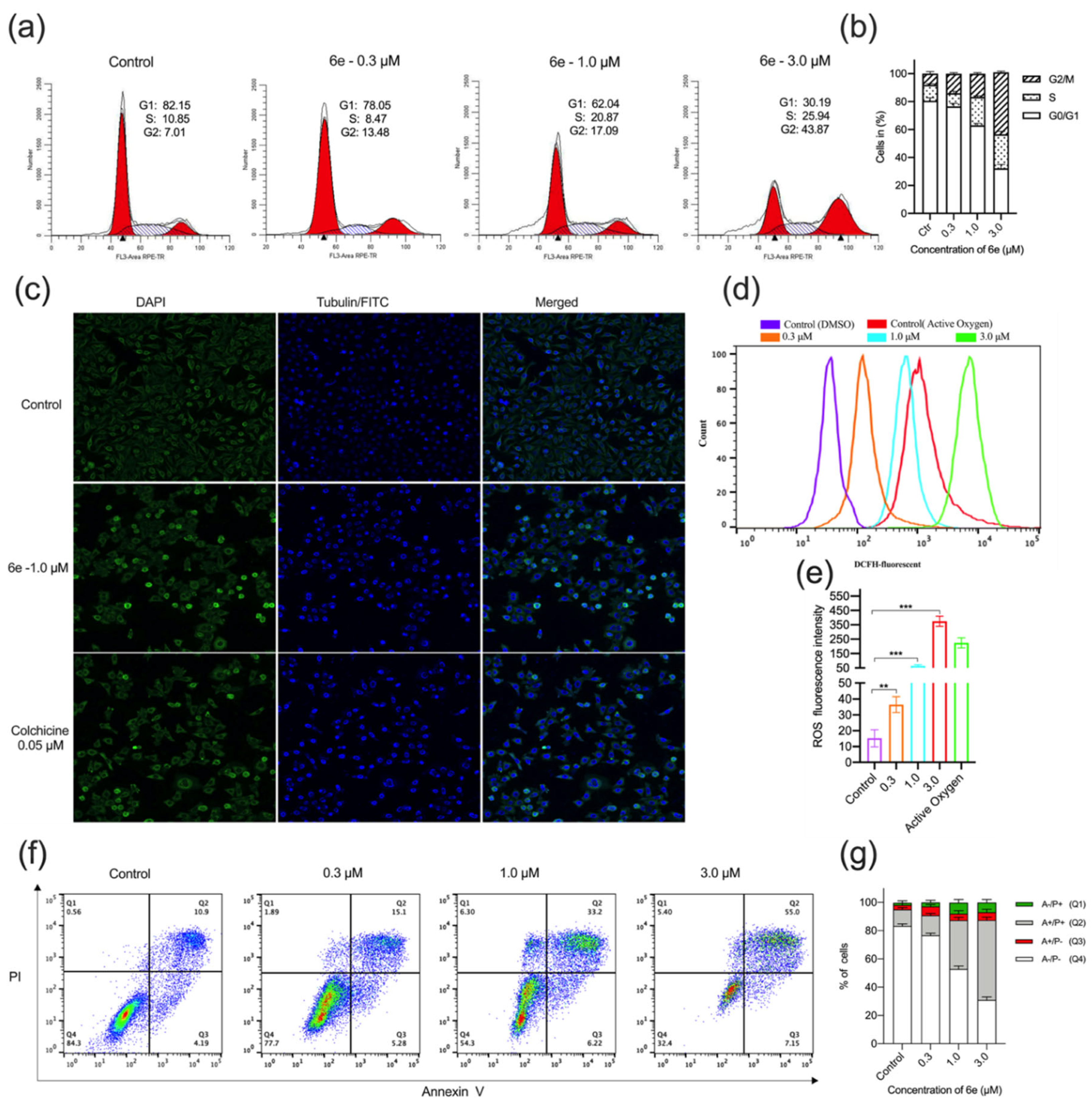
2.2.3. Cell-Cycle Arrest, Microtubule Structures, Intracellular ROS Accumulation, and Apoptosis in HeLa Cells by 6e
2.3. Molecular Docking
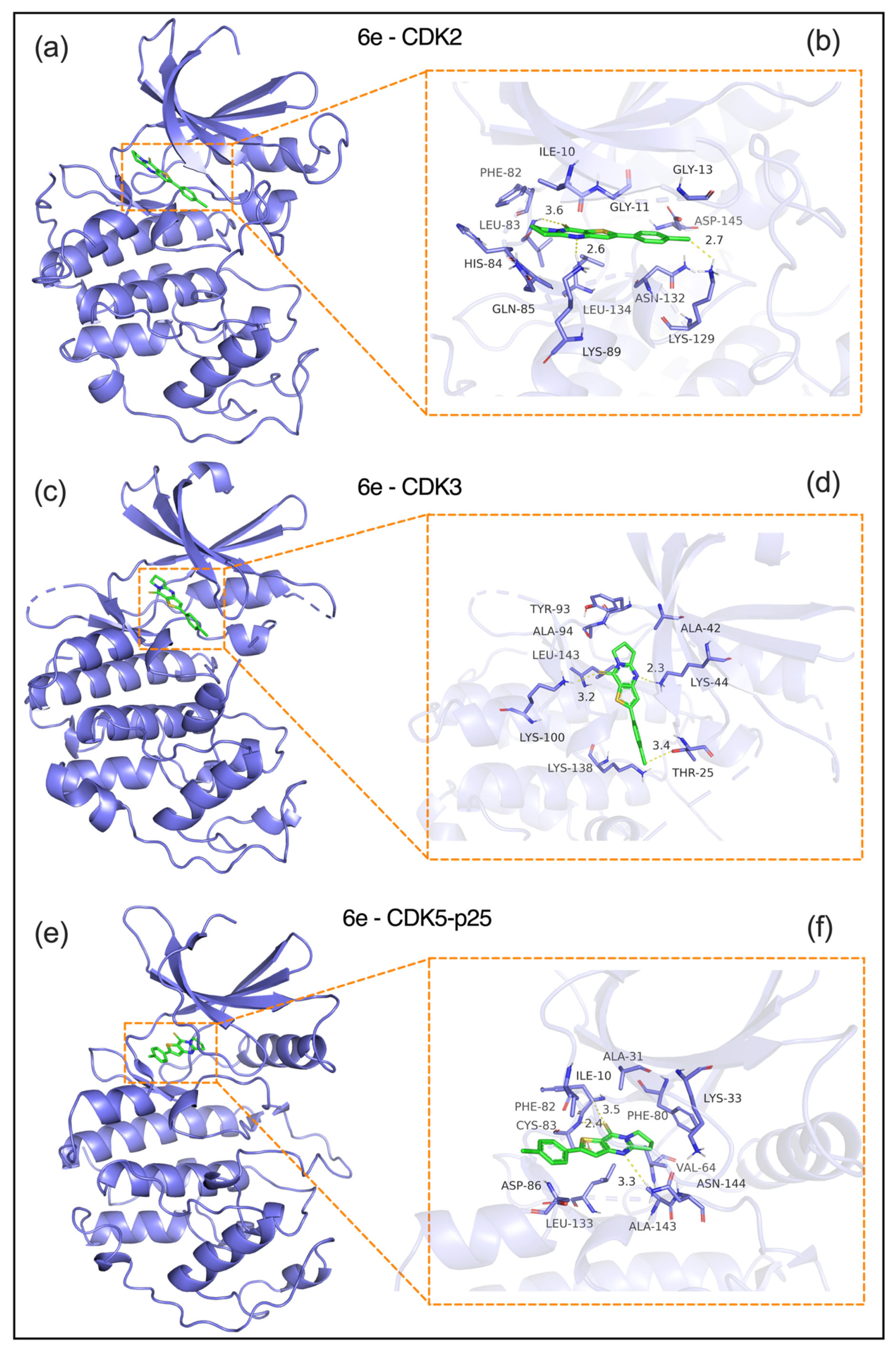
2.3.1. Analysis of Protein–Protein Interactions
- a) CDK2 (PDB: 1H15, Figure 6a,b): 6e was embedded in the ATP-binding pocket of CDK2, forming critical interactions and a CDOCKER energy of -8.2 kcal/mol. Hydrogen bond: A 2.6-Å bond with the ILE10 backbone carbonyl anchors the ligand. Hydrophobic interactions: Close contacts with PHE82 (2.7 Å), LEU83 (3.1 Å), and VAL104 (3.6 Å) via van der Waals forces. Electrostatic interaction: A 3.2 Å interaction with ASN132 stabilizes the ligand’s pyrimidine moiety. These interactions block CDK2-mediated phosphorylation of the Rb protein, which is consistent with the G1/S phase arrest observed in cellular assays.
- b) CDK3 (PDB: 1O37, Figure 6c,d): The ligand is bound to a conserved hydrophobic pocket in CDK3 (CDOCKER energy: -7.8 kcal/mol) through the following interactions. Hydrogen bond: A 2.3 Å bond between the ligand’s purine ring and the LYS44 side chain. Hydrophobic interactions: A 3.2-Å interaction with ALA42, a 3.4-Å interaction with LEU143, and π-π stacking with TYR93 at 3.6 Å. Electrostatic interaction: A 3.1 Å interaction with THR25 enhances binding specificity. This binding mode disrupts the formation of the CDK3-cyclin E complex, thereby inhibiting G1 phase progression.
- c) CDK5-p25 complex (PDB: 1UGS, Figure 6e,f): 6e engages the ATP-binding site of the CDK5-p25 complex (CDOCKER energy: −7.6 kcal/mol) via: Hydrogen bond: A 2.4-Å bond with the ALA31 backbone amide. Hydrophobic interactions: PHE80 (3.3 Å), ILE10 (3.5 Å), and CYS82 (3.4 Å); and π-π stacking with PHE80 (3.8 Å). An interaction with PHE80’s aromatic ring (3.8 Å) is critical for ligand orientation. These interactions may interfere with CDK5-p25-mediated phosphorylation of the tau protein, which is associated with reduced metastatic potential.
