Identifying New Loci and Genes Associated with Feed Efficiency in Broilers
Abstract
1. Introduction
2. Results
2.1. Descriptive Statistics of Feed Efficiency Traits
2.2. Annotation of Genotype Data After Quality Control
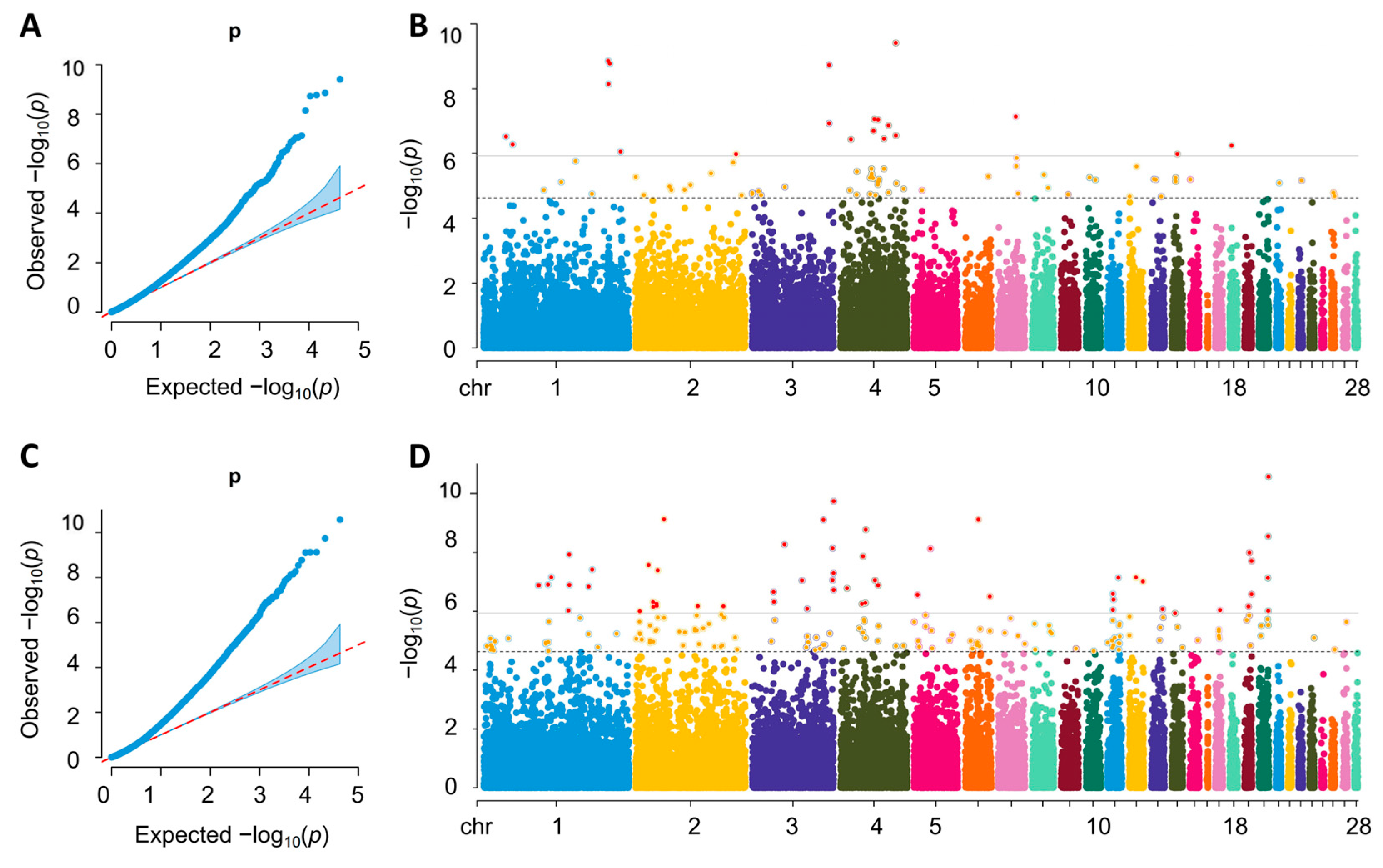
2.3. SNPs Identified by Single-Trait Genome-Wide Association Analysis
2.4. SNPs Identified by Genome-Wide Association Analysis of Longitudinal Traits
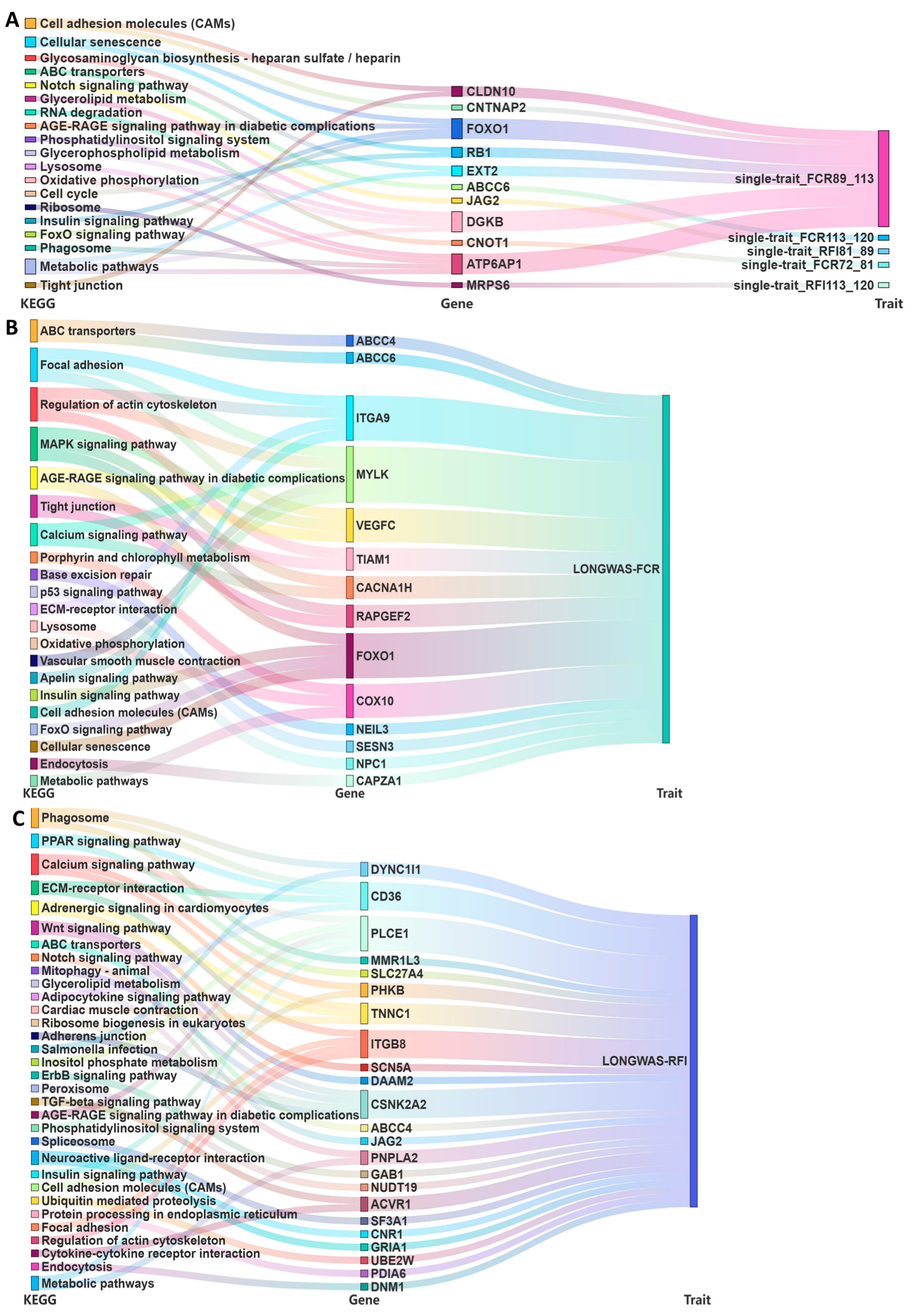
2.5. Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Candidate Genes Associated with Phenotype
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals and Sample Collection
4.3. Phenotyping
4.4. Genotyping and Quality Control
4.5. Single-Trait Genome-Wide Association Study
4.6. LONG Genome-Wide Association Study
4.7. Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ABCC4 | ATP-Binding Cassette Subfamily C Member 4 |
| ABCB5 | ATP-Binding Cassette Subfamily B Member 5 |
| ABCC6 | ATP-Binding Cassette Subfamily C Member 6 |
| ADFI | average daily feed efficiency |
| ADG | average daily gain |
| AGA | Aspartylglucosaminidase |
| chCADD | chicken Combined Annotation-Dependent Depletion |
| COX10 | Cytochrome C Oxidase Assembly Factor Heme A: Farnesyltransferase COX10 |
| CSNK2A2 | Casein Kinase 2 Alpha 2 |
| DAAM2 | Disheveled-Associated Activator of Morphogenesis 2 |
| DGKB | Diacylglycerol Kinase Beta |
| DYNC1I1 | Dynein Cytoplasmic 1 Intermediate Chain 1 |
| EPHA5 | EPH Receptor A5 |
| EXT2 | Exostosin Glycosyltransferase 2 |
| FCR | feed conversion ratio |
| FOXO1 | Forkhead Box O1 |
| FSTL4 | Follistatin Like 4 |
| GEC | Genetic Type 1 Error Calculator |
| GGA | Gallus gallus chromosome |
| GIF | genomic inflation factor |
| GTF2I | General Transcription Factor Iii |
| GWAS | genome-wide association |
| HS3ST3B1 | Heparan Sulfate-Glucosamine 3-Sulfotransferase 3B1 |
| ITGA9 | Integrin Subunit Alpha 9 |
| ITPKA | Inositol-Trisphosphate 3-Kinase A |
| ITPR2 | Inositol 1,4,5-Trisphosphate Receptor Type 2 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LMM | linear mixed model |
| LONG-GWAS | longitudinal GWAS |
| MAF | minimum minor allele frequency |
| MBOAT1 | Membrane-Bound O-Acyltransferase Domain-Containing 1 |
| MMR1L3 | Macrophage Mannose Receptor 1-like 3 |
| MYLK | Myosin Light Chain Kinase |
