Lentiviral Vectors: From Wild-Type Viruses to Efficient Multi-Functional Delivery Vectors
Abstract
1. Introduction
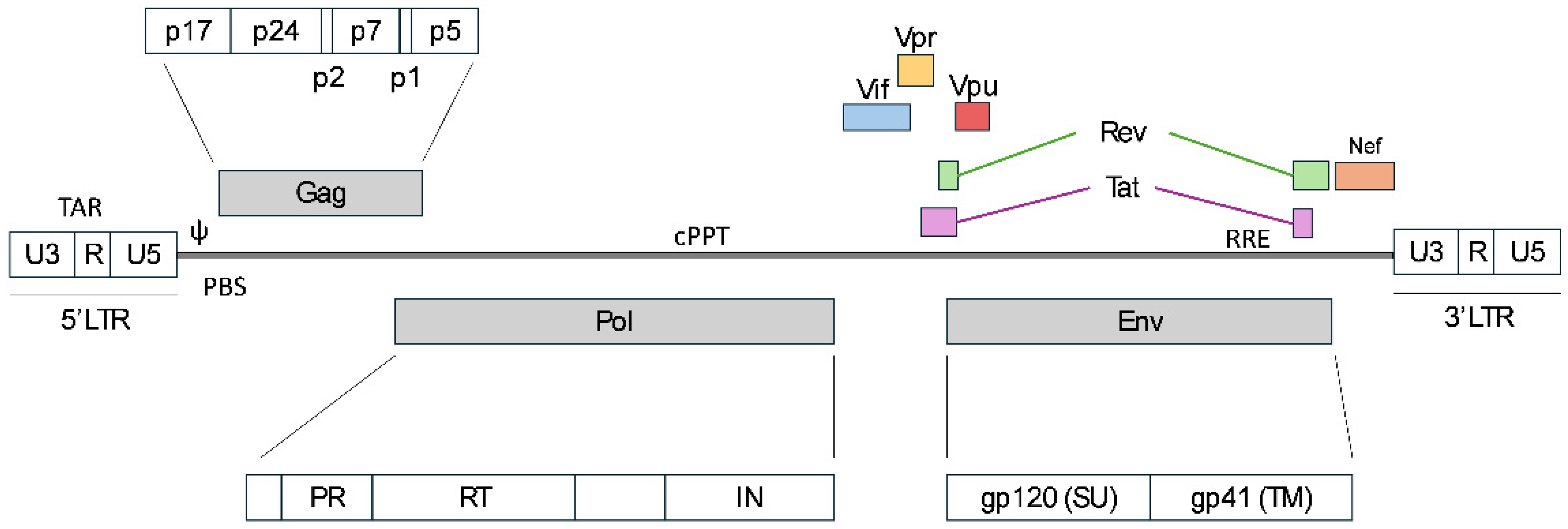
1.1. Overview of Lentiviruses Origin
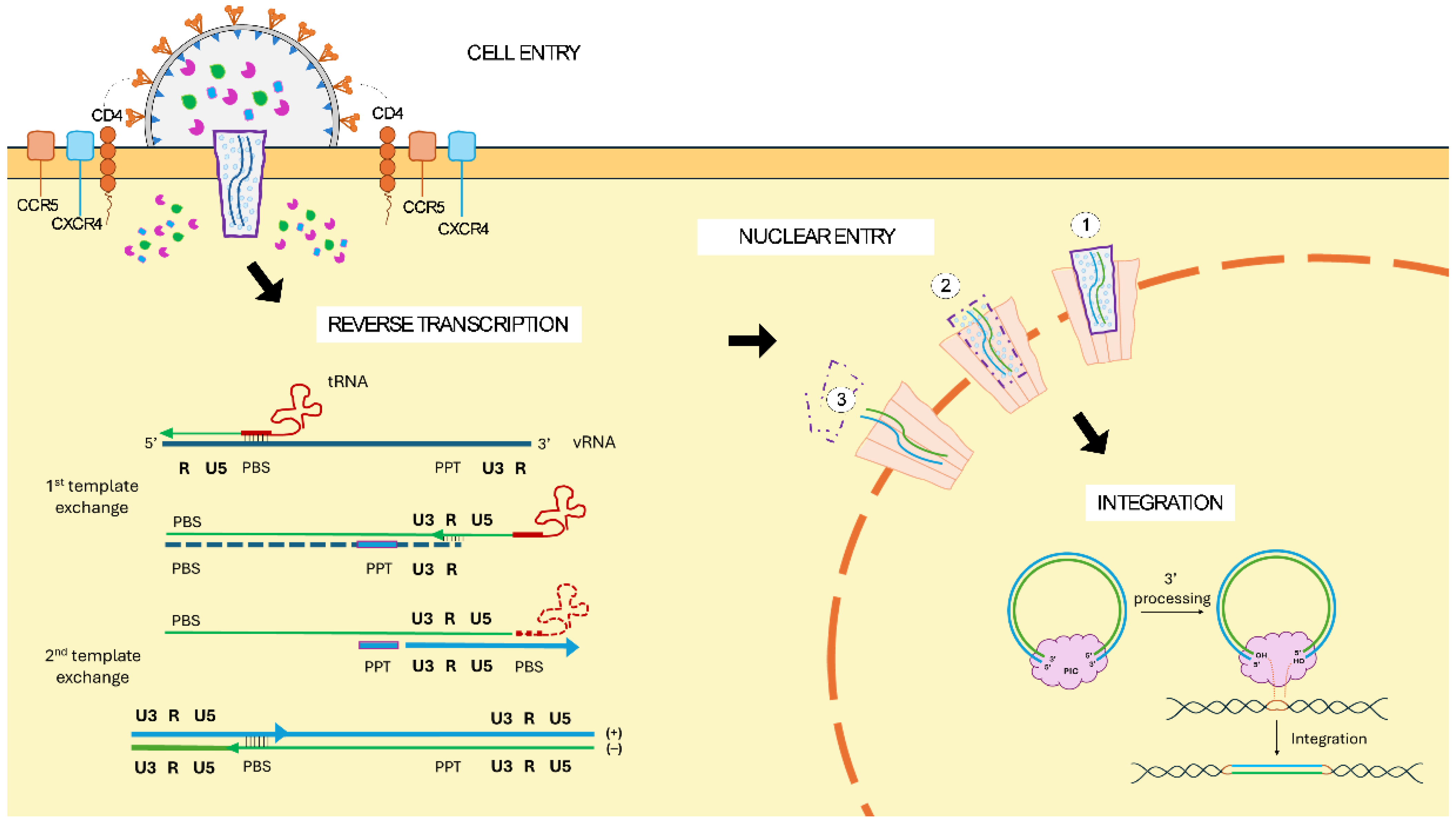
1.2. General Virological Properties of Lentiviruses
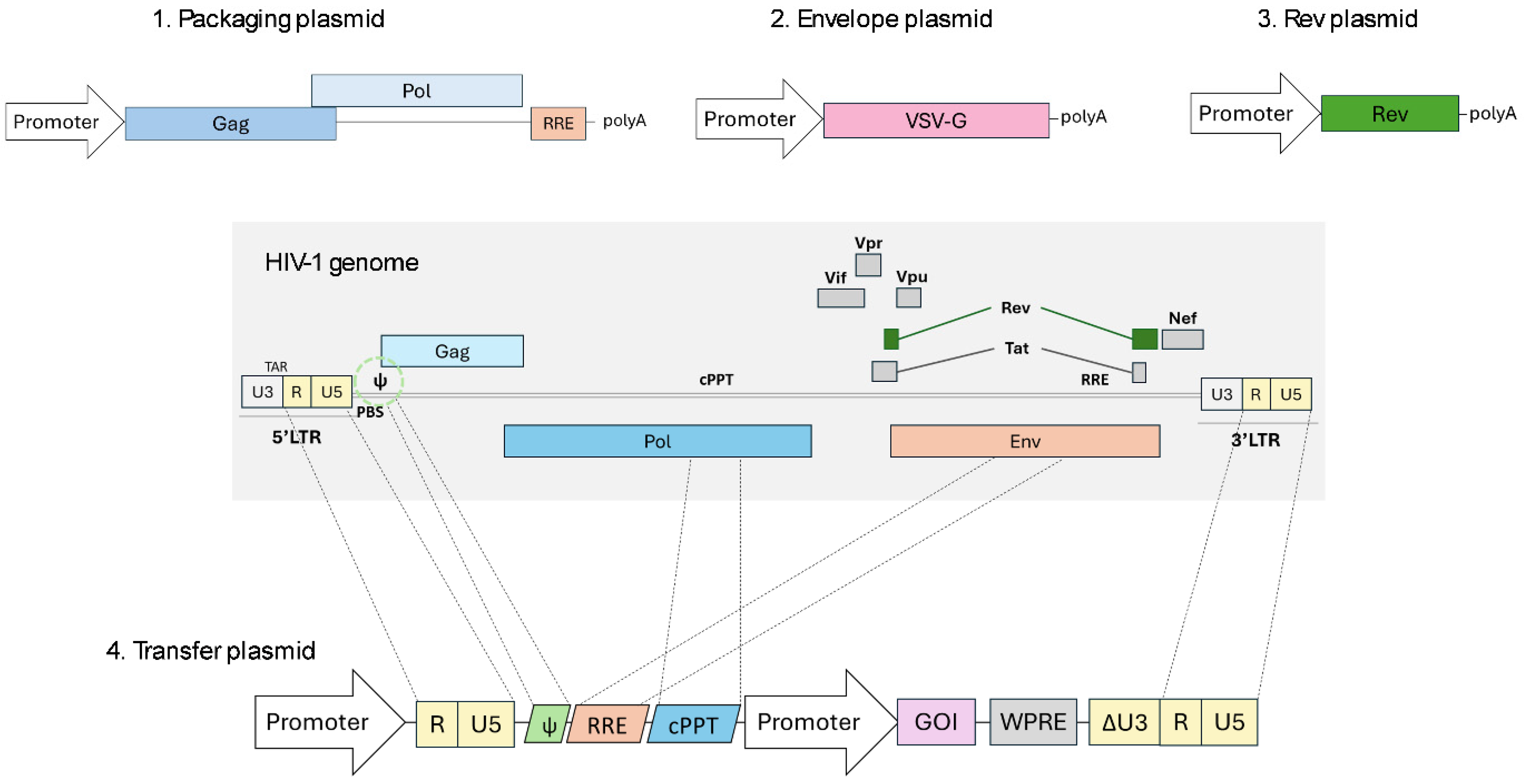
1.3. The Evolution from HIV-1 to 3rd Generation LVV
1.4. Current Situation of Lentiviral Vector in Gene Therapy and Research
2. State of the Art of Manufacturing
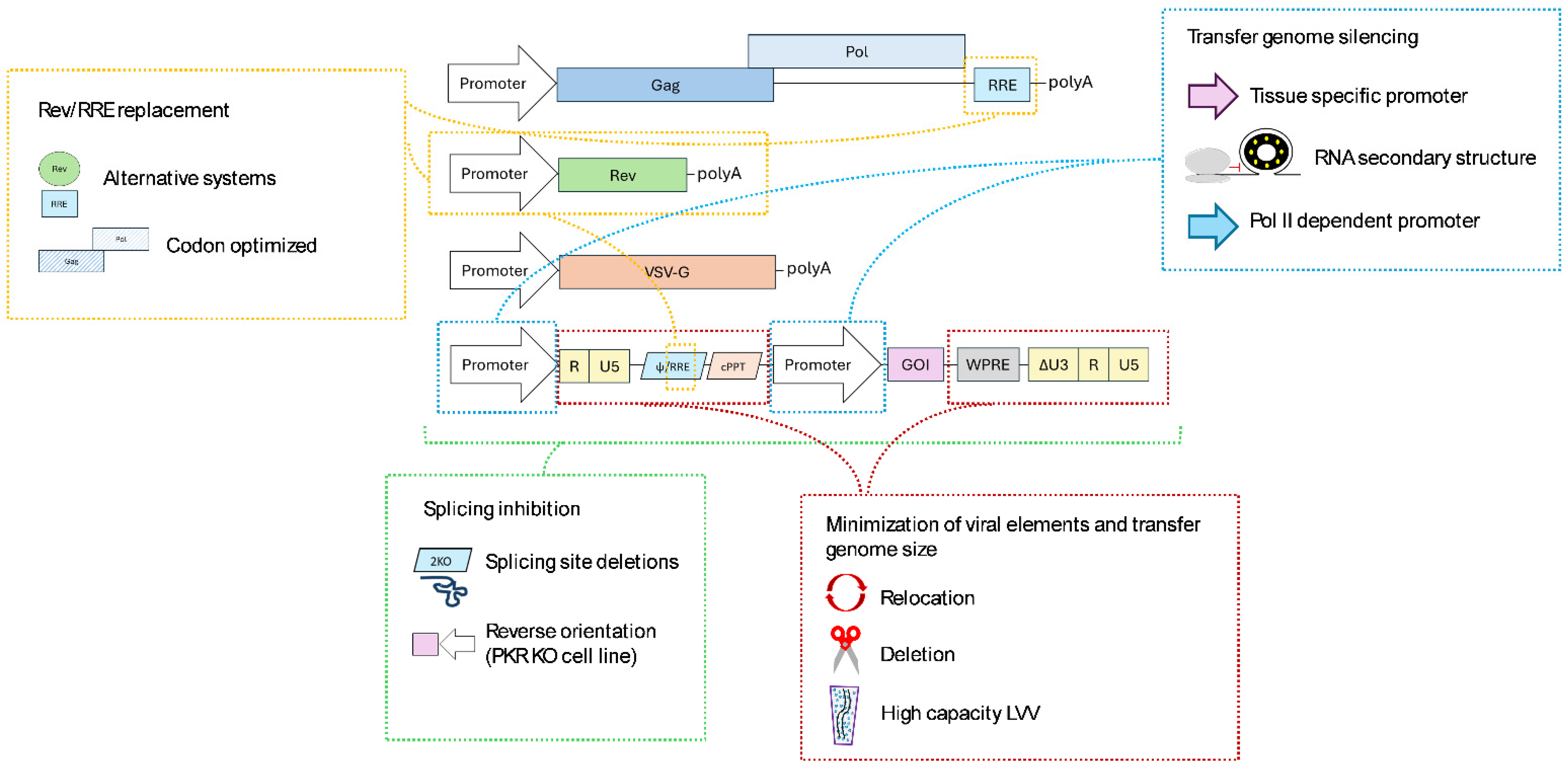
2.1. Optimizations in Lentiviral Constructs
2.1.1. Rev/RRE System Replacement
2.1.2. Minimization of Viral Elements and Transfer Genome Size
2.1.3. Transfer Genome Silencing
2.1.4. Splicing Escape
2.2. Optimizations in the Producer Cell Line
2.2.1. Retro-Transduction
2.2.2. Engineering of the Producer Cell Line
2.2.3. Packaging/Stable Producer Cell Lines
3. New Opportunities for LVV
3.1. LVV Pseudotyping for In Vivo Administration
3.2. Integrase Deficient Lentiviral Vectors (IDLVs) and Their Application in Vaccinology
3.3. Reverse Transcriptase Deficient LVV for RNA Delivery
3.4. Lentivirus Derived Nanoparticles (LVNPs) for Gene Editing
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AAV | Adeno-Associated Virus |
| ABE | Adenosine Base Editor |
| ALDP | Adenoleukodistrophy Protein |
| APC | Antigen-Presenting Cells |
| APOBEC | Apolipoprotein B MRNA Editing Enzyme |
| ASCT | Alanine, Serine, Cysteine Transporter |
| ATMP | Advanced Therapy Medicinal Product |
| Att | DNA Attachment Site |
| B2M | Β-2 Microglobulin |
| BaEv | Baboon Endogenous Virus |
| BPIFC | Bpi Fold-Containing Family C |
| CA | Capsid |
| CALD | Cerebral Adrenoleukodystrophy |
| CAR | Chimeric Antigen Receptor |
| CBE | Cytosine Base Editor |
| CDMO | Contract Development And Manufacturing Organization |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CTE | Constitutive Transport Element |
| DARPins | Designed Ankyrin Repeat Proteins |
| DC | Dendritic Cells |
| DDR | DNA Damage Responses |
| DIS | Dimerization Initiation Site |
| DMV | Dolphin Morbillivirus |
| DSB | Double-Strand Break |
| DSP | Downstream Processing |
| GaLV | Gibbon Ape Leukemia Virus |
| GBP3 | Guanylate-Binding Protein 3 |
| GMP | Good Manufacturing Practices |
| GOI | Gene Of Interest |
| HDR | Homology-Directed Repair |
| HIV-1 | Human Immunodeficiency Virus Type 1 |
| HPV | Human Papillomavirus |
| HSC | Hematopoietic Stem Cell |
| HSPC | Hematopoietic Stem And Progenitor Cell |
| IDLV | Integrase Deficient Lentiviral Vector |
| IN | Integrase |
| IRES | Internal Ribosome Entry Site |
| ITR | Inverted Terminal Repeat |
| LDAH | Lipid Droplet-Associated Hydrolase |
| LDLR | Low-Density Lipoprotein Receptor |
| LNP | Lipid Nanoparticle |
| LTR | Long Terminal Repeats |
| LVNP | Lentivirus-Derived Nanoparticles |
| LVV | Lentiviral Vectors |
| MA | Matrix Protein |
| MHC-1 | Major Histocompatibility Complexes Class 1 |
| MLD | Metachromatic Leukodystrophy |
| MPMV | Mason-Pfizer Monkey Virus |
| MSD | Major Splicing Donor |
| MV-H | Measles Virus Hemagglutinin Protein |
| NC | Nucleocapsid |
| Nef | Negative Factor |
| NHEJ | Non-Homologous End Joining |
| NHLRC1 | NHL Repeat-Containing E3 Ubiquitin Protein Ligase 1 |
| NPC | Nuclear Pore Complexes |
| OAS1 | 2′-5′-Oligoadenylate Synthetase 1 |
| ORF | Open Reading Frame |
| ORI | Origin Of Replication |
| PBS | Primer Binding Site |
| PH | Pleckstrin Homology |
| PIC | Preintegration Complex |
| PKR | Protein Kinase RNA-Activated |
| PPT | Polypurine Tract |
| PR | Protease |
| RCL | Replication Competent Lentivirus |
| RD114 | Feline Endogenous Retrovirus |
| Rev | Regulator Of Expression Of Virion Proteins |
| RF | Restriction Factor |
| RRE | Rev Responsive Element |
| RT | Reverse Transcriptase |
| RTE | RNA Transport Element |
| RV | Rabies Virus |
| S/MAR | Scaffold/Matrix Associated Region |
| SA | Splicing Acceptor |
| scFv | Single-Chain Variable Fragments |
| SD | Splicing Donor |
| SIN-LVV | Self-Inactivating Lentiviral Vectors |