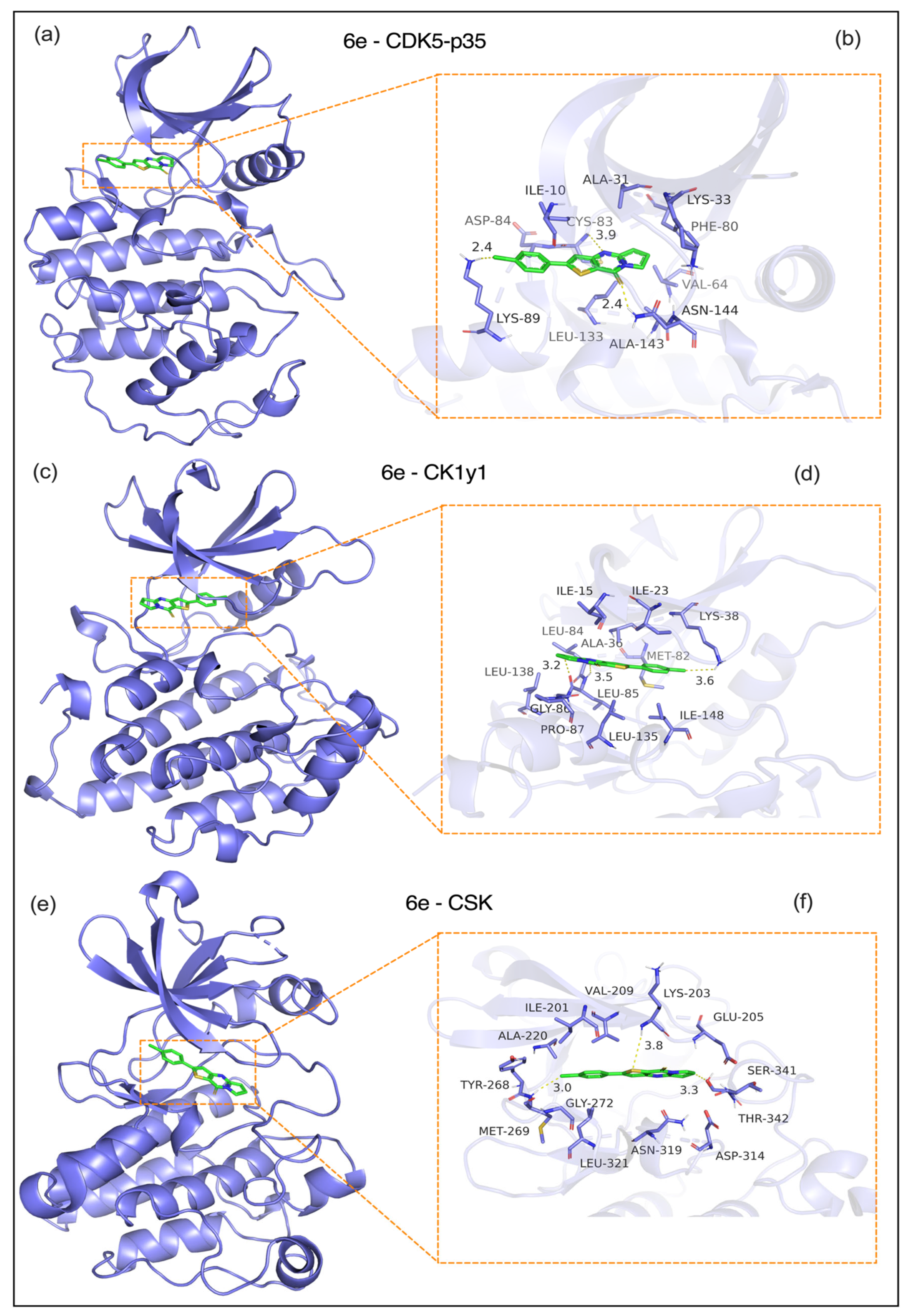
- d) CDK5-p35 complex (PDB: 2PH1, Figure 7a,b): 6e showed weaker binding to the CDK5-p35 complex (CDOCKER energy: -7.2 kcal/mol) through a hydrogen bond with the backbone carbonyl of GLY30. A 2.4 Å bond with the GLY30 backbone carbonyl. Hydrophobic interactions: Hydrophobic interactions with VAL32 (3.5 Å), ALA31 (3.5 Å), ILE10 (3.3 Å), and LEU83 (3.6 Å). Notably, the p35-specific loop (residues 140–150) showed minimal contact with 6e, suggesting a lower risk of off-target effects in neurons compared to p25.
- e) CK1γ1 (PDB: 3P70, Figure 7c,d): 6e bound to the CK1γ1 kinase domain (CDOCKER energy: -6.8 kcal/mol) via the following: Hydrogen bond: A 2.9-Å bond with the SER36 side chain, which is critical for ATP binding. Hydrophobic interactions: LEU85 (3.5 Å), ILE148 (3.6 Å), and VAL116 (3.4 Å); and electrostatic interactions: A 3.3 Å interaction with GLU63 stabilizes the ligand’s charged group. This binding mode may disrupt CK1γ1-mediated β-catenin phosphorylation and inhibit Wnt/β-catenin signaling.
- f) CSK (PDB: 1A1Q, Figure 7e,f): 6e interacts with the CSK SH2-kinase interface (CDOCKER energy: -6.5 kcal/mol) through the following. Hydrogen bond: A 2.8 Å bond with Tyr268, a key phosphorylation site. Hydrophobic interactions: ALA220 (3.3 Å), MET269 (3.8 Å), and LEU273 (3.5 Å). Electrostatic interaction: A 3.1 Å interaction with GLU222 enhances the binding affinity. These interactions may enhance CSK-mediated suppression of Src kinases, which aligns with reduced tumor cell survival.
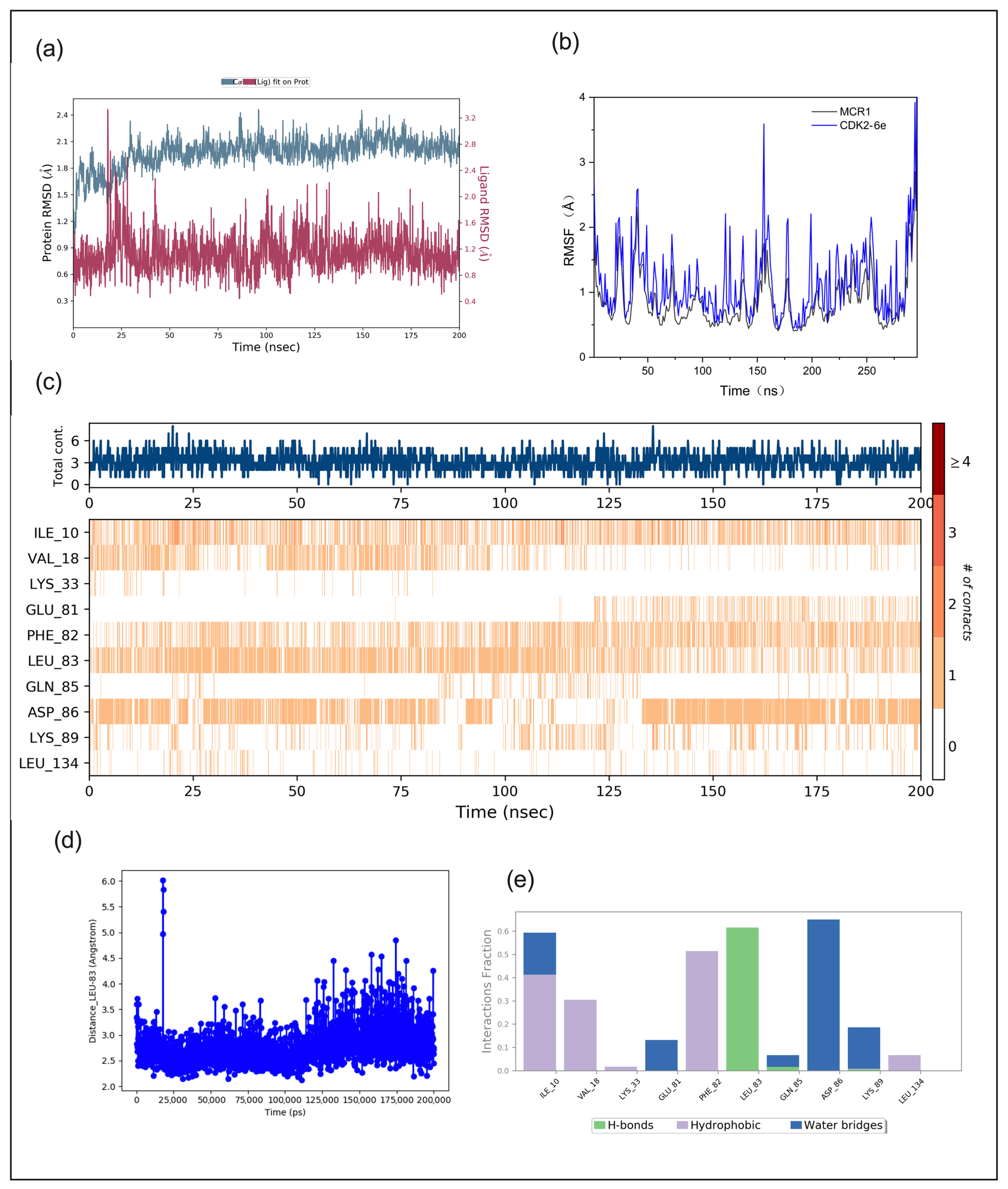
2.3.2. Molecular Dynamics Analysis
3. Materials and Methods
3.1. Chemistry
3.1.1. General Materials and Methods
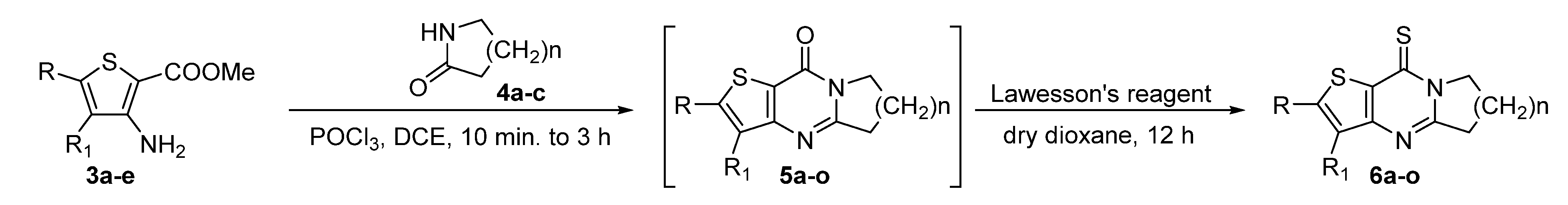
3.1.2. General Procedure for the Synthesis of Compounds 5a–5o
- 6,7-Dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-one (5a). Yield 92%, white solid, mp 154–155 °C; 1H NMR (400 MHz, CDCl3) δ 7.76 (d, J = 5.2 Hz, 1H), 7.26 (d, J = 5.2 Hz, 1H), 4.23 (t, J = 7.3 Hz, 2H), 3.19 (t, J = 7.9 Hz, 2H), 2.39–2.27 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 161.46, 158.40, 157.40, 134.14, 124.80, 121.46, 46.68, 32.33, 20.10. HRMS (ESI) calcd for C9H8N2OS[M+H]+ 193.0421, found 193.0426.