| P2RX5 | Purinergic Receptor P2X 5 |
| PFKP | Phosphofructokinase, Platelet |
| PHKB | Phosphorylase Kinase Regulatory Subunit Beta |
| PNPLA2 | Patatin-Like Phospholipase Domain-Containing 2 |
| POLD3 | DNA Polymerase Delta 3, Accessory Subunit |
| PPP1R12A | Protein Phosphatase 1 Regulatory Subunit 12A |
| QTL | quantitative trait locus |
| Q–Q | quantile–quantile |
| RB1 | RB Transcriptional Corepressor 1 |
| RFI | residual feed intake |
| SHC1 | SHC Adaptor Protein 1 |
| SIAH1 | Siah E3 Ubiquitin Protein Ligase 1 |
| SNPs | Single-nucleotide polymorphisms |
| TAF13. | TATA-Box-Binding Protein-Associated Factor 13 |
| TIAM1 | TIAM Rac1-Associated GEF 1 |
| TNNC1 | Troponin C1, Slow Skeletal and Cardiac Type |
| VEGFC | Vascular Endothelial Growth Factor C |
References
- Hu, Z.; Park, C.; Fritz-Waters, E.; Reecy, J. QTLdb: A Comprehensive Database Tool Building Bridges between Genotypes and Phenotypes. In Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, 1–6 August 2010. [Google Scholar]
- Ankra-Badu, G.A.; Le Bihan-Duval, E.; Mignon-Grasteau, S.; Pitel, F.; Beaumont, C.; Duclos, M.J.; Simon, J.; Carre, W.; Porter, T.E.; Vignal, A.; et al. Mapping QTL for growth and shank traits in chickens divergently selected for high or low body weight. Anim. Genet. 2010, 41, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, R.; Zheng, M.; Feng, F.; Liu, D.; Guo, Y.; Zhao, G.; Wen, J. New insights into the associations among feed efficiency, metabolizable efficiency traits and related QTL regions in broiler chickens. J. Anim. Sci. Biotechnol. 2020, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zheng, M.; Zhao, G.; Wang, J.; Liu, J.; Wang, S.; Feng, F.; Liu, D.; Zhu, D.; Li, Q.; et al. Identification of QTL regions and candidate genes for growth and feed efficiency in broilers. Genet. Sel. Evol. GSE 2021, 53, 13. [Google Scholar] [CrossRef]
- Xu, Z.; Ji, C.; Zhang, Y.; Zhang, Z.; Nie, Q.; Xu, J.; Zhang, D.; Zhang, X. Combination analysis of genome-wide association and transcriptome sequencing of residual feed intake in quality chickens. BMC Genom. 2016, 17, 594. [Google Scholar] [CrossRef]