| SPM | Secondary Primary Malignancies |
| SRV-1 | Simian Retrovirus Type 1 |
| SU | Surface Glycoprotein |
| TAA | Tumor-Associated Antigens |
| TALEN | Transcription Activator-Like Effector Nucleases |
| TAR | Transactivation-Reponse Element |
| Tat | Trans-Activator Of Transcription |
| TM | Transmembrane Glycoprotein |
| TRAP | Tryptophan RNA-Binding Attenuation Protein |
| USP | Upstream Processing |
| VHH | Variable Heavy Domain Of Heavy Chain |
| Vif | Virion Infectivity Factor |
| Vpr | Viral Protein R |
| Vpu | Vira Protein U |
| VSV-G | Vesicular Stomatitis Virus Glycoprotein |
| WHX | Woodchuck Hepatitis Virus X Protein |
| WPRE | Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element |
| ZFN | Zinc Finger Nuclease |
| ZNF425 | Zinc Finger Protein 425 |
References
- Johnson, N.M.; Alvarado, A.F.; Moffatt, T.N.; Edavettal, J.M.; Swaminathan, T.A.; Braun, S.E. HIV-Based Lentiviral Vectors: Origin and Sequence Differences. Mol. Ther. Methods Clin. Dev. 2021, 21, 451–465. [Google Scholar] [CrossRef]
- Liu, Y.P.; Berkhout, B. HIV-1-Based Lentiviral Vectors. Methods Mol. Biol. 2014, 1087, 273–284. [Google Scholar] [CrossRef]
- Sakuma, T.; Barry, M.A.; Ikeda, Y. Lentiviral Vectors: Basic to Translational. Biochem. J. 2012, 443, 603–618. [Google Scholar] [CrossRef]
- Clever, J.L.; Miranda, D.; Parslow, T.G. RNA Structure and Packaging Signals in the 5′ Leader Region of the Human Immunodeficiency Virus Type 1 Genome. J. Virol. 2002, 76, 12381–12387. [Google Scholar] [CrossRef]
- Swanson, C.M.; Malim, M.H. SnapShot: HIV-1 Proteins. Cell 2008, 133, 9–10. [Google Scholar] [CrossRef]
- Emery, A.; Swanstrom, R. Hiv-1: To Splice or Not to Splice, That Is the Question. Viruses 2021, 13, 181. [Google Scholar] [CrossRef]
- Skalka, A.M.; Flint, J.; Rall, G.F.; Racaniello, V.R. Principles of Virology, Volume I: Molecular Biology; American Society of Microbiology: Washington, WA, USA, 2015; ISBN 9781555819330. [Google Scholar]
- Chen, B. Molecular Mechanism of HIV-1 Entry. Trends Microbiol. 2019, 27, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.B.; Arrildt, K.T.; Sturdevant, C.B.; Swanstrom, R. HIV-1 Target Cells in the CNS. J. Neurovirol. 2015, 21, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Hu, W.S. HIV-1 RNA Dimerization: It Takes Two to Tango. AIDS Rev. 2009, 11, 91–102. [Google Scholar] [PubMed] [PubMed Central]
- Cara, A.; Klotman, M.E. Retroviral E-DNA: Persistence and Gene Expression in Nondividing Immune Cells. J. Leukoc. Biol. 2006, 80, 1013–1017. [Google Scholar] [CrossRef]
- Engelman, A.N. Hiv Capsid and Integration Targeting. Viruses 2021, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, M.A. Determinants for Lentiviral Infection of Non-Dividing Cells. Somat. Cell Mol. Genet. 2002, 26, 35–49. [Google Scholar] [CrossRef]
- Shen, Q.; Wu, C.; Freniere, C.; Tripler, T.N.; Xiong, Y. Nuclear Import of Hiv-1. Viruses 2021, 13, 2242. [Google Scholar] [CrossRef]
- Engelman, A.N.; Singh, P.K. Cellular and Molecular Mechanisms of HIV-1 Integration Targeting. Cell. Mol. Life Sci. 2018, 75, 2491–2507. [Google Scholar] [CrossRef]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L.; Pongor, S.; Luzzati, R.; Recchia, A.; Mavilio, F.; et al. Nuclear Architecture Dictates HIV-1 Integration Site Selection. Nature 2015, 521, 227–231. [Google Scholar] [CrossRef]
- Duvergé, A.; Negroni, M. Pseudotyping Lentiviral Vectors: When the Clothes Make the Virus. Viruses 2020, 12, 1311. [Google Scholar] [CrossRef] [PubMed]
- Cabot, M.; Kiessling, V.; White, J.M.; Tamm, L.K. Endosomes Supporting Fusion Mediated by Vesicular Stomatitis Virus Glycoprotein Have Distinctive Motion and Acidification. Traffic 2022, 23, 221–234. [Google Scholar] [CrossRef]
- Yao, Y.; Ghosh, K.; Epand, R.F.; Epand, R.M.; Ghosh, H.P. Membrane Fusion Activity of Vesicular Stomatitis Virus Glycoprotein G Is Induced by Low PH but Not by Heat or Denaturant. Virology 2003, 310, 319–332. [Google Scholar] [CrossRef]