- 3-Methyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-one (5b). Yield 97%, white solid, mp 179–180 °C; 1H NMR (400 MHz, CDCl3) δ 7.40 (q, J = 1.1 Hz, 1H), 4.22 (t, J = 7.3 Hz, 2H), 3.21 (t, J = 8.0 Hz, 2H), 2.38 (s, 3H), 2.37–2.25 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 161.30, 158.78, 157.00, 149.85, 122.93, 119.96, 46.70, 32.39, 20.03, 16.92. 13C NMR (100 MHz, CDCl3) δ 161.08, 157.62, 157.35, 133.72, 129.34, 121.57, 46.60, 32.47, 20.22, 13.26. HRMS (ESI) calcd for C10H10N2OS[M+H]+ 207.0578, found 207.0582.
- 2-Methyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-one (5c). Yield 91%, white solid, mp 172–174 °C; 1H NMR (400 MHz, CDCl3) δ 6.92 (q, J = 1.1 Hz, 1H), 4.20 (t, J = 7.3 Hz, 2H), 3.16 (t, J = 7.9 Hz, 2H), 2.60 (d, J = 1.1 Hz, 3H), 2.36–2.24 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 161.09, 158.58, 156.80, 149.65, 122.72, 119.76, 46.50, 32.18, 19.82, 16.71. HRMS (ESI) calcd for C10H10N2OS[M+H]+ 207.0575, found 207.0582.
- 2-Phenyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-one (5d). Yield 90%, light yellow solid, mp 205–206 °C; 1H NMR (400 MHz, CDCl3) δ 7.73–7.67 (m, 2H), 7.48–7.37 (m, 4H), 4.24 (t, J = 7.3 Hz, 2H), 3.19 (t, J = 8.0 Hz, 2H), 2.38–2.28 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 161.72, 158.96, 157.23, 152.51, 133.48, 129.63, 129.38, 126.60, 120.70, 120.23, 46.83, 32.48, 20.07. HRMS (ESI) calcd for C15H12N2OS[M+H]+ 269.0743, found 269.0745.
- 2-(4-Chlorophenyl)-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-one (5e). Yield 89%, light yellow solid, mp 242–244 °C; 1H NMR (400 MHz, CDCl3) δ 7.66–7.58 (m, 2H), 7.45–7.38 (m, 3H), 4.24 (t, J = 7.3 Hz, 2H), 3.19 (t, J = 8.0 Hz, 2H), 2.38–2.27 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 161.89, 158.91, 157.12, 150.95, 135.59, 131.99, 129.57, 127.77, 120.89, 120.57, 46.86, 32.47, 20.04. HRMS (ESI) calcd for C15H11ClN2OS[M+H]+ 303.0353, found 303.0357.
- 7,8-Dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-one (5f). Yield 88%, white solid, mp 103–105 °C; 1H NMR (400 MHz, CDCl3) δ 7.74 (d, J = 5.3 Hz, 1H), 7.24 (d, J = 5.2 Hz, 1H), 4.11 (t, J = 6.1 Hz, 2H), 3.01 (t, J = 6.7 Hz, 2H), 2.08–1.90 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 158.64, 156.59, 156.38, 134.31, 124.75, 121.14, 42.41, 31.87, 22.26, 19.48. HRMS (ESI) calcd for C10H10N2OS[M+H]+ 207.0587, found 207.0591.
- 3-Methyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-one (5g). Yield 93%, white solid, mp 99 °C; 1H NMR (400 MHz, CDCl3) δ 7.37 (d, J = 1.2 Hz, 1H), 4.11 (t, J = 6.1 Hz, 2H), 3.03 (t, J = 6.7 Hz, 2H), 2.38 (d, J = 1.1 Hz, 3H), 2.05–1.91 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 158.89, 156.23, 155.37, 133.75, 129.35, 121.24, 42.23, 31.99, 22.29, 19.53, 13.11. HRMS (ESI) calcd for C11H12N2OS[M+H]+ 221.0743, found 221.0745.
- 2-Methyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-one (5h). Yield 92%, white solid, mp 97–98 °C; 1H NMR (400 MHz, CDCl3) δ 6.89 (d, J = 1.1 Hz, 1H), 4.08 (t, J = 6.1 Hz, 2H), 2.98 (t, J = 6.7 Hz, 2H), 2.59 (d, J = 1.2 Hz, 3H), 2.06–1.88 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 158.13, 156.89, 156.51, 150.02, 122.80, 119.81, 42.34, 31.84, 22.29, 19.52, 16.90. HRMS (ESI) calcd for C11H12N2OS[M+H]+ 221.0743, found 221.0745.
- 2-Phenyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-one (5i). Yield 88%, light yellow solid, mp 166–167 °C; 1H NMR (400 MHz, CDCl3) δ 7.74–7.66 (m, 2H), 7.48–7.37 (m, 4H), 4.11 (t, J = 6.1 Hz, 2H), 3.01 (t, J = 6.7 Hz, 2H), 2.08–1.90 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 158.35, 156.99, 156.94, 152.60, 133.52, 129.62, 129.37, 126.64, 120.44, 120.09, 42.46, 31.91, 22.29, 19.51. HRMS (ESI) calcd for C16H14N2OS[M+H]+ 283.0900, found 283.0903.
- 2-(4-Chlorophenyl)-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-one (5j). Yield 87%, light yellow solid, mp 190–192 °C; 1H NMR (400 MHz, CDCl3) δ 7.66–7.58 (m, 2H), 7.45–7.35 (m, 3H), 4.11 (t, J = 6.1 Hz, 2H), 3.01 (t, J = 6.7 Hz, 2H), 2.08–1.91 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 158.25, 157.11, 156.95, 151.05, 135.58, 132.03, 129.56, 127.81, 120.62, 120.45, 42.50, 31.90, 22.26, 19.48. HRMS (ESI) calcd for C16H13ClN2OS[M+H]+ 317.0510, found 317.0515.
- 6,7,8,9-Tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-one (5k). Yield 86%, white solid, mp 111–112 °C; 1H NMR (400 MHz, CDCl3) δ 7.73 (d, J = 5.2 Hz, 1H), 7.24 (d, J = 5.3 Hz, 1H), 4.42 (t, J = 4.7 Hz, 2H), 3.08 (t, J = 5.8 Hz, 2H), 1.90–1.76 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 161.51, 158.41, 156.26, 134.36, 125.04, 121.53, 42.85, 37.65, 29.76, 27.98, 25.38. HRMS (ESI) calcd for C11H12N2OS[M+H]+ 221.0743, found 221.0748.
- 3-Methyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimid[1,2-a]azepin-11(5H)-one (5l). Yield 95%, white solid, mp 113 °C; 1H NMR (400 MHz, CDCl3) δ 7.36 (q, J = 1.0 Hz, 1H), 4.41 (t, J = 4.9 Hz, 2H), 3.10 (t, J = 5.8 Hz, 2H), 2.37 (d, J = 1.1 Hz, 3H), 1.91–1.76 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 161.02, 158.66, 155.22, 134.04, 129.35, 121.53, 42.76, 37.65, 29.80, 28.02, 25.53, 13.01. HRMS (ESI) calcd for C12H14N2OS[M+H]+ 235.0900, found 235.0911.
- 2-Methyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimid[1,2-a]azepin-11(5H)-one (5m). Yield 90%, white solid, mp 114–115 °C; 1H NMR (400 MHz, CDCl3) δ 6.92–6.87 (m, 1H), 4.39 (t, J = 4.7 Hz, 2H), 3.05 (t, J = 4.3 Hz, 2H), 2.59 (s, 3H), 1.90–1.75 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 161.45, 157.91, 156.76, 150.07, 123.08, 120.16, 42.77, 37.63, 29.79, 27.99, 25.39, 16.87. HRMS (ESI) calcd for C12H14N2OS[M+H]+ 235.0900, found 235.0909.
- 2-Phenyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimid[1,2-a]azepin-11(5H)-one (5n). Yield 89%, light yellow solid, mp 134–135 °C; 1H NMR (400 MHz, CDCl3) δ 7.69 (dt, J = 8.0, 1.4 Hz, 2H), 7.51–7.35 (m, 4H), 4.42 (t, J = 4.3 Hz, 2H), 3.08 (t, J = 4.3 Hz, 2H), 1.91–1.79 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 161.87, 158.13, 156.88, 152.69, 133.52, 129.60, 129.37, 126.63, 120.82, 120.40, 42.91, 37.71, 29.80, 28.01, 25.40. HRMS (ESI) calcd for C17H16N2OS[M+H]+ 297.1056, found 297.1060.