- Paganoni, B.; Rose, G.; Macleay, C.; Jones, C.; Brown, D.J.; Kearney, G.; Ferguson, M.; Thompson, A.N. More feed efficient sheep produce less methane and carbon dioxide when eating high-quality pellets. J. Anim. Sci. 2017, 95, 3839–3850. [Google Scholar] [CrossRef][Green Version]
- Løvendahl, P.; Difford, G.F.; Li, B.; Chagunda, M.G.G.; Huhtanen, P.; Lidauer, M.H.; Lassen, J.; Lund, P. Review: Selecting for improved feed efficiency and reduced methane emissions in dairy cattle. Anim. Int. J. Anim. Biosci. 2018, 12, s336–s349. [Google Scholar] [CrossRef] [PubMed]
- Dumont, B.; Groot, J.C.J.; Tichit, M. Review: Make ruminants green again—How can sustainable intensification and agroecology converge for a better future? Anim. Int. J. Anim. Biosci. 2018, 12, s210–s219. [Google Scholar] [CrossRef]
- Koch, R.M.; Swiger, L.A.; Chambers, D.; Gregory, K.E. Efficiency of Feed Use in Beef Cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- Byerly, T.C.; Kessler, J.W.; Gous, R.M.; Thomas, O.P. Feed Requirements for Egg Production1. Poult. Sci. 1980, 59, 2500–2507. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Cell Biology Symposium: Genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 2013, 91, 1594–1613. [Google Scholar] [CrossRef]
- Cammack, K.M.; Leymaster, K.A.; Jenkins, T.G.; Nielsen, M.K. Estimates of genetic parameters for feed intake, feeding behavior, and daily gain in composite ram lambs. J. Anim. Sci. 2005, 83, 777–785. [Google Scholar] [CrossRef]
- Gunsett, F.C. Linear Index Selection to Improve Traits Defined as Ratios. J. Anim. Sci. 1984, 59, 1185–1193. [Google Scholar] [CrossRef]
- Van der Werf, J.H.J. Is it useful to define residual feed intake as a trait in animal breeding programs? Aust. J. Exp. Agric. 2004, 44, 405–409. [Google Scholar] [CrossRef]
- Furlotte, N.A.; Eskin, E.; Eyheramendy, S. Genome-wide association mapping with longitudinal data. Genet. Epidemiol. 2012, 36, 463–471. [Google Scholar] [CrossRef]
- Das, K.; Li, J.; Wang, Z.; Tong, C.; Fu, G.; Li, Y.; Xu, M.; Ahn, K.; Mauger, D.; Li, R.; et al. A dynamic model for genome-wide association studies. Hum. Genet. 2011, 129, 629–639. [Google Scholar] [CrossRef]
- Kim, S.; Xing, E.P. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 2009, 5, e1000587. [Google Scholar] [CrossRef]