- Sun, X.; Yau, V.K.; Briggs, B.J.; Whittaker, G.R. Role of Clathrin-Mediated Endocytosis during Vesicular Stomatitis Virus Entry into Host Cells. Virology 2005, 338, 53–60. [Google Scholar] [CrossRef]
- Milone, M.C.; O’Doherty, U. Clinical Use of Lentiviral Vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef]
- Ferreira, M.V.; Cabral, E.T.; Coroadinha, A.S. Progress and Perspectives in the Development of Lentiviral Vector Producer Cells. Biotechnol. J. 2021, 16, 2000017. [Google Scholar] [CrossRef]
- Strebel, K. HIV Accessory Proteins versus Host Restriction Factors. Curr. Opin. Virol. 2013, 3, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-F.; Von Ruden, T.; Kantofft, P.W.; Garbert, C.; Seiberg, M.; Ruthert, U.; Andersont, W.F.; Wagnero, E.F.; Gilboa, E. Self-Inactivating Retroviral Vectors Designed for Transfer of Whole Genes into Mammalian Cells (Neomycin-Resistance Selection/c-Fos/Gene Therapy). Proc. Natl. Acad. Sci. USA 1986, 83, 3194–3198. [Google Scholar] [CrossRef] [PubMed]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A Third-Generation Lentivirus Vector with a Conditional Packaging System. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef]
- Kim, V.N.; Mitrophanous, K.; Kingsman, S.M.; Kingsman, A.J. Minimal Requirement for a Lentivirus Vector Based on Human Immunodeficiency Virus Type 1. J. Virol. 1998, 72, 811–816. [Google Scholar] [CrossRef]
- Van Maele, B.; De Rijck, J.; De Clercq, E.; Debyser, Z. Impact of the Central Polypurine Tract on the Kinetics of Human Immunodeficiency Virus Type 1 Vector Transduction. J. Virol. 2003, 77, 4685–4694. [Google Scholar] [CrossRef]
- Zufferey, R.; Donello, J.E.; Trono, D.; Hope, T.J. Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances Expression of Transgenes Delivered by Retroviral Vectors. J. Virol. 1999, 73, 2886–2892. [Google Scholar] [CrossRef]
- Brun, S.; Faucon-Biguet, N.; Mallet, J. Optimization of Transgene Expression at the Posttranscriptional Level in Neural Cells: Implications for Gene Therapy. Mol. Ther. 2003, 7, 782–789. [Google Scholar] [CrossRef]
- Zanta-Boussif, M.A.; Charrier, S.; Brice-Ouzet, A.; Martin, S.; Opolon, P.; Thrasher, A.J.; Hope, T.J.; Galy, A. Validation of a Mutated PRE Sequence Allowing High and Sustained Transgene Expression While Abrogating WHV-X Protein Synthesis: Application to the Gene Therapy of WAS. Gene Ther. 2009, 16, 605–619. [Google Scholar] [CrossRef]
- Schambach, A.; Galla, M.; Maetzig, T.; Loew, R.; Baum, C. Improving Transcriptional Termination of Self-Inactivating Gamma-Retroviral and Lentiviral Vectors. Mol. Ther. 2007, 15, 1167–1173. [Google Scholar] [CrossRef]
- Higashimoto, T.; Urbinati, F.; Perumbeti, A.; Jiang, G.; Zarzuela, A.; Chang, L.J.; Kohn, D.B.; Malik, P. The Woodchuck Hepatitis Virus Post-Transcriptional Regulatory Element Reduces Readthrough Transcription from Retroviral Vectors. Gene Ther. 2007, 14, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Kingsman, S.M.; Mitrophanous, K.; Olsen, J.C. Potential Oncogene Activity of the Woodchuck Hepatitis Post-Transcriptional Regulatory Element (WPRE). Gene Ther. 2005, 12, 3–4. [Google Scholar] [CrossRef]
- Schambach, A.; Bohne, J.; Baum, C.; Hermann, F.G.; Egerer, L.; von Laer, D.; Giroglou, T. Woodchuck Hepatitis Virus Post-Transcriptional Regulatory Element Deleted from X Protein and Promoter Sequences Enhances Retroviral Vector Titer and Expression. Gene Ther. 2006, 13, 641–645. [Google Scholar] [CrossRef]
- Razi Soofiyani, S.; Baradaran, B.; Lotfipour, F.; Kazemi, T.; Mohammadnejad, L. Gene Therapy, Early Promises, Subsequent Problems, and Recent Breakthroughs. Adv. Pharm. Bull. 2013, 3, 249–255. [Google Scholar] [CrossRef]
- Martínez-Molina, E.; Chocarro-Wrona, C.; Martínez-Moreno, D.; Marchal, J.A.; Boulaiz, H. Large-Scale Production of Lentiviral Vectors: Current Perspectives and Challenges. Pharmaceutics 2020, 12, 1051. [Google Scholar] [CrossRef]
- Food and Drug Administration Approved Cellular and Gene Therapy Products. Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products (accessed on 17 July 2025).