- 2-(4-Chlorophenyl)-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-one (5o). Yield 94%, light yellow solid, mp 165–166 °C; 1H NMR (400 MHz, CDCl3) δ 7.65–7.57 (m, 2H), 7.45–7.36 (m, 3H), 4.42 (t, J = 4.7 Hz, 2H), 3.08 (t, J = 4.6 Hz, 2H), 1.93–1.80 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 162.04, 158.05, 156.85, 151.16, 135.59, 132.04, 129.58, 127.81, 121.03, 120.76, 42.95, 37.70, 29.79, 27.99, 25.38. HRMS (ESI) calcd for C17H15ClN2OS[M+H]+ 331.0666, found 331.0671.
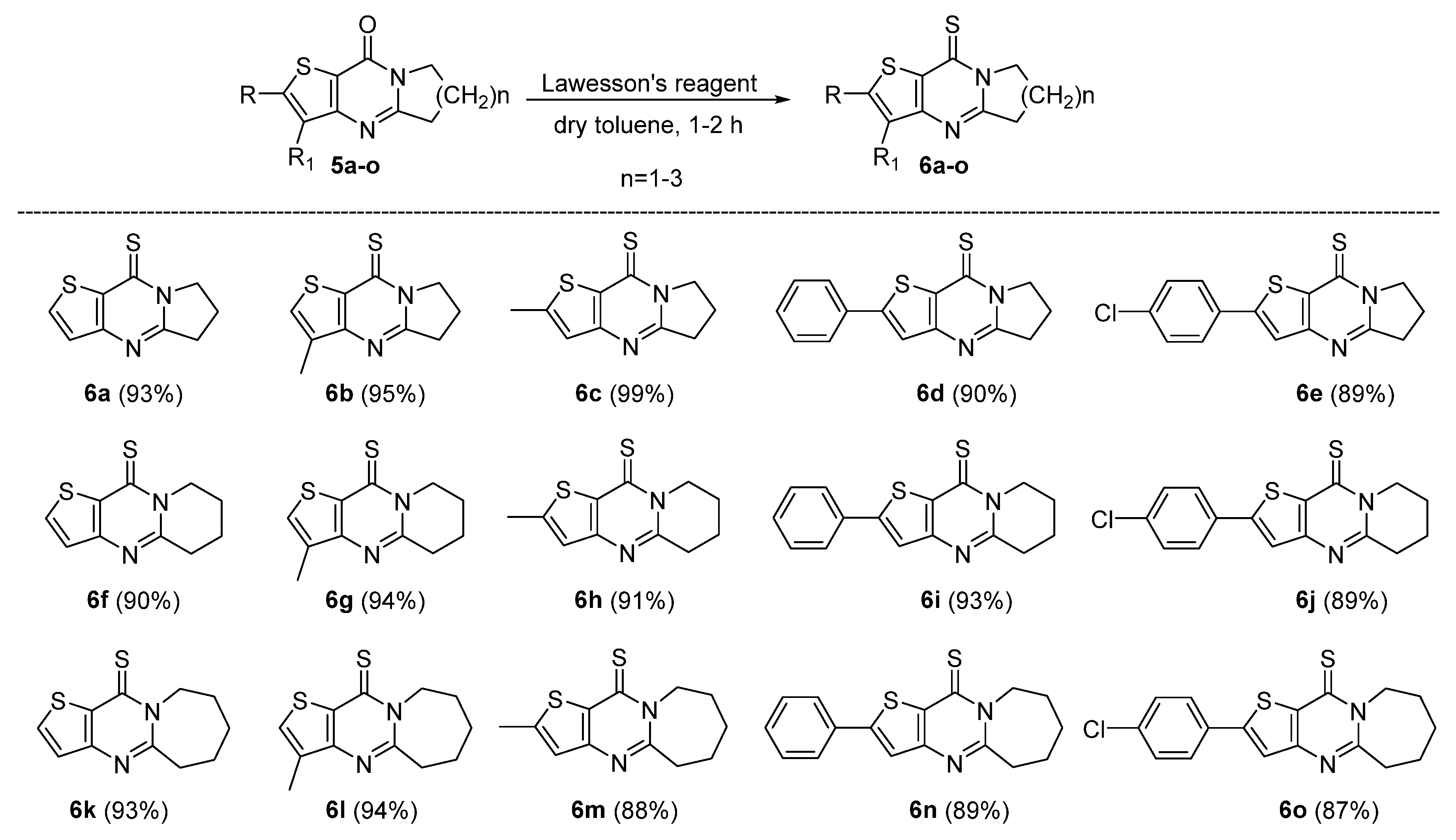
3.1.3. General Procedure for the Synthesis of Compounds 6a–6o
- 6,7-Dihydropyrrolo[1,2-a]thien[3,2-d]pyrimidin-9(5H)-thione (6a). Yield 93%, white solid, mp 148–150 °C; 1H NMR (400 MHz, CDCl3) δ 7.83 (d, J = 5.4 Hz, 1H), 7.28 (d, J = 5.4 Hz, 1H), 4.53 (t, J = 7.4 Hz, 2H), 3.33 (t, J = 8.0 Hz, 2H), 2.39 (p, J = 7.8 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 176.79, 160.21, 151.91, 137.31, 136.34, 125.03, 52.48, 32.96, 19.53. HRMS (ESI) calcd for C9H8N2S2[M+H]+ 209.0202, found 209.0206.
- 3-Methyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-thione (6b). Yield 95%, white solid, mp 166–167 °C; 1H NMR (400 MHz, CDCl3) δ 7.49 (d, J = 1.2 Hz, 1H), 4.55 (t, J = 7.4 Hz, 2H), 3.35 (t, J = 8.0 Hz, 2H), 2.44–2.32 (m, 5H). 13C NMR (100 MHz, CDCl3) δ 176.70, 159.94, 151.12, 136.53, 133.97, 132.65, 52.36, 33.04, 19.64, 13.22. HRMS (ESI) calcd for C10H10N2S2[M+H]+ 223.0358, found 223.0363.
- 2-Methyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-thione (6c). Yield 99%, white solid, mp 251–253 °C; 1H NMR (400 MHz, CDCl3) δ 6.96 (q, J = 1.1 Hz, 1H), 4.53 (t, J = 7.4 Hz, 2H), 3.30 (t, J = 8.0 Hz, 2H), 2.60 (d, J = 1.1 Hz, 3H), 2.42–2.29 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 175.42, 160.17, 153.63, 152.57, 135.39, 122.99, 52.46, 33.04, 19.49, 17.39. HRMS (ESI) calcd for C10H10N2S2[M+H]+ 223.0358, found 223.0361.
- 2-Phenyl-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-thione (6d). Yield 90%, white solid, mp 145–146 °C; 1H NMR (400 MHz, CDCl3) δ 7.75–7.68 (m, 2H), 7.50–7.37 (m, 4H), 4.54 (t, J = 7.4 Hz, 2H), 3.32 (t, J = 8.0 Hz, 2H), 2.44–2.31 (m, 2H).13C NMR (100 MHz, CDCl3) δ 175.81, 160.52, 155.36, 152.61, 135.74, 133.27, 130.03, 129.44, 126.61, 120.07, 52.52, 33.08, 19.48.HRMS (ESI) calcd for C15H12N2S2[M+H]+ 285.0515, found 285.0519
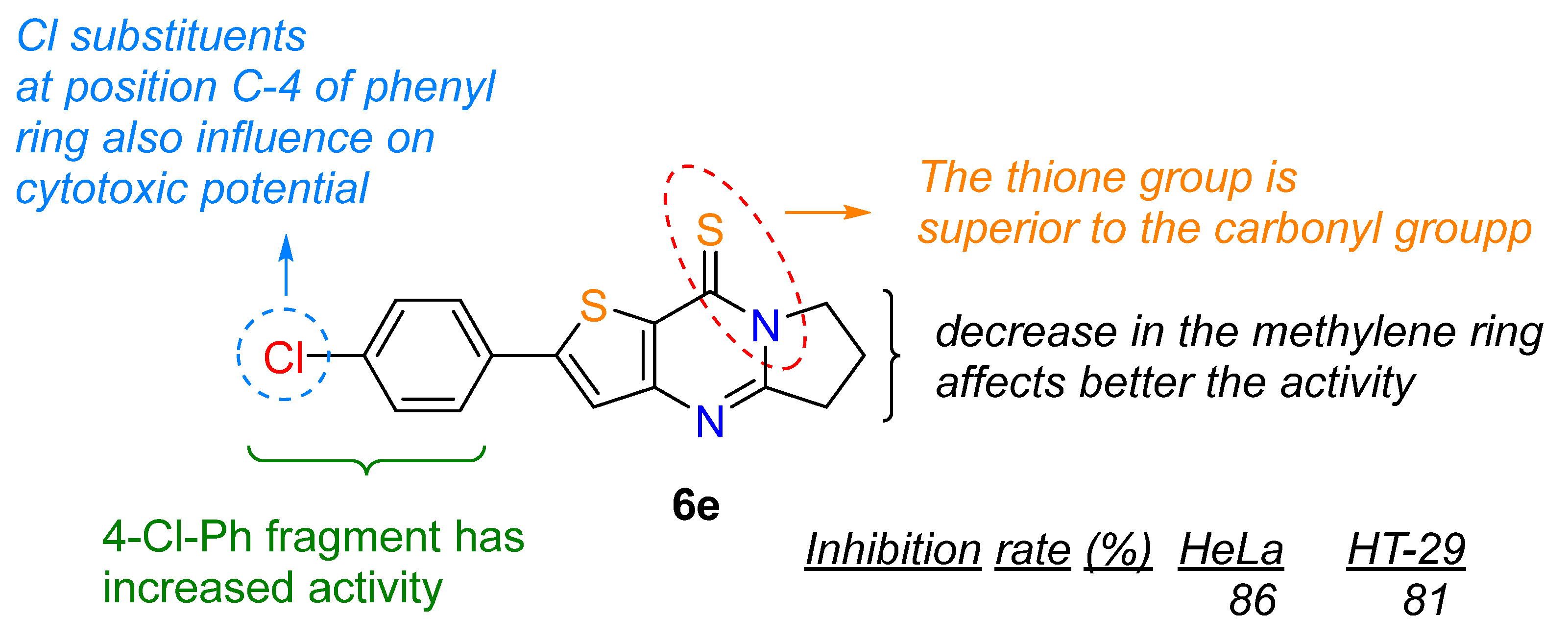
- 2-(4-Chlorophenyl)-6,7-dihydropyrrolo[1,2-a]thieno[3,2-d]pyrimidin-9(5H)-thione (6e). Yield 89%, white solid, mp 161–162 °C; 1H NMR (400 MHz, CDCl3) δ 7.65 (d, J = 8.2 Hz, 2H), 7.48–7.37 (m, 3H), 4.54 (t, J = 7.4 Hz, 2H), 3.33 (t, J = 8.0 Hz, 2H), 2.39 (p, J = 7.8 Hz, 2H). 13C NMR (100 MHz, CDC l3) δ 175.94, 160.67, 153.80, 152.55, 136.06, 135.98, 131.82, 129.70, 127.81, 120.46, 52.54, 33.10, 19.50. HRMS (ESI) calcd for C15H11ClN2S2[M+H]+ 319.0125, found 319.0132.