- Dekkers, J.C. Commercial application of marker- and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004, 82 (Suppl. E), E313–E328. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Zhou, L.; Wei, J.; Liu, Y.; Kang, H.; Zhang, S.; Zhou, X.; Xu, S.; Liu, J.F. Efficient multivariate analysis algorithms for longitudinal genome-wide association studies. Bioinformatics 2019, 35, 4879–4885. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Kang, H.; Zhou, L.; Wang, D.; Wang, H.; Wang, A.; Fu, J.; Zhang, S.; Liu, J. Performance Gains in Genome-Wide Association Studies for Longitudinal Traits via Modeling Time-varied effects. Sci. Rep. 2017, 7, 590. [Google Scholar] [CrossRef] [PubMed]
- Auclair, D.; Garrel, D.R.; Chaouki Zerouala, A.; Ferland, L.H. Activation of the ubiquitin pathway in rat skeletal muscle by catabolic doses of glucocorticoids. Am. J. Physiol. 1997, 272, C1007–C1016. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Cho, J.E.; Fournier, M.; Da, X.; Lewis, M.I. Time course expression of Foxo transcription factors in skeletal muscle following corticosteroid administration. J. Appl. Physiol. 2010, 108, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, X.; Chen, D.; Yu, B.; Huang, Z. FoxO1: A novel insight into its molecular mechanisms in the regulation of skeletal muscle differentiation and fiber type specification. Oncotarget 2017, 8, 10662–10674. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.; Candau, R.B.; Bernardi, H. FoxO transcription factors: Their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 2014, 71, 1657–1671. [Google Scholar] [CrossRef] [PubMed]
- Kamei, Y.; Miura, S.; Suzuki, M.; Kai, Y.; Mizukami, J.; Taniguchi, T.; Mochida, K.; Hata, T.; Matsuda, J.; Aburatani, H.; et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004, 279, 41114–41123. [Google Scholar] [CrossRef]
- Kitamura, T.; Kitamura, Y.I.; Funahashi, Y.; Shawber, C.J.; Castrillon, D.H.; Kollipara, R.; DePinho, R.A.; Kitajewski, J.; Accili, D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 2007, 117, 2477–2485. [Google Scholar] [CrossRef]
- Léger, B.; Cartoni, R.; Praz, M.; Lamon, S.; Dériaz, O.; Crettenand, A.; Gobelet, C.; Rohmer, P.; Konzelmann, M.; Luthi, F.; et al. Akt signalling through GSK-3beta, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J. Physiol. 2006, 576, 923–933. [Google Scholar] [CrossRef]