- White, M.; Whittaker, R.; Gándara, C.; Stoll, E.A. A Guide to Approaching Regulatory Considerations for Lentiviral-Mediated Gene Therapies. Hum. Gene Ther. Methods 2017, 28, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Srinivasakumar, N.; Schuening, F.G. A Lentivirus Packaging System Based on Alternative RNA Transport Mechanisms To Express Helper and Gene Transfer Vector RNAs and Its Use To Study the Requirement of Accessory Proteins for Particle Formation and Gene Delivery. J. Virol. 1999, 73, 9589–9598. [Google Scholar] [CrossRef]
- Zolotukhin, A.S.; Valentin, A.; Pavlakis, G.N.; Felber, B.K. Continuous Propagation of RRE(-) and Rev(-)RRE(-) Human Immunodeficiency Virus Type 1 Molecular Clones Containing a Cis-Acting Element of Simian Retrovirus Type 1 in Human Peripheral Blood Lymphocytes. J. Virol. 1994, 68, 7944–7952. [Google Scholar] [CrossRef]
- Bray, M.; Prasad, S.; Dubay, J.W.; Hunter, E.; Jeang, K.T.; Rekosh, D.; Hammarskjöld, M.L. A Small Element from the Mason-Pfizer Monkey Virusgenome Makes Human Immunodeficiency Virus Type 1 Expression and Replication Rev-Independent. Proc. Natl. Acad. Sci. USA 1994, 91, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.M.; Puffer, B.A.; Ahmad, K.M.; Doms, R.W.; Malim, M.H. Retroviral MRNA Nuclear Export Elements Regulate Protein Function and Virion Assembly. EMBO J. 2004, 23, 2632–2640. [Google Scholar] [CrossRef]
- Nappi, F.; Schneider, R.; Zolotukhin, A.; Smulevitch, S.; Michalowski, D.; Bear, J.; Felber, B.K.; Pavlakis, G.N. Identification of a Novel Posttranscriptional Regulatory Element by Using a Rev- and RRE-Mutated Human Immunodeficiency Virus Type 1 DNA Proviral Clone as a Molecular Trap. J. Virol. 2001, 75, 4558–4569. [Google Scholar] [CrossRef]
- Reddy, T.R.; Xu, W.; Mau, J.K.L.; Goodwin, C.D.; Suhasini, M.; Tang, H.; Frimpong, K.; Rose, D.W.; Wong-Staal, F. Inhibition of HIV Replication by Dominant Negative Mutants of Sam68, a Functional Homolog of HIV-1 Rev. Nat. Med. 1999, 5, 635–642, Erratum in Nat. Med. 1999, 5, 849. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Campbell, M.; Nasioulas, G.; Felber, B.K.; Pavlakis, G.N. Inactivation of the Human Immunodeficiency Virus Type 1 Inhibitory Elements Allows Rev-Independent Expression of Gag and Gag/Protease and Particle Formation. J. Virol. 1997, 71, 4892–4903. [Google Scholar] [CrossRef] [PubMed]
- D’agostino, D.M.; Felber, B.K.; Harrison, J.E.; Pavlakis, G.N. The Rev Protein of Human Immunodeficiency Virus Type 1 Promotes Polysomal Association and Translation of Gag/Pol and Vpu/Env MRNAs. Mol. Cell. Biol. 1992, 12, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, S.J.; Chen, I.S. Rev Is Necessary for Translation but Not Cytoplasmic Accumulation of HIV-1 Vif, Vpr, and Env/Vpu 2 RNAs. Genes Dev. 1991, 5, 808–819. [Google Scholar] [CrossRef]
- Haas, J.; Park, E.-C.; Seed, B. Codon Usage Limitation in the Expression of HIV-1 Envelope Glycoprotein. Curr. Biol. 1996, 6, 315–324. [Google Scholar] [CrossRef]
- Schwartz, S.; Felber, B.K.; Pavlakis1, G.N. Distinct RNA Sequences in the Gag Region of Human Immunodeficiency Virus Type 1 Decrease RNA Stability and Inhibit Expression in the Absence of Rev Protein. J. Virol. 1992, 66, 150–159. [Google Scholar] [CrossRef]
- Kotsopoulou, E.; Kim, V.N.; Kingsman, A.J.; Kingsman, S.M.; Mitrophanous, K.A. A Rev-Independent Human Immunodeficiency Virus Type 1 (HIV-1)-Based Vector That Exploits a Codon-Optimized HIV-1 Gag-Pol Gene. J. Virol. 2000, 74, 4839–4852. [Google Scholar] [CrossRef]
- Wagner, R.; Graf, M.; Bieler, K.; Wolf, H.; Grunwald, T.; Foley, P.; Überla, K. Rev-Independent Expression of Synthetic Gag-Pol Genes of Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus: Implications for the Safety of Lentiviral Vectors. Hum. Gene Ther. 2000, 11, 2403–2413. [Google Scholar] [CrossRef]
- Kumar, M.; Keller, B.; Makalou, N.; Sutton, R.E. Systematic Determination of the Packaging Limit of Lentiviral Vectors. Hum. Gene Ther. 2001, 12, 1893–1905. [Google Scholar] [CrossRef]
- Sweeney, N.P.; Vink, C.A. The Impact of Lentiviral Vector Genome Size and Producer Cell Genomic to Gag-Pol MRNA Ratios on Packaging Efficiency and Titre. Mol. Ther. Methods Clin. Dev. 2021, 21, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Sertkaya, H.; Ficarelli, M.; Sweeney, N.P.; Parker, H.; Vink, C.A.; Swanson, C.M. HIV-1 Sequences in Lentiviral Vector Genomes Can Be Substantially Reduced without Compromising Transduction Efficiency. Sci. Rep. 2021, 11, 12067. [Google Scholar] [CrossRef] [PubMed]
- Vink, C.A.; Counsell, J.R.; Perocheau, D.P.; Karda, R.; Buckley, S.M.K.; Brugman, M.H.; Galla, M.; Schambach, A.; McKay, T.R.; Waddington, S.N.; et al. Eliminating HIV-1 Packaging Sequences from Lentiviral Vector Proviruses Enhances Safety and Expedites Gene Transfer for Gene Therapy. Mol. Ther. 2017, 25, 1790–1804. [Google Scholar] [CrossRef]
- Fischbach, M.; Caliando, B.; Filsinger, G. High Capacity Lentiviral Vectors. International Patent Application No. PCT/US2023/020566, 1 May 2023. Available online: https://patentscope.wipo.int/search/en/WO2023212396 (accessed on 25 August 2025).
- Tran, T.; Liu, Y.; Marchant, J.; Monti, S.; Seu, M.; Zaki, J.; Yang, A.L.; Bohn, J.; Ramakrishnan, V.; Singh, R.; et al. Conserved Determinants of Lentiviral Genome Dimerization. Retrovirology 2015, 12, 83. [Google Scholar] [CrossRef]
- Sakuragi, J.; Sakuragi, S.; Shioda, T. Minimal Region Sufficient for Genome Dimerization in the Human Immunodeficiency Virus Type 1 Virion and Its Potential Roles in the Early Stages of Viral Replication. J. Virol. 2007, 81, 7985–7992. [Google Scholar] [CrossRef]
- Sakuragi, J.-I.; Ueda, S.; Iwamoto, A.; Shioda, T. Possible Role of Dimerization in Human Immunodeficiency Virus Type 1 Genome RNA Packaging. J. Virol. 2003, 77, 4060–4069. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.-S. High Efficiency of HIV-1 Genomic RNA Packaging and Heterozygote Formation Revealed by Single Virion Analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef]
- Moore, M.D.; Fu, W.; Nikolaitchik, O.; Chen, J.; Ptak, R.G.; Hu, W.-S. Dimer Initiation Signal of Human Immunodeficiency Virus Type 1: Its Role in Partner Selection during RNA Copackaging and Its Effects on Recombination. J. Virol. 2007, 81, 4002–4011. [Google Scholar] [CrossRef]
- Cordes, N.; Kolbe, C.; Lock, D.; Holzer, T.; Althoff, D.; Schäfer, D.; Blaeschke, F.; Kotter, B.; Karitzky, S.; Rossig, C.; et al. Anti-CD19 CARs Displayed at the Surface of Lentiviral Vector Particles Promote Transduction of Target-Expressing Cells. Mol. Ther. Methods Clin. Dev. 2021, 21, 42–53. [Google Scholar] [CrossRef]
- Maunder, H.E.; Wright, J.; Kolli, B.R.; Vieira, C.R.; Mkandawire, T.T.; Tatoris, S.; Kennedy, V.; Iqball, S.; Devarajan, G.; Ellis, S.; et al. Enhancing Titres of Therapeutic Viral Vectors Using the Transgene Repression in Vector Production (TRiP) System. Nat. Commun. 2017, 8, 14834. [Google Scholar] [CrossRef]
- Wright, J.; Alberts, B.; Hood, A.; Nogueira, C.; Miskolczi, Z.; Vieira, C.; Chipchase, D.; Lamont, C.; Goodyear, O.; Moyce, L.; et al. Improved Production and Quality of Lentiviral Vectors by Major-Splice-Donor Mutation and Co-Expression of a Novel U1 SnRNA-Based Enhancer. Heliyon 2024. [Google Scholar] [CrossRef]
- Inche, A.; Anderson, L. Expression Construct. International Patent Application No. PCT/GB2024/050776, 22 March 2024. Available online: https://patentscope.wipo.int/search/en/WO2024194651 (accessed on 25 August 2025).