- 7,8-Dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-thione (6f). Yield 90%, white solid, mp 131–132 °C; 1H NMR (400 MHz, CDCl3) δ 7.83 (d, J = 5.4 Hz, 1H), 7.27 (d, J = 5.4 Hz, 1H), 4.62 (t, J = 6.2 Hz, 2H), 3.11 (t, J = 6.8 Hz, 2H), 2.15–1.94 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 179.62, 156.42, 149.56, 138.21, 137.38, 124.94, 49.16, 32.67, 22.65, 19.17. HRMS (ESI) calcd for C10H10N2S2[M+H]+ 223.0358, found 223.0367.
- 3-Methyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-thione (6g). Yield 94%, white solid, mp 163–164 °C; 1H NMR (400 MHz, CDCl3) δ 7.48 (q, J = 1.1 Hz, 1H), 4.65 (t, J = 6.2 Hz, 2H), 3.13 (t, J = 6.8 Hz, 2H), 2.38 (d, J = 1.1 Hz, 3H), 2.13–2.04 (m, 2H), 2.04–1.95 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 179.44, 156.15, 148.88, 137.47, 133.95, 133.35, 48.88, 32.73, 22.62, 19.19, 13.07. HRMS (ESI) calcd for C11H12N2S2[M+H]+ 237.0515, found 237.0521.
- 2-Methyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-thione (6h). Yield 91%, white solid, mp 240–241 °C; 1H NMR (400 MHz, CDCl3) δ 6.93 (s, 1H), 4.60 (t, J = 6.2 Hz, 2H), 3.08 (t, J = 6.8 Hz, 2H), 2.60 (d, J = 0.9 Hz, 3H), 2.08 (p, J = 6.4 Hz, 2H), 1.98 (p, J = 6.6 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 178.09, 156.49, 154.46, 150.36, 136.54, 122.77, 49.02, 32.63, 22.65, 19.19, 17.33.
- 2-Phenyl-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-thione (6i). Yield 93%, white solid, mp 195–196 °C; 1H NMR (400 MHz, CDCl3) δ 7.77–7.69 (m, 2H), 7.50–7.38 (m, 4H), 4.62 (t, J = 6.2 Hz, 2H), 3.11 (t, J = 6.8 Hz, 2H), 2.10 (p, J = 6.0 Hz, 2H), 2.01 (p, J = 6.7 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 178.47, 156.82, 156.14, 150.32, 136.83, 133.32, 130.04, 129.44, 126.65, 119.87, 49.11, 32.68, 22.67, 19.18. HRMS (ESI) calcd for C16H14N2S2[M+H]+ 299.0671, found 299.0675.
- 2-(4-Chlorophenyl)-7,8-dihydro-5H-pyrido[1,2-a]thieno[3,2-d]pyrimidin-10(6H)-thione (6j). Yield 89%, white solid, mp 216–217 °C; 1H NMR (400 MHz, cdcl3) δ 7.69–7.61 (m, 2H), 7.47–7.39 (m, 3H), 4.61 (t, J = 6.2 Hz, 2H), 3.11 (t, J = 6.8 Hz, 2H), 2.16–2.06 (m, 2H), 2.05–1.95 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 178.58, 156.98, 154.55, 150.21, 137.02, 136.07, 131.85, 129.69, 127.83, 120.25, 49.18, 32.69, 22.67, 19.18. HRMS (ESI) calcd for C16H13ClN2S2[M+H]+ 333.0281, found 333.0285.
- 6,7,8,9-Tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-thione (6k). Yield 93%, white solid, mp 118–119 °C; 1H NMR (400 MHz, CDCl3) δ 7.84 (d, J = 5.4 Hz, 1H), 7.26 (s, 1H), 5.08 (t, J = 5.0 Hz, 2H), 3.23 (t, J = 5.2 Hz, 2H), 1.94–1.87 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 179.86, 160.83, 149.49, 138.78, 137.57, 125.25, 50.29, 38.14, 29.36, 26.63, 25.30. HRMS (ESI) calcd for C11H12N2S2[M+H]+ 237.0515, found 237.0519.
- 3-Methyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-thione (6l). Yield 94%, white solid, mp 137–138 °C; 1H NMR (400 MHz, CDCl3) δ 7.49 (d, J = 1.1 Hz, 1H), 5.09 (t, J = 5.4 Hz, 2H), 3.24 (t, J = 6.2 Hz, 2H), 2.37 (d, J = 1.1 Hz, 3H), 1.92–1.88 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 179.77, 160.47, 148.84, 137.67, 134.34, 133.88, 50.13, 38.15, 29.41, 26.69, 25.45, 13.03. HRMS (ESI) calcd for C12H14N2S2[M+H]+ 251.0671, found 251.06776.
- 2-Methyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-thione (6m). Yield 88%, white solid, mp 255–256 °C; 1H NMR (400 MHz, CDCl3) δ 6.93 (q, J = 1.0 Hz, 1H), 5.06 (t, J = 5.8 Hz, 2H), 3.19 (t, J = 4.6 Hz, 2H), 2.59 (d, J = 1.2 Hz, 3H), 1.91–1.85 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 178.32, 160.90, 155.05, 150.28, 136.68, 123.08, 50.17, 38.12, 29.36, 26.65, 25.27, 17.32. HRMS (ESI) calcd for C12H14N2S2[M+H]+ 251.0671, found 251.0675.
- 2-Phenyl-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-thione (6n). Yield 89%, white solid, mp 202–203 °C; 1H NMR (400 MHz, CDCl3) δ 7.76–7.68 (m, 2H), 7.50–7.38 (m, 4H), 5.07 (t, J = 5.5 Hz, 2H), 3.22 (t, J = 4.7 Hz, 2H), 1.93–1.88 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 178.75, 161.24, 156.72, 150.27, 137.03, 133.32, 130.05, 129.46, 126.64, 120.20, 50.23, 38.16, 29.37, 26.69, 25.30. HRMS (ESI) calcd for C17H16N2S2[M+H]+ 313.0828, found 313.0833.
- 2-(4-Chlorophenyl)-6,7,8,9-tetrahydrothieno[3′,2′:4,5]pyrimido[1,2-a]azepin-11(5H)-thione (6o). Yield 87%, white solid, mp 230–231 °C; 1H NMR (400 MHz, cdcl3) δ 7.68–7.60 (m, 2H), 7.46–7.38 (m, 3H), 5.06 (t, J = 5.8 Hz, 2H), 3.21 (t, J = 6.0 Hz, 2H), 1.93–1.88 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 178.83, 161.39, 155.10, 150.15, 137.21, 136.07, 131.82, 129.69, 127.80, 120.56, 50.26, 38.15, 29.36, 26.67, 25.29. HRMS (ESI) calcd for C17H15ClN2S2[M+H]+ 347.0438, found 347.0443.