- Yuan, Y.; Shi, X.E.; Liu, Y.G.; Yang, G.S. FoxO1 regulates muscle fiber-type specification and inhibits calcineurin signaling during C2C12 myoblast differentiation. Mol. Cell. Biochem. 2011, 348, 77–87. [Google Scholar] [CrossRef]
- O’Neill, B.T.; Bhardwaj, G.; Penniman, C.M.; Krumpoch, M.T.; Suarez Beltran, P.A.; Klaus, K.; Poro, K.; Li, M.; Pan, H.; Dreyfuss, J.M.; et al. FoxO Transcription Factors Are Critical Regulators of Diabetes-Related Muscle Atrophy. Diabetes 2018, 68, 556–570. [Google Scholar] [CrossRef]
- Caricasole, A.; Bettini, E.; Sala, C.; Roncarati, R.; Kobayashi, N.; Caldara, F.; Goto, K.; Terstappen, G.C. Molecular cloning and characterization of the human diacylglycerol kinase beta (DGKbeta) gene: Alternative splicing generates DGKbeta isotypes with different properties. J. Biol. Chem. 2002, 277, 4790–4796. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Süveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Trip, M.D.; Smulders, Y.M.; Wegman, J.J.; Hu, X.; Boer, J.M.; ten Brink, J.B.; Zwinderman, A.H.; Kastelein, J.J.; Feskens, E.J.; Bergen, A.A. Frequent mutation in the ABCC6 gene (R1141X) is associated with a strong increase in the prevalence of coronary artery disease. Circulation 2002, 106, 773–775. [Google Scholar] [CrossRef]
- Willis, S.A.; Bawden, S.J.; Malaikah, S.; Sargeant, J.A.; Stensel, D.J.; Aithal, G.P.; King, J.A. The role of hepatic lipid composition in obesity-related metabolic disease. Liver Int. Off. J. Int. Assoc. Study Liver 2021, 41, 2819–2835. [Google Scholar] [CrossRef]
- Tsukita, S.; Furuse, M.; Itoh, M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001, 2, 285–293. [Google Scholar] [CrossRef]
- Fox, C.S.; Heard-Costa, N.; Cupples, L.A.; Dupuis, J.; Vasan, R.S.; Atwood, L.D. Genome-wide association to body mass index and waist circumference: The Framingham Heart Study 100K project. BMC Med. Genet. 2007, 8 (Suppl. 1), S18. [Google Scholar] [CrossRef]
- Yang, Y.; Ahn, Y.H.; Gibbons, D.L.; Zang, Y.; Lin, W.; Thilaganathan, N.; Alvarez, C.A.; Moreira, D.C.; Creighton, C.J.; Gregory, P.A.; et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J. Clin. Investig. 2011, 121, 1373–1385. [Google Scholar] [CrossRef]