- He, Z.; Kwee, E.J.; Cleveland, M.H.; Cole, K.D.; Lin-Gibson, S.; He, H.J. Quantitation and Integrity Evaluation of RNA Genome in Lentiviral Vectors by Direct Reverse Transcription-Droplet Digital PCR (Direct RT-DdPCR). Sci. Rep. 2023, 13, 14470. [Google Scholar] [CrossRef] [PubMed]
- Wentz, M.P.; Moore, B.E.; Cloyd, M.W.; Berget, S.M.; Donehower, L.A. A Naturally Arising Mutation of a Potential Silencer of Exon Splicing in Human Immunodeficiency Virus Type 1 Induces Dominant Aberrant Splicing and Arrests Virus Production. J. Virol. 1997, 71, 8542–8551. [Google Scholar] [CrossRef]
- Purcell, D.F.J.; Martin, M.A. Alternative Splicing of Human Immunodeficiency Virus Type 1 MRNA Modulates Viral Protein Expression, Replication, and Infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [CrossRef]
- Shaul, O. How Introns Enhance Gene Expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Rose, A.B. Introns as Gene Regulators: A Brick on the Accelerator. Front. Genet. 2019, 9, 672. [Google Scholar] [CrossRef]
- Cooper, A.R.; Lill, G.R.; Gschweng, E.H.; Kohn, D.B. Rescue of Splicing-Mediated Intron Loss Maximizes Expression in Lentiviral Vectors Containing the Human Ubiquitin C Promoter. Nucleic Acids Res. 2015, 43, 682–690. [Google Scholar] [CrossRef]
- Poling, B.C.; Tsai, K.; Kang, D.; Ren, L.; Kennedy, E.M.; Cullen, B.R. A Lentiviral Vector Bearing a Reverse Intron Demonstrates Superior Expression of Both Proteins and MicroRNAs. RNA Biol. 2017, 14, 1570–1579. [Google Scholar] [CrossRef]
- Bauler, M.; Ferrara, F.; Lowe, B.; Beard, J.A.; Wincek, C.; Wielgosz, M.M.; Park, J.J.; Shang, N.; Nandy, S.; Li, C.; et al. Genetic Alteration of SJ293TS Cells and Modification of Serum-Free Media Enhances Lentiviral Vector Production. Mol. Ther. Methods Clin. Dev. 2024, 32, 101270. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, M.; Shioda, T.; Sakuragi, J. ichi Retro-Transduction by Virus Pseudotyped with Glycoprotein of Vesicular Stomatitis Virus. Virology 2007, 362, 131–138. [Google Scholar] [CrossRef]
- Banos-Mateos, S.; Lopez-Robles, C.; Yubero, M.E.; Jurado, A.; Arbelaiz-Sarasola, A.; Lamsfus-Calle, A.; Arrasate, A.; Albo, C.; Ramírez, J.C.; Fertin, M.J. Abolishing Retro-Transduction of Producer Cells in Lentiviral Vector Manufacturing. Viruses 2024, 16, 1216. [Google Scholar] [CrossRef]
- Han, J.; Tam, K.; Tam, C.; Hollis, R.P.; Kohn, D.B. Improved Lentiviral Vector Titers from a Multi-Gene Knockout Packaging Line. Mol. Ther. Oncolytics 2021, 23, 582–592. [Google Scholar] [CrossRef]
- Otahal, A.; Fuchs, R.; Al-Allaf, F.A.; Blaas, D. Release of Vesicular Stomatitis Virus Spike Protein G-Pseudotyped Lentivirus from the Host Cell Is Impaired upon Low-Density Lipoprotein Receptor Overexpression. J. Virol. 2015, 89, 11723–11726. [Google Scholar] [CrossRef]
- Williams-Fegredo, T.; Davies, L.; Knevelman, C.; Miskin, J.; Mitrophanous, K.; Rafiq, Q.A. Auto-Transduction in Lentiviral Vector Bioprocessing: A Quantitative Assessment and a Novel Inhibition Strategy. Biotechnol. Bioeng. 2024, 121, 3728–3741. [Google Scholar] [CrossRef]
- Gama-Norton, L.; Botezatu, L.; Herrmann, S.; Schweizer, M.; Alves, P.M.; Hauser, H.; Wirth, D. Lentivirus Production Is Influenced by Sv40 Large T-Antigen and Chromosomal Integration of the Vector in Hek293 Cells. Hum. Gene Ther. 2011, 22, 1269–1279. [Google Scholar] [CrossRef]
- Xinyue, Z.; Li, S.; Yujie, W.; Yingcai, D.; Changhao, B.; Xueli, Z. Engineering of HEK293T Cell Factory for Lentiviral Production by High-Throughput Selected Genes. CRISPR J. 2024, 7, 272–282. [Google Scholar] [CrossRef]
- Swanson, J.; Tonne, J.; Sangsuwannukul, T.; Thompson, J.; Kendall, B.; Liseth, O.; Metko, M.; Vile, R. APOBEC3B Expression in 293T Lentiviral Producer Cells Drives Mutations in Chimeric Antigen Receptors and Reduces CAR T Cell Efficacy. Mol. Ther. Oncol. 2024, 32, 200873. [Google Scholar] [CrossRef]
- Coroadinha, A.S. Host Cell Restriction Factors Blocking Efficient Vector Transduction: Challenges in Lentiviral and Adeno-Associated Vector Based Gene Therapies. Cells 2023, 12, 732. [Google Scholar] [CrossRef]
- Kolegraff, K.; Bostik, P.; Ansari, A.A. Characterization and Role of Lentivirus-Associated Host Proteins. Exp. Biol. Med. 2006, 231, 252–263. [Google Scholar] [CrossRef]
- Milani, M.; Annoni, A.; Bartolaccini, S.; Biffi, M.; Russo, F.; Di Tomaso, T.; Raimondi, A.; Lengler, J.; Holmes, M.C.; Scheiflinger, F.; et al. Genome Editing for Scalable Production of Alloantigen-free Lentiviral Vectors for in Vivo Gene Therapy. EMBO Mol. Med. 2017, 9, 1558–1573. [Google Scholar] [CrossRef]
- Milani, M.; Annoni, A.; Moalli, F.; Liu, T.; Cesana, D.; Calabria, A.; Bartolaccini, S.; Biffi, M.; Russo, F.; Visigalli, I.; et al. Phagocytosis-Shielded Lentiviral Vectors Improve Liver Gene Therapy in Nonhuman Primates. Sci. Transl. Med. 2019, 11, eaav7325. [Google Scholar] [CrossRef]
- Leinonen, H. Commercial-Scale Lentiviral Vector Manufacturing: Is the Myth Busted? Cell Gene Ther. Insights 2022, 8, 3–13. [Google Scholar] [CrossRef]
- Tomás, H.A.; Rodrigues, A.F.; Carrondo, M.J.T.; Coroadinha, A.S. LentiPro26: Novel Stable Cell Lines for Constitutive Lentiviral Vector Production. Sci. Rep. 2018, 8, 5271. [Google Scholar] [CrossRef]
- Sang, Y.; Xie, K.; Mu, Y.; Lei, Y.; Zhang, B.; Xiong, S.; Chen, Y.; Qi, N. Salt Ions and Related Parameters Affect PEI–DNA Particle Size and Transfection Efficiency in Chinese Hamster Ovary Cells. Cytotechnology 2015, 67, 67–74. [Google Scholar] [CrossRef]
- Klimpel, M.; Terrao, M.; Ching, N.; Climenti, V.; Noll, T.; Pirzas, V.; Laux, H. Development of a Perfusion Process for Continuous Lentivirus Production Using Stable Suspension Producer Cell Lines. Biotechnol. Bioeng. 2023, 120, 2622–2638. [Google Scholar] [CrossRef]
- Manceur, A.P.; Kim, H.; Misic, V.; Andreev, N.; Dorion-Thibaudeau, J.; Lanthier, S.; Bernier, A.; Tremblay, S.; Gélinas, A.M.; Broussau, S.; et al. Scalable Lentiviral Vector Production Using Stable HEK293SF Producer Cell Lines. Hum. Gene Ther. Methods 2017, 28, 330–339. [Google Scholar] [CrossRef]
- Arrasate, A.; Bravo, I.; Lopez-Robles, C.; Arbelaiz-Sarasola, A.; Ugalde, M.; Meijueiro, M.L.; Zuazo, M.; Valero, A.; Banos-Mateos, S.; Ramirez, J.C.; et al. Establishment and Characterization of a Stable Producer Cell Line Generation Platform for the Manufacturing of Clinical-Grade Lentiviral Vectors. Biomedicines 2024, 12, 2265. [Google Scholar] [CrossRef]
- Poeschla, E.; Corbeau, P.; Wong-Staal, F. Development of HIV Vectors for Anti-HIV Gene Therapy. Proc. Natl. Acad. Sci. USA 1996, 93, 11395–11399. [Google Scholar] [CrossRef]
- Yu, H.; Rabson, A.B.; Kaul, M.; Ron, Y.; Dougherty, J.P. Inducible Human Immunodeficiency Virus Type 1 Packaging Cell Lines. J. Virol. 1996, 70, 4530–4537. [Google Scholar] [CrossRef]
- Kaul, M.; Yu, H.; Ron, Y.; Dougherty, J.P. Regulated Lentiviral Packaging Cell Line Devoid of Most Viralcis-Acting Sequences. Virology 1998, 249, 167–174. [Google Scholar] [CrossRef][Green Version]
- Gutierrez-Guerrero, A.; Cosset, F.L.; Verhoeyen, E. Lentiviral Vector Pseudotypes: Precious Tools to Improve Gene Modification of Hematopoietic Cells for Research and Gene Therapy. Viruses 2020, 12, 1016. [Google Scholar] [CrossRef]
- Dautzenberg, I.J.C.; Rabelink, M.J.W.E.; Hoeben, R.C. The Stability of Envelope-Pseudotyped Lentiviral Vectors. Gene Ther. 2021, 28, 89–104. [Google Scholar] [CrossRef]
- Burns, J.C.; Friedmann, T.; Driever, W.; Burrascano, M.; Yee, J.K. Vesicular Stomatitis Virus G Glycoprotein Pseudotyped Retroviral Vectors: Concentration to Very High Titer and Efficient Gene Transfer into Mammalian and Nonmammalian Cells. Proc. Natl. Acad. Sci. USA 1993, 90, 8033–8037. [Google Scholar] [CrossRef]
- Ikeda, Y.; Takeuchi, Y.; Martin, F.; Cosset, F.L.; Mitrophanous, K.; Collins, M. Continuous High-Titer HIV-1 Vector Production. Nat. Biotechnol. 2003, 21, 569–572. [Google Scholar] [CrossRef]
- Stornaiuolo, A.; Piovani, B.M.; Bossi, S.; Zucchelli, E.; Corna, S.; Salvatori, F.; Mavilio, F.; Bordignon, C.; Rizzardi, G.P.; Bovolenta, C. RD2-Molpack-Chim3, a Packaging Cell Line for Stable Production of Lentiviral Vectors for Anti-HIV Gene Therapy. Hum. Gene Ther. Methods 2013, 24, 228–240. [Google Scholar] [CrossRef]