3.2. Biology
3.2.1. Cell Lines and Culture Conditions
3.2.2. MTT Assay
3.2.3. Effect of Compound 6e on Morphological Changes in HeLa Cells
3.2.4. Colony Formation Assay
3.2.5. Wound Healing Assay
3.2.6. Transwell Migration Assay
3.2.7. Cell Cycle Analysis by Flow Cytometry
3.2.8. Effect of Compound 6e on Microtubule Network
3.2.9. Analysis of Reactive Oxygen Species (ROS) Levels
3.2.10. Apoptosis Analysis by Flow Cytometry
3.3. Molecular Docking Study of Compound 6e
3.3.1. Preparation of Target Protein Structure
3.3.2. Molecular Docking Analysis
3.3.3. Screening and Analysis of Docking Results
3.3.4. Molecular Dynamics (MD) Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmad, E.; Ali, A.; Nimisha; Sharma, A.K.; Ahmed, F.; Dar, G.M.; Singh, A.M.; Apurva; Kumar, A.; Athar, A.; et al. Molecular approaches in cancer. Clin. Chim. Acta 2022, 537, 60–73. [Google Scholar] [CrossRef]
- Hulvat, M.C. Cancer Incidence and Trends. Surg. Clin. N. Am. 2020, 100, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.P.; Lin, J.J.; Shah, S.C.; Kim, M.K.; Montminy, E.M.; Rustgi, S.D.; Karlitz, J.J.; Itzkowitz, S.H. Geographic Variation in Colorectal Cancer Incidence Among Asian Americans: A Population-Based Analysis 2006–2016. Clin. Gastroenterol. Hepatol. 2023, 21, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, D.; Tatiparti, K.; Gavande, N.S.; Sau, S.; Iyer, A.K. Nanomedicine for overcoming therapeutic and diagnostic challenges associated with pancreatic cancer. Drug Discov. Today 2022, 27, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cashman, J.R. PAWI-2: A novel inhibitor for eradication of cancer. Med. Chem. Res. 2020, 29, 1147–1159. [Google Scholar] [CrossRef]
- Khaled, M.; Belaaloui, G.; Jiang, Z.-Z.; Zhu, X.; Zhang, L.-Y. Deoxypodophyllotoxin, a semi-synthetic compound from Dysosma versipellis, induces selective cell death in human breast cancer cell lines. Med. Chem. Res. 2017, 26, 1241–1258. [Google Scholar] [CrossRef]
- Kumar, R.; Shin, W.S.; Sunwoo, K.; Kim, W.Y.; Koo, S.; Bhuniya, S.; Kim, J.S. Small conjugate-based theranostic agents: An encouraging approach for cancer therapy. Chem. Soc. Rev. 2015, 44, 6670–6683. [Google Scholar] [CrossRef]
- Dickens, E.; Ahmed, S. Principles of cancer treatment by chemotherapy. Surgery 2021, 39, 215–220. [Google Scholar]
- Rong, X.; Liu, C.; Li, X.; Zhu, H.; Wang, K.; Zhu, B. Recent advances in chemotherapy-based organic small molecule theranostic reagents. Coord. Chem. Rev. 2022, 473, 214808. [Google Scholar] [CrossRef]
- Murtazaeva, Z.; Nasrullaev, A.; Buronov, A.; Gaybullaev, S.; Nie, L.; Numonov, S.; Khushnazarov, Z.; Turgunov, D.; Kuryazov, R.; Zhao, J.; et al. Imidazole Hybrids: A Privileged Class of Heterocycles in Medicinal Chemistry with New Insights into Anticancer Activity. Molecules 2025, 30, 2245. [Google Scholar] [CrossRef]
- Schneider, G.; Neidhart, W.; Giller, T.; Schmid, G. “Scaffold-Hopping” by Topological Pharmacophore Search: A Contribution to Virtual Screening. Angew. Chem. Int. Ed. 1999, 38, 2894–2896. [Google Scholar] [CrossRef]
- Hessler, G.; Baringhaus, K.-H. The scaffold hopping potential of pharmacophores. Drug Discov. Today Technol. 2010, 7, e263–e269. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Stumpfe, D.; Bajorath, J. Recent Advances in Scaffold Hopping. J. Med. Chem. 2017, 60, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.-J.; Flohr, A.; Stahl, M. Scaffold hopping. Drug Discov. Today Technol. 2004, 1, 217–224. [Google Scholar] [CrossRef]
- Sun, H.; Tawa, G.; Wallqvist, A. Classification of scaffold-hopping approaches. Drug Discov. Today 2012, 17, 310–324. [Google Scholar] [CrossRef]
- Yang, C.; Xu, C.; Li, Z.; Chen, Y.; Wu, T.; Hong, H.; Lu, M.; Jia, Y.; Yang, Y.; Liu, X.; et al. Bioisosteric replacements of the indole moiety for the development of a potent and selective PI3Kδ inhibitor: Design, synthesis and biological evaluation. Eur. J. Med. Chem. 2021, 223, 113661. [Google Scholar] [CrossRef]
- Zeng, Y.; Nie, L.; Niu, C.; Mamatjan, A.; Bozorov, K.; Zhao, J.; Aisa, H.A. Synthesis and Biological Activities of Dihydrooxazolo [5,4-d]-pyrrolo [1,2-a]pyrimidinones. Chin. J. Org. Chem. 2022, 42, 543–556. [Google Scholar] [CrossRef]
- Vasina, M.; Velecký, J.; Planas-Iglesias, J.; Marques, S.M.; Skarupova, J.; Damborsky, J.; Bednar, D.; Mazurenko, S.; Prokop, Z. Tools for computational design and high-throughput screening of therapeutic enzymes. Adv. Drug Del. Rev. 2022, 183, 114143. [Google Scholar] [CrossRef]
- Moinul, M.; Khatun, S.; Amin, S.A.; Jha, T.; Gayen, S. Recent trends in fragment-based anticancer drug design strategies against different targets: A mini-review. Biochem. Pharmacol. 2022, 206, 115301. [Google Scholar] [CrossRef]
- Yin, L.J.; bin Ahmad Kamar, A.K.D.; Fung, G.T.; Liang, C.T.; Avupati, V.R. Review of anticancer potentials and structure-activity relationships (SAR) of rhodanine derivatives. Biomed. Pharmacother. 2022, 145, 112406. [Google Scholar] [CrossRef]
- Guo, H.; Nie, L.; Bozorov, K.; Aisa, H.A.; Zhao, J. Synthesis and Antitumor Activity of Novel Linear Tricyclic Compounds Derived from Purine. Heterocycles 2022, 104, 1085–1097. [Google Scholar]
- Nasrullaev, A.; Bozorov, K.; Bobakulov, K.; Zhao, J.; Nie, L.F.; Turgunov, K.K.; Elmuradov, B.; Aisa, H.A. Synthesis, characterization, and antimicrobial activity of novel hydrazone-bearing tricyclic quinazolines. Res. Chem. Intermed. 2019, 45, 2287–2300. [Google Scholar] [CrossRef]
- Elmuradov, B.Z.; Bozorov, K.A.; Shakhidoyatov, K.M. Thieno [2,3-d]pyrimidin-4-ones 1. Condensation of 2,3-dimethyl- and 2,3-tri-, 2,3-tetra-, and 2,3-pentamethylene-7,8-dihydro-pyrrolo [1,2-a]thieno [2,3-d]pyriminidin-4(6H)-ones with aromatic aldehydes and furfural. Chem. Heterocycl. Compd. 2011, 46, 1393–1399. [Google Scholar] [CrossRef]
- Liu, F.; Hou, X.; Nie, L.F.; Bozorov, K.; Decker, M.; Huang, G. A Convenient One-pot Synthesis of 2,3-Disubstituted Thieno [2,3-d]pyrimidin-4(3H)-ones from 2H-Thieno [2,3-d][1,3]oxazine-2,4(1H)-diones, Aromatic Aldehydes and Amines. SynOpen 2018, 02, 0207–0212. [Google Scholar] [CrossRef]
- Song, B.; Nie, L.; Bozorov, K.; Kuryazov, R.; Aisa, H.A.; Zhao, J. Parallel synthesis of condensed pyrimidine-thiones and their antitumor activities. Res. Chem. Intermed. 2023, 49, 1327–1348. [Google Scholar] [CrossRef]
- Zeng, Y.; Nie, L.; Liu, L.; Niu, C.; Li, Y.; Bozorov, K.; Zhao, J.; Shen, J.; Aisa, H.A. Design, synthesis, in vitro evaluation of a new pyrrolo [1,2-a]thiazolo [5,4-d]pyrimidinone derivatives as cholinesterase inhibitors against Alzheimer’s disease. J. Heterocycl. Chem. 2022, 59, 1086–1101. [Google Scholar] [CrossRef]
- Ma, F.; Du, H. Novel deoxyvasicinone derivatives as potent multitarget-directed ligands for the treatment of Alzheimer’s disease: Design, synthesis, and biological evaluation. Eur. J. Med. Chem. 2017, 140, 118–127. [Google Scholar] [CrossRef]
- Jaouen, J.; Bailly, C. Alkaloids from Mackinlaya species and synthetic mackinazolinone derivatives: An overview. Biorg. Med. Chem. 2025, 117, 118018. [Google Scholar] [CrossRef]
- Gutti, U.; Komati, J.K.; Kotipalli, A.; Saladi, R.G.V.; Gutti, R.K. Justicia adhatoda induces megakaryocyte differentiation through mitochondrial ROS generation. Phytomedicine 2018, 43, 135–139. [Google Scholar] [CrossRef]
- Jain, M.P.; Koul, S.K.; Dhar, K.L.; Atal, C.K. Novel nor-harmal alkaloid from Adhatoda vasica. Phytochemistry 1980, 19, 1880–1882. [Google Scholar] [CrossRef]
- Song, B.; Nie, L.; Bozorov, K.; Niu, C.; Kuryazov, R.; Akber Aisa, H.; Zhao, J. Furo [2,3-d]pyrimidines as Mackinazolinone/Isaindigotone Analogs: Synthesis, Modification, Antitumor Activity, and Molecular Docking Study. Chem. Biodivers. 2023, 20, e202201059. [Google Scholar] [CrossRef]
- Kuryazov, R.S.; Mukhamedov, N.S.; Dushamov, D.A.; Okmanov, R.Y.; Shakhidoyatov, K.M.; Tashkhodzaev, B. Quinazolines. 3*. synthesis of 6-bromo-8-chloro-sulfonylquinazoline-2,4(1H,3H)-dione and its interaction with nucleophilic reagents. Chem. Heterocycl. Compd. 2010, 46, 585–591. [Google Scholar] [CrossRef]