- Remke, M.; Hielscher, T.; Korshunov, A.; Northcott, P.A.; Bender, S.; Kool, M.; Westermann, F.; Benner, A.; Cin, H.; Ryzhova, M.; et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 3852–3861. [Google Scholar] [CrossRef]
- Dos Santos, F.C.; Peixoto, M.G.; Fonseca, P.A.; Pires, M.F.; Ventura, R.V.; Rosse, I.D.; Bruneli, F.A.; Machado, M.A.; Carvalho, M.R. Identification of Candidate Genes for Reactivity in Guzerat (Bos indicus) Cattle: A Genome-Wide Association Study. PLoS ONE 2017, 12, e0169163. [Google Scholar] [CrossRef]
- Herd, R.M.; Arthur, P.F. Physiological basis for residual feed intake1. J. Anim. Sci. 2009, 87, E64–E71. [Google Scholar] [CrossRef] [PubMed]
- Nkrumah, J.D.; Basarab, J.A.; Wang, Z.; Li, C.; Price, M.A.; Okine, E.K.; Crews, D.H., Jr.; Moore, S.S. Genetic and phenotypic relationships of feed intake and measures of efficiency with growth and carcass merit of beef cattle1. J. Anim. Sci. 2007, 85, 2711–2720. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Yu, Q.; Hui, R.; Reuhl, K.; Gale, N.W.; Zhou, R. EphA5 and EphA6: Regulation of neuronal and spine morphology. Cell Biosci. 2016, 6, 48. [Google Scholar] [CrossRef]
- D’Occhio, M.J.; Baruselli, P.S.; Campanile, G. Influence of nutrition, body condition, and metabolic status on reproduction in female beef cattle: A review. Theriogenology 2019, 125, 277–284. [Google Scholar] [CrossRef]
- Zampiga, M.; Flees, J.; Meluzzi, A.; Dridi, S.; Sirri, F. Application of omics technologies for a deeper insight into quali-quantitative production traits in broiler chickens: A review. J. Anim. Sci. Biotechnol. 2018, 9, 61. [Google Scholar] [CrossRef]
- Liu, R.; Xing, S.; Wang, J.; Zheng, M.; Cui, H.; Crooijmans, R.; Li, Q.; Zhao, G.; Wen, J. A new chicken 55K SNP genotyping array. BMC Genom. 2019, 20, 410. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinform. 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Chai, S.; Yang, Z.; Zhang, Q.; Xin, H.; Xu, Y.; Lin, S.; Chen, X.; Yao, Z.; et al. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat. Commun. 2022, 13, 682. [Google Scholar] [CrossRef]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Zheng, X.; Zhang, Q.; Zhang, S.; Mrode, R.; Liu, J.F. Eigen decomposition expedites longitudinal genome-wide association studies for milk production traits in Chinese Holstein. Genet. Sel. Evol. GSE 2018, 50, 12. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Mackay, T.F.D.S. Falconer and Introduction to quantitative genetics. Genetics 2004, 167, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]