- Humbert, O.; Gisch, D.W.; Wohlfahrt, M.E.; Adams, A.B.; Greenberg, P.D.; Schmitt, T.M.; Trobridge, G.D.; Kiem, H.P. Development of Third-Generation Cocal Envelope Producer Cell Lines for Robust Lentiviral Gene Transfer into Hematopoietic Stem Cells and t-Cells. Mol. Ther. 2016, 24, 1237–1246. [Google Scholar] [CrossRef]
- Bryson, P.D.; Zhang, C.; Lee, C.L.; Wang, P. A Tetracycline-Regulated Cell Line Produces High-Titer Lentiviral Vectors That Specifically Target Dendritic Cells. J. Vis. Exp. 2013, 76, e50606. [Google Scholar] [CrossRef]
- Chen, Y.H.; Pallant, C.; Sampson, C.J.; Boiti, A.; Johnson, S.; Brazauskas, P.; Hardwicke, P.; Marongiu, M.; Marinova, V.M.; Carmo, M.; et al. Rapid Lentiviral Vector Producer Cell Line Generation Using a Single DNA Construct. Mol. Ther. Methods Clin. Dev. 2020, 19, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Broussau, S.; Jabbour, N.; Lachapelle, G.; Durocher, Y.; Tom, R.; Transfiguracion, J.; Gilbert, R.; Massie, B. Inducible Packaging Cells for Large-Scale Production of Lentiviral Vectors in Serum-Free Suspension Culture. Mol. Ther. 2008, 16, 500–507. [Google Scholar] [CrossRef]
- Sparacio, S.; Pfeiffer, T.; Schaal, H.; Bosch, V. Generation of a Flexible Cell Line with Regulatable, High-Level Expression of HIV Gag/Pol Particles Capable of Packaging HIV-Derived Vectors. Mol. Ther. 2001, 3, 602–612. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene Therapy Comes of Age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Abordo-Adesida, E.; Follenzi, A.; Barcia, C.; Sciascia, S.; Castro, M.G.; Naldini, L.; Lowenstein, P.R. Stability of Lentiviral Vector-Mediated Transgene Expression in the Brain in the Presence of Systemic Antivector Immune Responses. Hum. Gene Ther. 2005, 16, 741–751. [Google Scholar] [CrossRef]
- Blömer, U.; Naldini, L.; Kafri, T.; Trono, D.; Verma, I.M.; Gage, F.H. Highly Efficient and Sustained Gene Transfer in Adult Neurons with a Lentivirus Vector. J. Virol. 1997, 71, 6641–6649. [Google Scholar] [CrossRef] [PubMed]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-Inactivating Lentivirus Vector for Safe and Efficient In Vivo Gene Delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar] [CrossRef]
- Klatt, D.; Sereni, L.; Liu, B.; Genovese, P.; Schambach, A.; Verhoeyen, E.; Williams, D.A.; Brendel, C. Engineered Packaging Cell Line for the Enhanced Production of Baboon-Enveloped Retroviral Vectors. Mol. Ther. Nucleic Acids 2024, 35, 102389. [Google Scholar] [CrossRef]
- Stitz, J. Development of HIV-1 Vectors Pseudotyped with Envelope Proteins of Other Retroviruses. Virology 2025, 602, 110300. [Google Scholar] [CrossRef]
- Tomás, H.A.; Mestre, D.A.; Rodrigues, A.F.; Guerreiro, M.R.; Carrondo, M.J.T.; Coroadinha, A.S. Improved GaLV-TR Glycoproteins to Pseudotype Lentiviral Vectors: Impact of Viral Protease Activity in the Production of LV Pseudotypes. Mol. Ther. Methods Clin. Dev. 2019, 15, 1–8. [Google Scholar] [CrossRef]
- Cantore, A.; Ranzani, M.; Bartholomae, C.C.; Volpin, M.; Valle, P.D.; Sanvito, F.; Sergi, L.S.; Gallina, P.; Benedicenti, F.; Bellinger, D.; et al. Liver-Directed Lentiviral Gene Therapy in a Dog Model of Hemophilia B. Sci. Transl. Med. 2015, 7, 277ra28. [Google Scholar] [CrossRef]
- Cantore, A.; Nair, N.; Della Valle, P.; Di Matteo, M.; Màtrai, J.; Sanvito, F.; Brombin, C.; Di Serio, C.; D’Angelo, A.; Chuah, M.; et al. Hyperfunctional Coagulation Factor IX Improves the Efficacy of Gene Therapy in Hemophilic Mice. Blood 2012, 120, 4517–4520. [Google Scholar] [CrossRef]
- Matsumoto, H.; Kimura, T.; Haga, K.; Kasahara, N.; Anton, P.; McGowan, I. Effective in Vivo and Ex Vivogene Transfer to Intestinal Mucosa by VSV-G-Pseudotyped Lentiviral Vectors. BMC Gastroenterol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- DePolo, N.J.; Reed, J.D.; Sheridan, P.L.; Townsend, K.; Sauter, S.L.; Jolly, D.J.; Dubensky, T.W. VSV-G Pseudotyped Lentiviral Vector Particles Produced in Human Cells Are Inactivated by Human Serum. Mol. Ther. 2000, 2, 218–222. [Google Scholar] [CrossRef]
- Trobridge, G.D.; Wu, R.A.; Hansen, M.; Ironside, C.; Watts, K.L.; Olsen, P.; Beard, B.C.; Kiem, H.P. Cocal-Pseudotyped Lentiviral Vectors Resist Inactivation by Human Serum and Efficiently Transduce Primate Hematopoietic Repopulating Cells. Mol. Ther. 2010, 18, 725–733. [Google Scholar] [CrossRef]
- Hindi, S.M.; Petrany, M.J.; Greenfeld, E.; Focke, L.C.; Cramer, A.A.W.; Whitt, M.A.; Prasad, V.; Chamberlain, J.S.; Podbilewicz, B.; Millay, D.P. Enveloped Viruses Pseudotyped with Mammalian Myogenic Cell Fusogens Target Skeletal Muscle for Gene Delivery. Cell 2023, 186, 2062–2077.e17. [Google Scholar] [CrossRef] [PubMed]
- Mazarakis, N.D. Rabies Virus Glycoprotein Pseudotyping of Lentiviral Vectors Enables Retrograde Axonal Transport and Access to the Nervous System after Peripheral Delivery. Hum. Mol. Genet. 2001, 10, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Arduini, A.; Katiyar, H.; Liang, C. Progress in Pseudotyping Lentiviral Vectors Towards Cell-Specific Gene Delivery In Vivo. Viruses 2025, 17, 802. [Google Scholar] [CrossRef]
- Mears, K.S.; Ibrahim, K.; Allen, P.M.; Chinai, J.M.; Avila, O.I.; Muscato, A.J.; Lane-Reticker, S.K.; Rojas, A.; Knudsen, N.H.; Chao, C.-C.; et al. In Vivo Generation of Chimeric Antigen Receptor T Cells Using Optimally Retargeted and Functionalized Lentiviral Vectors with Reduced Immune Clearance. bioRxiv 2025. [Google Scholar] [CrossRef]
- ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/search?term=lentiviral%20vector (accessed on 17 July 2025).
- Chong, E.A.; Ruella, M.; Schuster, S.J. Five-Year Outcomes for Refractory B-Cell Lymphomas with CAR T-Cell Therapy. N. Engl. J. Med. 2021, 384, 673–674. [Google Scholar] [CrossRef]
- Zhao, W.H.; Wang, B.Y.; Chen, L.J.; Fu, W.J.; Xu, J.; Liu, J.; Jin, S.W.; Chen, Y.X.; Cao, X.M.; Yang, Y.; et al. Four-Year Follow-up of LCAR-B38M in Relapsed or Refractory Multiple Myeloma: A Phase 1, Single-Arm, Open-Label, Multicenter Study in China (LEGEND-2). J. Hematol. Oncol. 2022, 15, 86. [Google Scholar] [CrossRef]
- Cordeiro, A.; Bezerra, E.D.; Hirayama, A.V.; Hill, J.A.; Wu, Q.V.; Voutsinas, J.; Sorror, M.L.; Turtle, C.J.; Maloney, D.G.; Bar, M. Late Events after Treatment with CD19-Targeted Chimeric Antigen Receptor Modified T Cells. Biol. Blood Marrow Transplant. 2020, 26, 26–33. [Google Scholar] [CrossRef]
- Steffin, D.H.M.; Muhsen, I.N.; Hill, L.C.; Ramos, C.A.; Ahmed, N.; Hegde, M.; Wang, T.; Wu, M.; Gottschalk, S.; Whittle, S.B.; et al. Long-Term Follow-up for the Development of Subsequent Malignancies in Patients Treated with Genetically Modified IECs. Blood 2022, 140, 16–24. [Google Scholar] [CrossRef]
- Cappell, K.M.; Sherry, R.M.; Yang, J.C.; Goff, S.L.; Vanasse, D.A.; McIntyre, L.; Rosenberg, S.A.; Kochenderfer, J.N. Long-Term Follow-Up of Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy. J. Clin. Oncol. 2020, 38, 3805–3815. [Google Scholar] [CrossRef]
- Ghilardi, G.; Fraietta, J.A.; Gerson, J.N.; Van Deerlin, V.M.; Morrissette, J.J.D.; Caponetti, G.C.; Paruzzo, L.; Harris, J.C.; Chong, E.A.; Susanibar Adaniya, S.P.; et al. T Cell Lymphoma and Secondary Primary Malignancy Risk after Commercial CAR T Cell Therapy. Nat. Med. 2024, 30, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Aiuti, A.; Biasco, L.; Scaramuzza, S.; Ferrua, F.; Cicalese, M.P.; Baricordi, C.; Dionisio, F.; Calabria, A.; Giannelli, S.; Castiello, M.C.; et al. Lentiviral Hematopoietic Stem Cell Gene Therapy in Patients with Wiskott-Aldrich Syndrome. Science 2013, 341, 1233151. [Google Scholar] [CrossRef] [PubMed]
- Giménez, Y.; Palacios, M.; Sánchez-Domínguez, R.; Zorbas, C.; Peral, J.; Puzik, A.; Ugalde, L.; Alberquilla, O.; Villanueva, M.; Río, P.; et al. Lentivirus-Mediated Gene Therapy Corrects Ribosomal Biogenesis and Shows Promise for Diamond Blackfan Anemia. JCI Insight 2024, 9, e171650. [Google Scholar] [CrossRef]
- Shah, N.N.; Qin, H.; Yates, B.; Su, L.; Shalabi, H.; Raffeld, M.; Ahlman, M.A.; Stetler-Stevenson, M.; Yuan, C.; Guo, S.; et al. Clonal Expansion of CAR T Cells Harboring Lentivector Integration in the CBL Gene Following Anti-CD22 CAR T-Cell Therapy. Blood Adv. 2019, 3, 2317–2322. [Google Scholar] [CrossRef]