- Turgunov, D.; Nie, L.; Nasrullaev, A.; Murtazaeva, Z.; Wang, B.; Kholmurodova, D.; Kuryazov, R.; Zhao, J.; Bozorov, K.; Aisa, H.A. Synthesis of Novel 7-Phenyl-2,3-Dihydropyrrolo [2,1-b]Quinazolin-9(1H)-ones as Cholinesterase Inhibitors Targeting Alzheimer’s Disease Through Suzuki–Miyaura Cross-Coupling Reaction. Molecules 2025, 30, 2791. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Murtazaeva, Z.; Nie, L.; Kuryazov, R.; Gaybullaev, S.; Niu, C.; Bozorov, K.; Aisa, H.A.; Zhao, J. Pyrrolopyrimidines: Design, Synthesis and Antitumor Properties of Novel Tricyclic Pyrrolo [2,3-d]pyrimidine Derivatives. Molecules 2025, 30, 2917. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Nie, L.; Tang, D.; Bozorov, K.; Zhao, J.; Aisa, H.A. Synthesis of tricyclic pyrazolopyrimidine arylidene ester derivatives and their cytotoxic and molecular docking evaluations. J. Heterocycl. Chem. 2024, 61, 651–668. [Google Scholar] [CrossRef]
- Ruzi, Z.; Bozorov, K.; Nie, L.; Zhao, J.; Akber Aisa, H. Discovery of novel (E)-1-methyl-9-(3-methylbenzylidene)-6,7,8,9-tetrahydropyrazolo [3,4-d]pyrido [1,2-a]pyrimidin-4(1H)-one as DDR2 kinase inhibitor: Synthesis, molecular docking, and anticancer properties. Bioorg. Chem. 2023, 135, 106506. [Google Scholar] [CrossRef]
- Zeng, Y.; Nie, L.; Liu, L.; Bozorov, K.; Zhao, J. Design, synthesis, biological evaluation of a new tricyclicthiazolopy-rimidinone derivatives as acetylcholinesterase inhibitors. J. Heterocycl. Chem. 2024, 61, 1542–1553. [Google Scholar] [CrossRef]
- Hu, H.; Dong, Y.; Li, M.; Wang, R.; Zhang, X.; Gong, P.; Zhao, Y. Design, synthesis and biological evaluation of novel thieno [3,2-d]pyrimidine and quinazoline derivatives as potent antitumor agents. Bioorg. Chem. 2019, 90, 103086. [Google Scholar] [CrossRef]
- Romagnoli, R.; Prencipe, F.; Oliva, P.; Baraldi, S.; Baraldi, P.G.; Schiaffino Ortega, S.; Chayah, M.; Kimatrai Salvador, M.; Lopez-Cara, L.C.; Brancale, A.; et al. Design, Synthesis, and Biological Evaluation of 6-Substituted Thieno [3,2-d]pyrimidine Analogues as Dual Epidermal Growth Factor Receptor Kinase and Microtubule Inhibitors. J. Med. Chem. 2019, 62, 1274–1290. [Google Scholar] [CrossRef]
- Toolabi, M.; Moghimi, S.; Bakhshaiesh, T.O.; Salarinejad, S.; Aghcheli, A.; Hasanvand, Z.; Nazeri, E.; Khalaj, A.; Esmaeili, R.; Foroumadi, A. 6-Cinnamoyl-4-arylaminothienopyrimidines as highly potent cytotoxic agents: Design, synthesis and structure-activity relationship studies. Eur. J. Med. Chem. 2020, 185, 111786. [Google Scholar] [CrossRef]
- Aly, H.M.; Saleh, N.M.; Elhady, H.A. Design and synthesis of some new thiophene, thienopyrimidine and thienothiadiazine derivatives of antipyrine as potential antimicrobial agents. Eur. J. Med. Chem. 2011, 46, 4566–4572. [Google Scholar] [CrossRef] [PubMed]
- Makker, V.; Recio, F.O.; Ma, L.; Matulonis, U.A.; Lauchle, J.O.; Parmar, H.; Gilbert, H.N.; Ware, J.A.; Zhu, R.; Lu, S.; et al. A multicenter, single-arm, open-label, phase 2 study of apitolisib (GDC-0980) for the treatment of recurrent or persistent endometrial carcinoma (MAGGIE study). Cancer 2016, 122, 3519–3528. [Google Scholar] [CrossRef] [PubMed]
- Rizk, O.H.; Shaaban, O.G.; El-Ashmawy, I.M. Design, synthesis and biological evaluation of some novel thienopyrimidines and fused thienopyrimidines as anti-inflammatory agents. Eur. J. Med. Chem. 2012, 55, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Han, Y.; Wang, R.; Yan, P.; Chen, S.; Hou, Y.; Zhao, Y. Design, synthesis and biological evaluation of novel 2,4-bismorpholinothieno [3,2-d]pyrimidine and 2-morpholinothieno [3,2-d]pyrimidinone derivatives as potent antitumor agents. Bioorg. Chem. 2020, 99, 103796. [Google Scholar] [CrossRef]
- Yang, X.; Deng, M.; Zhang, X.; Wang, Y.; Song, K.; Cong, R.; Meng, L.; Zhang, J. Design, synthesis, and biological evaluation of thieno [3,2-d]pyrimidine derivatives as potential simplified phosphatidylinositol 3-kinase alpha inhibitors. Chem. Biol. Drug Des. 2019, 94, 2013–2022. [Google Scholar] [CrossRef]
- Franco, L.C.; Morales, F.; Boffo, S.; Giordano, A. CDK9: A key player in cancer and other diseases. J. Cell. Biochem. 2018, 119, 1273–1284. [Google Scholar] [CrossRef]
- Gong, Y.; Li, H. CDK7 in breast cancer: Mechanisms of action and therapeutic potential. Cell Commun. Signal. 2024, 22, 226. [Google Scholar] [CrossRef]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Anshabo, A.T.; Milne, R.; Wang, S.; Albrecht, H. CDK9: A Comprehensive Review of Its Biology, and Its Role as a Potential Target for Anti-Cancer Agents. Front. Oncol. 2021, 11, 678559. [Google Scholar] [CrossRef]
- Santo, L.; Siu, K.T.; Raje, N. Targeting Cyclin-Dependent Kinases and Cell Cycle Progression in Human Cancers. Semin. Oncol. 2015, 42, 788–800. [Google Scholar] [CrossRef]
- Muste Sadurni, M.; Saponaro, M. Deregulations of RNA Pol II Subunits in Cancer. Appl. Biosci. 2023, 2, 459–476. [Google Scholar] [CrossRef]
- Zhu, Y.; Alqahtani, S.; Hu, X. Aromatic Rings as Molecular Determinants for the Molecular Recognition of Protein Kinase Inhibitors. Molecules 2021, 26, 1776. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Hawro, I.; Derewenda, U. C–H⋯O hydrogen bonds in kinase-inhibitor interfaces. IUBMB Life 2020, 72, 1233–1242. [Google Scholar] [CrossRef]
- Johnson, L.N. Protein kinase inhibitors: Contributions from structure to clinical compounds. Q. Rev. Biophys. 2009, 42, 1–40. [Google Scholar] [CrossRef] [PubMed]
- El-Meligie, S.E.M.; Khalil, N.A.; El-Nassan, H.B.; Ibraheem, A.A.M. New synthetic approaches to thieno [3,2-d]pyrimidine and thieno [3,4-b]pyridine derivatives. Chem. Pap. 2020, 74, 2501–2514. [Google Scholar] [CrossRef]
- Perspicace, E.; Jouan-Hureaux, V.; Ragno, R.; Ballante, F.; Sartini, S.; La Motta, C.; Da Settimo, F.; Chen, B.; Kirsch, G.; Schneider, S.; et al. Design, synthesis and biological evaluation of new classes of thieno [3,2-d]pyrimidinone and thieno [1,2,3]triazine as inhibitor of vascular endothelial growth factor receptor-2 (VEGFR-2). Eur. J. Med. Chem. 2013, 63, 765–781. [Google Scholar] [CrossRef]
- Kandeel, M.; Abdelhameid, M.K.; Eman, K.; Labib, M.B. Synthesis of Some Novel Thieno [3,2-d]pyrimidines as Potential Cytotoxic Small Molecules against Breast Cancer. Chem. Pharm. Bull. 2013, 61, 637–647. [Google Scholar] [CrossRef]
- Das, A.; Ashraf, M.W.; Banik, B.K. Thione Derivatives as Medicinally Important Compounds. ChemistrySelect 2021, 6, 9069–9100. [Google Scholar] [CrossRef]
- Akrivos, P.D. Recent studies in the coordination chemistry of heterocyclic thiones and thionates. Coord. Chem. Rev. 2001, 213, 181–210. [Google Scholar] [CrossRef]
- Supino, R. MTT Assays. In In Vitro Toxicity Testing Protocols; O’Hare, S., Atterwill, C.K., Eds.; Humana Press: Totowa, NJ, USA, 1995; pp. 137–149. [Google Scholar]
- Eskiocak, U.; İşeri, O.D.; Kars, M.D.; Biçer, A.; Gunduz, U. Effect of Doxorubicin on Telomerase Activity and Apoptotic Gene Expression in Doxorubicin-Resistant and -Sensitive MCF-7 Cells: An Experimental Study. Chemotherapy 2008, 54, 209–216. [Google Scholar] [CrossRef]
- Bortner, C.D.; Cidlowski, J.A. Cell shrinkage and monovalent cation fluxes: Role in apoptosis. Arch. Biochem. Biophys. 2007, 462, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Dini, L.; Coppola, S.; Ruzittu, M.T.; Ghibelli, L. Multiple Pathways for Apoptotic Nuclear Fragmentation. Exp. Cell Res. 1996, 223, 340–347. [Google Scholar] [CrossRef]
- KASS, G.E.N.; ERIKSSON, J.E.; WEIS, M.; ORRENIUS, S.; CHOW, S.C. Chromatin condensation during apoptosis requires ATP. Biochem. J. 1996, 318, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Rello, S.; Stockert, J.C.; Moreno, V.; Gámez, A.; Pacheco, M.; Juarranz, A.; Cañete, M.; Villanueva, A. Morphological criteria to distinguish cell death induced by apoptotic and necrotic treatments. Apoptosis 2005, 10, 201–208. [Google Scholar] [CrossRef]