| Traits (g/d) | N | Mean | SD | Max | Min | CV (%) |
|---|---|---|---|---|---|---|
| 72–81 ADG | 788 | 18.32 | 5.28 | 34.67 | 10 | 29 |
| 72–81 ADFI | 937 | 83.02 | 14.35 | 125 | 27.44 | 17 |
| 72–81 FCR | 787 | 4.92 | 1.31 | 11.36 | 1.44 | 27 |
| 72–81 RFI | 787 | −6.35 × 10−11 | 11.69 | 42.53 | −53.32 | / |
| 81–89 ADG | 1540 | 16.54 | 4.20 | 31.56 | 10 | 25 |
| 81–89 ADFI | 1877 | 73.18 | 10.23 | 104.67 | 41.67 | 14 |
| 81–89 FCR | 1530 | 4.74 | 1.20 | 9.36 | 1.83 | 25 |
| 81–89 RFI | 1530 | −1.05 × 10−11 | 8.27 | 31.33 | −33.97 | / |
| 89–113 ADG | 3698 | 32.57 | 13.78 | 74.67 | 6.65 | 42 |
| 89–113 ADFI | 3592 | 173.98 | 66.11 | 316.11 | 44.65 | 38 |
| 89–113 FCR | 3592 | 5.48 | 1.70 | 19.35 | 2.47 | 31 |
| 89–113 RFI | 3592 | 1.84 × 10−11 | 19.17 | 98.72 | −102.23 | / |
| 113–120 ADG | 1482 | 19.47 | 6.61 | 40.44 | 10 | 34 |
| 113–120 ADFI | 2071 | 79.93 | 25.53 | 155.78 | 17.67 | 32 |
| 113–120 FCR | 1481 | 4.75 | 1.31 | 13.47 | 0.89 | 28 |
| 113–120 RFI | 1481 | 2.30 × 10−11 | 17.33 | 70.56 | −85.70 | / |
| Type (Alphabetical Order) | Count | Percentage |
|---|---|---|
| 3_prime_UTR_variant | 782 | 0.009 |
| 5_prime_UTR_premature_start_codon_gain_variant | 25 | 0.00029 |
| 5_prime_UTR_variant | 125 | 0.00144 |
| downstream_gene_variant | 8382 | 0.09648 |
| intergenic_region | 16,828 | 0.1937 |
| intron_variant | 48,581 | 0.55919 |
| missense_variant | 268 | 0.00308 |
| non_coding_transcript_exon_variant | 558 | 0.00642 |
| splice_acceptor_variant | 3 | 0.00003 |
| splice_donor_variant | 1 | 0.00001 |
| splice_region_variant | 296 | 0.00341 |
| stop_gained | 2 | 0.00002 |
| synonymous_variant | 2832 | 0.0326 |
| upstream_gene_variant | 8195 | 0.09433 |
| Traits | GGA | Position | Allele | AF | Beta | p | PVE | Gene | Ensembl ID | CADD |
|---|---|---|---|---|---|---|---|---|---|---|
| 72–81 FCR | 11 | 1446218 | T/A | 0.039 | −0.7419951 | 3.14011 × 10−5 | 0.025887031 | CNOT1 | ENSGALG00000002301 | 4.12349 |
| 72–81 FCR | 1 | 67628914 | T/C | 0.35 | −0.3458966 | 2.14648 × 10−6 | 0.035913685 | ITPR2 | ENSGALG00000014071 | 0.00395 |