- Fraietta, J.A.; Nobles, C.L.; Sammons, M.A.; Lundh, S.; Carty, S.A.; Reich, T.J.; Cogdill, A.P.; Morrissette, J.J.D.; DeNizio, J.E.; Reddy, S.; et al. Disruption of TET2 Promotes the Therapeutic Efficacy of CD19-Targeted T Cells. Nature 2018, 558, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Olvera, J.M.; Yoder, K.E.; Mitchell, R.S.; Butler, S.L.; Lieber, M.; Martin, S.L.; Bushman, F.D. Role of the Non-Homologous DNA End Joining Pathway in the Early Steps of Retroviral Infection. EMBO J. 2001, 20, 3272–3281. [Google Scholar] [CrossRef]
- Farnet, C.M.; Haseltine, W.A. Circularization of Human Immunodeficiency Virus Type 1 DNA in Vitro. J. Virol. 1991, 65, 6942–6952. [Google Scholar] [CrossRef]
- Zhu, K.; Dobard, C.; Chow, S.A. Requirement for Integrase during Reverse Transcription of Human Immunodeficiency Virus Type 1 and the Effect of Cysteine Mutations of Integrase on Its Interactions with Reverse Transcriptase. J. Virol. 2004, 78, 5045–5055. [Google Scholar] [CrossRef]
- Gallay, P.; Hope, T.; Chin, D.; Trono, D. HIV-1 Infection of Nondividing Cells through the Recognition of Integrase by the Importin/Karyopherin Pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 9825–9830. [Google Scholar] [CrossRef] [PubMed]
- Wanisch, K.; Yáñez-Muñoz, R.J. Integration-Deficient Lentiviral Vectors: A Slow Coming of Age. Mol. Ther. 2009, 17, 1316–1332. [Google Scholar] [CrossRef]
- Engelman, A.; Craigie, R. Identification of Conserved Amino Acid Residues Critical for Human Immunodeficiency Virus Type 1 Integrase Function in Vitro. J. Virol. 1992, 66, 6361–6369. [Google Scholar] [CrossRef]
- Leavitt, A.D.; Robles, G.; Alesandro, N.; Varmus, H.E. Human Immunodeficiency Virus Type 1 Integrase Mutants Retain in Vitro Integrase Activity yet Fail to Integrate Viral DNA Efficiently during Infection. J. Virol. 1996, 70, 721–728. [Google Scholar] [CrossRef]
- Wiskerchen, M.; Muesing, M.A. Human Immunodeficiency Virus Type 1 Integrase: Effects of Mutations on Viral Ability to Integrate, Direct Viral Gene Expression from Unintegrated Viral DNA Templates, and Sustain Viral Propagation in Primary Cells. J. Virol. 1995, 69, 376–386. [Google Scholar] [CrossRef]
- Ansari-Lari, M.A.; Donehower, L.A.; Gibbs, R.A. Analysis of Human Immunodeficiency Virus Type 1 Integrase Mutants. Virology 1995, 211, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, A.D.; Shiue, L.; Varmus, H.E. Site-Directed Mutagenesis of HIV-1 Integrase Demonstrates Differential Effects on Integrase Functions in Vitro. J. Biol. Chem. 1993, 268, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Philpott, N.J.; Thrasher, A.J. Use of Nonintegrating Lentiviral Vectors for Gene Therapy. Hum. Gene Ther. 2007, 18, 483–489. [Google Scholar] [CrossRef]
- Yáñez-Muñoz, R.J.; Balaggan, K.S.; MacNeil, A.; Howe, S.J.; Schmidt, M.; Smith, A.J.; Buch, P.; MacLaren, R.E.; Anderson, P.N.; Barker, S.E.; et al. Effective Gene Therapy with Nonintegrating Lentiviral Vectors. Nat. Med. 2006, 12, 348–353. [Google Scholar] [CrossRef]
- Philippe, S.; Sarkis, C.; Barkats, M.; Mammeri, H.; Ladroue, C.; Petit, C.; Mallet, J.; Serguera, C. Lentiviral Vectors with a Defective Integrase Allow Efficient and Sustained Transgene Expression in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 17684–17689. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.; Gusella, G.L.; Najfeld, V.; Klotman, M.E.; Cara, A. Novel Integrase-Defective Lentiviral Episomal Vectors for Gene Transfer. Hum. Gene Ther. 2004, 15, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Saenz, D.T.; Loewen, N.; Peretz, M.; Whitwam, T.; Barraza, R.; Howell, K.G.; Holmes, J.M.; Good, M.; Poeschla, E.M. Unintegrated Lentivirus DNA Persistence and Accessibility to Expression in Nondividing Cells: Analysis with Class I Integrase Mutants. J. Virol. 2004, 78, 2906–2920. [Google Scholar] [CrossRef]
- Brussel, A.; Sonigo, P. Evidence for Gene Expression by Unintegrated Human Immunodeficiency Virus Type 1 DNA Species. J. Virol. 2004, 78, 11263–11271. [Google Scholar] [CrossRef] [PubMed]
- Torres Ruiz, R.; Ramírez Martínez, J.; Garcia Torralba, A. Stable Episomes Base don Non-Integrative Lentiviral Vectors. U.S. Patent Application No. 15100110, 21 May 2019. Available online: https://patentscope.wipo.int/search/en/WO2015078999 (accessed on 25 August 2025).
- Cerundolo, V.; Hermans, I.F.; Salio, M. Dendritic Cells: A Journey from Laboratory to Clinic. Nat. Immunol. 2004, 5, 7–10. [Google Scholar] [CrossRef]
- Nemirov, K.; Bourgine, M.; Anna, F.; Wei, Y.; Charneau, P.; Majlessi, L. Lentiviral Vectors as a Vaccine Platform against Infectious Diseases. Pharmaceutics 2023, 15, 846. [Google Scholar] [CrossRef]
- Ku, M.-W.; Charneau, P.; Majlessi, L. Use of Lentiviral Vectors in Vaccination. Expert Rev. Vaccines 2021, 20, 1571–1586. [Google Scholar] [CrossRef]
- Dullaers, M.; Thielemans, K. From Pathogen to Medicine: HIV-1-Derived Lentiviral Vectors as Vehicles for Dendritic Cell Based Cancer Immunotherapy. J. Gene Med. 2006, 8, 3–17. [Google Scholar] [CrossRef]
- Goyvaerts, C.; De Groeve, K.; Dingemans, J.; Van Lint, S.; Robays, L.; Heirman, C.; Reiser, J.; Zhang, X.-Y.; Thielemans, K.; De Baetselier, P.; et al. Development of the Nanobody Display Technology to Target Lentiviral Vectors to Antigen-Presenting Cells. Gene Ther. 2012, 19, 1133–1140. [Google Scholar] [CrossRef]
- Ciré, S.; Da Rocha, S.; Yao, R.; Fisson, S.; Buchholz, C.J.; Collins, M.K.; Galy, A. Immunization of Mice with Lentiviral Vectors Targeted to MHC Class II+ Cells Is Due to Preferential Transduction of Dendritic Cells In Vivo. PLoS ONE 2014, 9, e101644. [Google Scholar] [CrossRef]
- Dai, B.; Yang, L.; Yang, H.; Hu, B.; Baltimore, D.; Wang, P. HIV-1 Gag-Specific Immunity Induced by a Lentivector-Based Vaccine Directed to Dendritic Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 20382–20387. [Google Scholar] [CrossRef] [PubMed]
- Ku, M.W.; Anna, F.; Souque, P.; Petres, S.; Prot, M.; Simon-Loriere, E.; Charneau, P.; Bourgine, M. A Single Dose of NILV-Based Vaccine Provides Rapid and Durable Protection against Zika Virus. Mol. Ther. 2020, 28, 1772–1782. [Google Scholar] [CrossRef] [PubMed]
- Gallinaro, A.; Borghi, M.; Bona, R.; Grasso, F.; Calzoletti, L.; Palladino, L.; Cecchetti, S.; Vescio, M.F.; Macchia, D.; Morante, V.; et al. Integrase Defective Lentiviral Vector as a Vaccine Platform for Delivering Influenza Antigens. Front. Immunol. 2018, 9, 171. [Google Scholar] [CrossRef]
- Ku, M.W.; Bourgine, M.; Authié, P.; Lopez, J.; Nemirov, K.; Moncoq, F.; Noirat, A.; Vesin, B.; Nevo, F.; Blanc, C.; et al. Intranasal Vaccination with a Lentiviral Vector Protects against SARS-CoV-2 in Preclinical Animal Models. Cell Host Microbe 2021, 29, 236–249.e6. [Google Scholar] [CrossRef]
- Douguet, L.; Fert, I.; Lopez, J.; Vesin, B.; Le Chevalier, F.; Moncoq, F.; Authié, P.; Nguyen, T.; Noirat, A.; Névo, F.; et al. Full Eradication of Pre-clinical Human Papilloma Virus-induced Tumors by a Lentiviral Vaccine. EMBO Mol. Med. 2023, 15, e17723. [Google Scholar] [CrossRef]
- Kymäläinen, H.; Appelt, J.U.; Giordano, F.A.; Davies, A.F.; Ogilvie, C.M.; Ahmed, S.G.; Laufs, S.; Schmidt, M.; Bode, J.; Yáñez-Muñoz, R.J.; et al. Long-Term Episomal Transgene Expression from Mitotically Stable Integration-Deficient Lentiviral Vectors. Hum. Gene Ther. 2014, 25, 428–442. [Google Scholar] [CrossRef]
- Wang, C.; Pan, C.; Yong, H.; Wang, F.; Bo, T.; Zhao, Y.; Ma, B.; He, W.; Li, M. Emerging Non-Viral Vectors for Gene Delivery. J. Nanobiotechnology 2023, 21, 272. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Gao, Y.-G.; Lin, X.; Li, Y.; Dang, K.; Tian, Y.; Zhang, W.-J.; Jiang, S.-F.; Qadir, A.; Qian, A.-R. The Development of Functional Non-Viral Vectors for Gene Delivery. Int. J. Mol. Sci. 2019, 20, 5491. [Google Scholar] [CrossRef]
- Guo, X.; Huang, L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012, 45, 971–979. [Google Scholar] [CrossRef]
- Wittrup, A.; Ai, A.; Liu, X.; Hamar, P.; Trifonova, R.; Charisse, K.; Manoharan, M.; Kirchhausen, T.; Lieberman, J. Visualizing Lipid-Formulated SiRNA Release from Endosomes and Target Gene Knockdown. Nat. Biotechnol. 2015, 33, 870–876. [Google Scholar] [CrossRef]
- Galla, M.; Schambach, A.; Baum, C. Retrovirus-Based MRNA Transfer for Transient Cell Manipulation. Methods Mol. Biol. 2013, 969, 139–161. [Google Scholar] [CrossRef] [PubMed]