- Majidpoor, J.; Mortezaee, K. Steps in metastasis: An updated review. Med. Oncol. 2021, 38, 3. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Zervantonakis, I.K.; Kamm, R.D. Tumor cell migration in complex microenvironments. Cell. Mol. Life Sci. 2013, 70, 1335–1356. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, H.; Duan, Y.; Tang, Q.; Huang, H.; Bi, F. Survival mechanisms of circulating tumor cells and their implications for cancer treatment. Cancer Metastasis Rev. 2024, 43, 941–957. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, J.; Ren, Y.; Liu, S.; Ba, Y.; Zuo, A.; Luo, P.; Cheng, Q.; Xu, H.; Han, X. Multi-stage mechanisms of tumor metastasis and therapeutic strategies. Signal Transduct. Target. Ther. 2024, 9, 270. [Google Scholar] [CrossRef]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, 92, e51998. [Google Scholar]
- Rashid, S. Hallmarks of Cancer Cell. In Cancer and Chemoprevention: An Overview; Rashid, S., Ed.; Springer: Singapore, 2017; pp. 3–13. [Google Scholar]
- Stewart, Z.A.; Westfall, M.D.; Pietenpol, J.A. Cell-cycle dysregulation and anticancer therapy. Trends Pharmacol. Sci. 2003, 24, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Patra, D.; Bhavya, K.; Ramprasad, P.; Kalia, M.; Pal, D. Chapter Eleven—Anti-cancer drug molecules targeting cancer cell cycle and proliferation. In Anti-Cancer Drug Molecules Targeting Cancer Cell Cycle and Proliferation; Advances in Protein Chemistry and Structural, Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2023; Volume 135, pp. 343–395. [Google Scholar]
- Skladanowski, A.; Bozko, P.; Sabisz, M. DNA Structure and Integrity Checkpoints during the Cell Cycle and Their Role in Drug Targeting and Sensitivity of Tumor Cells to Anticancer Treatment. Chem. Rev. 2009, 109, 2951–2973. [Google Scholar] [CrossRef] [PubMed]
- Binarová, P.; Tuszynski, J. Tubulin: Structure, Functions and Roles in Disease. Cells 2019, 8, 1294. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Peng, Y.; Tao, X.; Ding, X.; Li, R.; Jiang, Y.; Zuo, W. Microtubule Organization Is Essential for Maintaining Cellular Morphology and Function. Oxid. Med. Cell. Longev. 2022, 2022, 1623181. [Google Scholar] [CrossRef]
- Simon, H.U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef]
- Waris, G.; Ahsan, H. Reactive oxygen species: Role in the development of cancer and various chronic conditions. J. Carcinog. 2006, 5, 14. [Google Scholar] [CrossRef]
- Eruslanov, E.; Kusmartsev, S. Identification of ROS Using Oxidized DCFDA and Flow-Cytometry. In Advanced Protocols in Oxidative Stress II; Armstrong, D., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 57–72. [Google Scholar]
- Darzynkiewicz, Z.; Bedner, E.; Smolewski, P. Flow cytometry in analysis of cell cycle and apoptosis. Semin. Hematol. 2001, 38, 179–193. [Google Scholar] [CrossRef]
- Wlodkowic, D.; Skommer, J.; Darzynkiewicz, Z. Flow Cytometry-Based Apoptosis Detection. In Apoptosis: Methods and Protocols, 2nd ed.; Erhardt, P., Toth, A., Eds.; Humana Press: Totowa, NJ, USA, 2009; pp. 19–32. [Google Scholar]
- Teo, T.; Kasirzadeh, S.; Albrecht, H.; Sykes, M.J.; Yang, Y.; Wang, S. An overview of CDK3 in cancer: Clinical significance and pharmacological implications. Pharmacol. Res. 2022, 180, 106249. [Google Scholar] [CrossRef]
- Łukasik, P.; Załuski, M.; Gutowska, I. Cyclin-Dependent Kinases (CDK) and Their Role in Diseases Development–Review. Int. J. Mol. Sci. 2021, 22, 2935. [Google Scholar] [CrossRef]
- Roskoski, R. Cyclin-dependent protein kinase inhibitors including palbociclib as anticancer drugs. Pharmacol. Res. 2016, 107, 249–275. [Google Scholar] [CrossRef]
- Peyressatre, M.; Prével, C.; Pellerano, M.; Morris, M.C. Targeting Cyclin-Dependent Kinases in Human Cancers: From Small Molecules to Peptide Inhibitors. Cancers 2015, 7, 179–237. [Google Scholar] [CrossRef]
- Okada, M. Regulation of the SRC family kinases by Csk. Int. J. Biol. Sci. 2012, 8, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, H.; Ranjan, K.; Zhang, D. Regulation, targets and functions of CSK. Front. Cell Dev. Biol. 2023, 11, 1206539. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, H.; Leyton, L. CSK-mediated signalling by integrins in cancer. Front. Cell Dev. Biol. 2023, 11, 1214787. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Arya, G.; Kumari, R.M.; Gupta, N.; Nimesh, S. Evaluation of anticancer activity of silver nanoparticles on the A549 human lung carcinoma cell lines through alamar blue assay. Bio-Protoc. 2019, 9, e3131. [Google Scholar] [CrossRef]
- Du, F.; Zhao, X.; Fan, D. Soft Agar Colony Formation Assay as a Hallmark of Carcinogenesis. Bio Protoc. 2017, 7, e2351. [Google Scholar] [CrossRef]
- Chen, Y. Scratch wound healing assay. Bio-Protoc. 2012, 2, e100. [Google Scholar] [CrossRef]
- Schneider, M.A. In vitro Tumor Cell Migration Assay Using ThinCertsTM (Transwells). Bio-Protoc. 2016, 6, e1830. [Google Scholar] [CrossRef]
- Zhu, Y.; Tan, W.; Lee, W.L. An in vitro Microscopy-based Assay for Microtubule-binding and Microtubule-crosslinking by Budding Yeast Microtubule-associated Protein. Bio. Protoc. 2018, 8, e3110. [Google Scholar] [CrossRef]
- Chen, X.; Liu, H.; Xie, W.; Yang, Y.; Wang, Y.; Fan, Y.; Hua, Y.; Zhu, L.; Zhao, J.; Lu, T.; et al. Investigation of Crystal Structures in Structure-Based Virtual Screening for Protein Kinase Inhibitors. J. Chem. Inf. Model. 2019, 59, 5244–5262. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
 | ||||
|---|---|---|---|---|
| Entry | Substrate | Cyclization conditions | Product | Yield (%) |
| 1 | 3a | POCl3, DCM, 45 °C, 5 h | 5a | 42 |
| 2 | 3a | POCl3, DCE, 80 °C, 2 h | 5a | 79 |
| 3 | 3a | POCl3, Dioxane, 100 °C, 8 h | 5a | 77 |
| 4 | 3a | POCl3, Toluene, 120 °C, 20 h | 5a | 28 |
| 5 | 3a | POCl3, solvent-free, 100 °C, 3 h | 5a | 68 |
| 6 | 3a | POCl3, solvent-free, 140 °C, 2 h | 5a | 81 |
| Compound | Structure | Target | Pdbid | Cdocker Energy (kcal/mol) | Combination Type |
|---|---|---|---|---|---|
| 6e | 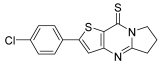 | CDK2 | 2C5Y | −9.4959 | Hydrogen bond, Hydrophobic interaction, Electrostatic interaction |
| CDK3 | 3GC0 | 0.114814 | Hydrogen bond, Hydrophobic interaction, Electrostatic interaction, π-interaction | ||
| CDK5-p25 | 1UNG | −5.79453 | Hydrogen bond, Hydrophobic interaction, Electrostatic interaction, π-interaction | ||
| CDK5-p35 | 1UNH | −8.30506 | Hydrogen bond, Hydrophobic interaction, Electrostatic interaction, π-interaction | ||
| CK1y1 | 6F1W | −7.23817 | Hydrogen bond, Hydrophobic interaction, Vander Waals forces, Electrostatic interaction | ||
| CSK | 1BYG | −2.28896 | Hydrogen bond, Hydrophobic interaction, Electrostatic interaction, π-interaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruzi, Z.; Buronov, A.; Nie, L.; Nasrullaev, A.; Murtazaeva, Z.; Kuryazov, R.; Zhao, J.; Efferth, T.; Aisa, H.A.; Bozorov, K. Scaffold-Hopping Design and Synthesis of Thieno[3,2-d]pyrimidines: Anticancer Activity, Apoptosis Induction, and In Silico Inhibition of CDKs. Int. J. Mol. Sci. 2025, 26, 8528. https://doi.org/10.3390/ijms26178528
Ruzi Z, Buronov A, Nie L, Nasrullaev A, Murtazaeva Z, Kuryazov R, Zhao J, Efferth T, Aisa HA, Bozorov K. Scaffold-Hopping Design and Synthesis of Thieno[3,2-d]pyrimidines: Anticancer Activity, Apoptosis Induction, and In Silico Inhibition of CDKs. International Journal of Molecular Sciences. 2025; 26(17):8528. https://doi.org/10.3390/ijms26178528
Chicago/Turabian StyleRuzi, Zukela, Anvarjon Buronov, Lifei Nie, Azizbek Nasrullaev, Zarifa Murtazaeva, Rustamkhon Kuryazov, Jiangyu Zhao, Thomas Efferth, Haji Akber Aisa, and Khurshed Bozorov. 2025. "Scaffold-Hopping Design and Synthesis of Thieno[3,2-d]pyrimidines: Anticancer Activity, Apoptosis Induction, and In Silico Inhibition of CDKs" International Journal of Molecular Sciences 26, no. 17: 8528. https://doi.org/10.3390/ijms26178528
APA StyleRuzi, Z., Buronov, A., Nie, L., Nasrullaev, A., Murtazaeva, Z., Kuryazov, R., Zhao, J., Efferth, T., Aisa, H. A., & Bozorov, K. (2025). Scaffold-Hopping Design and Synthesis of Thieno[3,2-d]pyrimidines: Anticancer Activity, Apoptosis Induction, and In Silico Inhibition of CDKs. International Journal of Molecular Sciences, 26(17), 8528. https://doi.org/10.3390/ijms26178528