- Schott, J.W.; Morgan, M.; Galla, M.; Schambach, A. Viral and Synthetic RNA Vector Technologies and Applications. Mol. Ther. 2016, 24, 1513–1527. [Google Scholar] [CrossRef]
- Mock, U.; Riecken, K.; Berdien, B.; Qasim, W.; Chan, E.; Cathomen, T.; Fehse, B. Novel Lentiviral Vectors with Mutated Reverse Transcriptase for MRNA Delivery of TALE Nucleases. Sci. Rep. 2014, 4, 6409. [Google Scholar] [CrossRef]
- Counsell, J.R.; De Brabandere, G.; Karda, R.; Moore, M.; Greco, A.; Bray, A.; Diaz, J.A.; Perocheau, D.P.; Mock, U.; Waddington, S.N. Re-Structuring Lentiviral Vectors to Express Genomic RNA via Cap-Dependent Translation. Mol. Ther. Methods Clin. Dev. 2021, 20, 357–365. [Google Scholar] [CrossRef]
- Prel, A.; Caval, V.; Gayon, R.; Ravassard, P.; Duthoit, C.; Payen, E.; Maouche-Chretien, L.; Creneguy, A.; Nguyen, T.H.; Martin, N.; et al. Highly Efficient in Vitro and in Vivo Delivery of Functional RNAs Using New Versatile MS2-Chimeric Retrovirus-like Particles. Mol. Ther. Methods Clin. Dev. 2015, 2, 15039. [Google Scholar] [CrossRef]
- Dong, W.; Kantor, B. Lentiviral Vectors for Delivery of Gene-Editing Systems Based on Crispr/Cas: Current State and Perspectives. Viruses 2021, 13, 1288. [Google Scholar] [CrossRef]
- Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. Targeted Chromosomal Cleavage and Mutagenesis in Drosophila Using Zinc-Finger Nucleases. Genetics 2002, 161, 1169–1175. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Lamsfus-Calle, A.; Daniel-Moreno, A.; Ureña-Bailén, G.; Raju, J.; Antony, J.S.; Handgretinger, R.; Mezger, M. Hematopoietic Stem Cell Gene Therapy: The Optimal Use of Lentivirus and Gene Editing Approaches. Blood Rev. 2020, 40, 100641. [Google Scholar] [CrossRef]
- Chandrasegaran, S.; Carroll, D. Origins of Programmable Nucleases for Genome Engineering. J. Mol. Biol. 2016, 428, 963–989. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and Evolution of Class 2 CRISPR–Cas Systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lee, B.; Lee, A.Y.-F.; Modzelewski, A.J.; He, L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol. Chem. 2016, 291, 14457–14467. [Google Scholar] [CrossRef]
- Qin, W.; Dion, S.L.; Kutny, P.M.; Zhang, Y.; Cheng, A.W.; Jillette, N.L.; Malhotra, A.; Geurts, A.M.; Chen, Y.-G.; Wang, H. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics 2015, 200, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Arai, Y.; Yamazaki, M.; Morita, S.; Kimura, M.; Itoh, M.; Abe, Y.; Hatada, I. Validation of Microinjection Methods for Generating Knockout Mice by CRISPR/Cas-Mediated Genome Engineering. Sci. Rep. 2014, 4, 4513. [Google Scholar] [CrossRef]
- Musunuru, K.; Grandinette, S.A.; Wang, X.; Hudson, T.R.; Briseno, K.; Berry, A.M.; Hacker, J.L.; Hsu, A.; Silverstein, R.A.; Hille, L.T.; et al. Patient-Specific In Vivo Gene Editing to Treat a Rare Genetic Disease. N. Engl. J. Med. 2025, 392, 2235–2243. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering Crispr: A Review of the Challenges and Approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Truong, D.-J.J.; Kühner, K.; Kühn, R.; Werfel, S.; Engelhardt, S.; Wurst, W.; Ortiz, O. Development of an Intein-Mediated Split–Cas9 System for Gene Therapy. Nucleic Acids Res. 2015, 43, 6450–6458. [Google Scholar] [CrossRef]
- Wang, J.; Exline, C.M.; DeClercq, J.J.; Llewellyn, G.N.; Hayward, S.B.; Li, P.W.-L.; Shivak, D.A.; Surosky, R.T.; Gregory, P.D.; Holmes, M.C.; et al. Homology-Driven Genome Editing in Hematopoietic Stem and Progenitor Cells Using ZFN MRNA and AAV6 Donors. Nat. Biotechnol. 2015, 33, 1256–1263. [Google Scholar] [CrossRef]
- Rai, R.; Romito, M.; Rivers, E.; Turchiano, G.; Blattner, G.; Vetharoy, W.; Ladon, D.; Andrieux, G.; Zhang, F.; Zinicola, M.; et al. Targeted Gene Correction of Human Hematopoietic Stem Cells for the Treatment of Wiskott-Aldrich Syndrome. Nat. Commun. 2020, 11, 4034. [Google Scholar] [CrossRef]
- Dever, D.P.; Bak, R.O.; Reinisch, A.; Camarena, J.; Washington, G.; Nicolas, C.E.; Pavel-Dinu, M.; Saxena, N.; Wilkens, A.B.; Mantri, S.; et al. CRISPR/Cas9 β-Globin Gene Targeting in Human Haematopoietic Stem Cells. Nature 2016, 539, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Jacob, A.; Cesana, D.; Laugel, M.; Beretta, S.; Varesi, A.; Unali, G.; Conti, A.; Canarutto, D.; Albano, L.; et al. Choice of Template Delivery Mitigates the Genotoxic Risk and Adverse Impact of Editing in Human Hematopoietic Stem Cells. Cell Stem Cell 2022, 29, 1428–1444.e9. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Xiao, H.; Kim, J.; Seshaiah, P.; Natsoulis, G.; Boeke, J.D.; Hahn, B.H.; Kappes, J.C. Targeting Foreign Proteins to Human Immunodeficiency Virus Particles via Fusion with Vpr and Vpx. J. Virol. 1995, 69, 3389–3398. [Google Scholar] [CrossRef]
- Haldrup, J.; Andersen, S.; LaVilla Labial, A.R.; Wolff, J.H.; Frandsen, F.P.; Skov, T.W.; Rovsing, A.B.; Nielsen, I.; Jakobsen, T.S.; Askou, A.L.; et al. Engineered Lentivirus-Derived Nanoparticles (LVNPs) for Delivery of CRISPR/Cas Ribonucleoprotein Complexes Supporting Base Editing, Prime Editing and in Vivo Gene Modification. Nucleic Acids Res. 2023, 51, 10059–10074. [Google Scholar] [CrossRef]
- Cai, Y.; Bak, R.O.; Krogh, L.B.; Staunstrup, N.H.; Moldt, B.; Corydon, T.J.; Schrøder, L.D.; Mikkelsen, J.G. DNA Transposition by Protein Transduction of the PiggyBac Transposase from Lentiviral Gag Precursors. Nucleic Acids Res. 2014, 42, e28. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Bak, R.O.; Mikkelsen, J.G. Targeted Genome Editing by Lentiviral Protein Transduction of Zinc-Finger and TAL-Effector Nucleases. eLife 2014, 3, e01911. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, Y.; Laustsen, A.; Zhou, Y.; Sun, C.; Anderson, M.V.; Li, S.; Uldbjerg, N.; Luo, Y.; Jakobsen, M.R.; Mikkelsen, J.G. Targeted, Homology-Driven Gene Insertion in Stem Cells by ZFN-Loaded ‘All-in-One’ Lentiviral Vectors. eLife 2016, 5, e12213. [Google Scholar] [CrossRef]
- Thomsen, E.A.; Skipper, K.A.; Andersen, S.; Haslund, D.; Skov, T.W.; Mikkelsen, J.G. CRISPR-Cas9-Directed Gene Tagging Using a Single Integrase-Defective Lentiviral Vector Carrying a Transposase-Based Cas9 off Switch. Mol. Ther. Nucleic Acids 2022, 29, 563–576. [Google Scholar] [CrossRef]
- Skipper, K.A.; Nielsen, M.G.; Andersen, S.; Ryø, L.B.; Bak, R.O.; Mikkelsen, J.G. Time-Restricted PiggyBac DNA Transposition by Transposase Protein Delivery Using Lentivirus-Derived Nanoparticles. Mol. Ther. Nucleic Acids 2018, 11, 253–262. [Google Scholar] [CrossRef]
- Nielsen, I.H.; Rovsing, A.B.; Janns, J.H.; Thomsen, E.A.; Ruzo, A.; Bøggild, A.; Nedergaard, F.; Møller, C.T.; Boesen, T.; Degn, S.E.; et al. Cell-Targeted Gene Modification by Delivery of CRISPR/Cas9 Ribonucleoprotein Complexes in Pseudotyped Lentivirus-Derived Nanoparticles. Mol. Ther. Nucleic Acids 2024, 35, 102318. [Google Scholar] [CrossRef] [PubMed]
- Janns, J.H.; Mikkelsen, J.G. Gene Editing by Ferrying of CRISPR/Cas Ribonucleoprotein Complexes in Enveloped Virus-Derived Particles. Hum. Gene Ther. 2024, 35, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of T to G C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arrasate, A.; Lopez-Robles, C.; Zuazo, M.; Banos-Mateos, S.; Martin, C.; Lamsfus-Calle, A.; Fertin, M.J. Lentiviral Vectors: From Wild-Type Viruses to Efficient Multi-Functional Delivery Vectors. Int. J. Mol. Sci. 2025, 26, 8497. https://doi.org/10.3390/ijms26178497
Arrasate A, Lopez-Robles C, Zuazo M, Banos-Mateos S, Martin C, Lamsfus-Calle A, Fertin MJ. Lentiviral Vectors: From Wild-Type Viruses to Efficient Multi-Functional Delivery Vectors. International Journal of Molecular Sciences. 2025; 26(17):8497. https://doi.org/10.3390/ijms26178497
Chicago/Turabian StyleArrasate, Ane, Carlos Lopez-Robles, Miren Zuazo, Soledad Banos-Mateos, Cesar Martin, Andrés Lamsfus-Calle, and Marie J. Fertin. 2025. "Lentiviral Vectors: From Wild-Type Viruses to Efficient Multi-Functional Delivery Vectors" International Journal of Molecular Sciences 26, no. 17: 8497. https://doi.org/10.3390/ijms26178497
APA StyleArrasate, A., Lopez-Robles, C., Zuazo, M., Banos-Mateos, S., Martin, C., Lamsfus-Calle, A., & Fertin, M. J. (2025). Lentiviral Vectors: From Wild-Type Viruses to Efficient Multi-Functional Delivery Vectors. International Journal of Molecular Sciences, 26(17), 8497. https://doi.org/10.3390/ijms26178497