| 89–113 FCR | 6 | 22856589 | T/C | 0.071 | −0.3941262 | 1.37804 × 10−7 | 0.008497121 | ADGRA1 | ENSGALG00000007045 | 0.41839 |
| 89–113 FCR | 7 | 6630777 | G/T | 0.165 | 0.3401085 | 1.60176 × 10−8 | 0.009312092 | AHR1B | ENSGALG00000004322 | 0.16682 |
| 89–113 FCR | 5 | 3550350 | G/T | 0.384 | −0.4573349 | 2.92904 × 10−46 | 0.072729824 | ANO3 | ENSGALG00000013311 | 12.51524 |
| 89–113 FCR | 1 | 22514424 | A/C | 0.069 | 0.4141535 | 9.66446 × 10−7 | 0.007064396 | ATP6AP1 | ENSGALG00000008836 | 3.72873 |
| 89–113 FCR | 1 | 147455082 | G/A | 0.484 | 0.1868363 | 2.58236 × 10−6 | 0.006392649 | CLDN10 | ENSGALG00000019114 | 0.81705 |
| 89–113 FCR | 2 | 27469326 | A/C | 0.39 | −0.4518481 | 5.48257 × 10−45 | 0.07039052 | DGKB | ENSGALG00000033561 | 2.15319 |
| 89–113 FCR | 5 | 21980526 | G/A | 0.357 | 0.2408767 | 5.26196 × 10−8 | 0.008563368 | EXT2 | ENSGALG00000031542 | 0.77067 |
| 89–113 FCR | 1 | 171954946 | T/G | 0.159 | 0.2839152 | 9.51733 × 10−6 | 0.00574995 | FOXO1 | ENSGALG00000017034 | 2.88924 |
| 89–113 FCR | 1 | 26392940 | T/C | 0.461 | −0.3662863 | 6.27082 × 10−17 | 0.022365852 | FOXP2 | ENSGALG00000009424 | 1.46472 |
| 89–113 FCR | 1 | 177838774 | G/A | 0.388 | −0.4592473 | 1.88762 × 10−46 | 0.073054161 | GPR12 | ENSGALG00000017102 | 6.93379 |
| 89–113 FCR | 14 | 14152014 | A/G | 0.419 | −0.4196301 | 2.26874 × 10−37 | 0.056652498 | IFT140 | ENSGALG00000009318 | 5.986 |
| 89–113 FCR | 5 | 57035557 | T/C | 0.436 | −0.4399652 | 6.21205 × 10−41 | 0.062907477 | MDGA2 | ENSGALG00000012228 | 1.62758 |
| 89–113 FCR | 1 | 183820939 | A/C | 0.436 | 0.1851372 | 2.7826 × 10−6 | 0.006350488 | MMP27 | ENSGALG00000019060 | 1.43259 |
| 89–113 FCR | 6 | 28586738 | A/G | 0.169 | 0.2801326 | 2.83443 × 10−6 | 0.006415069 | NHLRC2 | ENSGALG00000008946 | 3.70314 |
| 89–113 FCR | 5 | 36810052 | C/T | 0.3 | 0.2674278 | 1.98998 × 10−8 | 0.009124893 | NKX2−1 | ENSGALG00000037632 | 1.05414 |
| 89–113 FCR | 1 | 196418182 | G/A | 0.259 | 0.3011632 | 4.69091 × 10−10 | 0.011214515 | P2RY6 | ENSGALG00000017327 | 1.49934 |
| 89–113 FCR | 5 | 298547 | A/G | 0.073 | −0.3716725 | 8.55429 × 10−6 | 0.006026163 | PGA5 | ENSGALG00000039242 | 2.29974 |
| 89–113 FCR | 1 | 195873238 | A/C | 0.408 | −0.4398747 | 1.07204 × 10−41 | 0.064351973 | POLD3 | ENSGALG00000017307 | 2.20877 |
| 89–113 FCR | 1 | 196207597 | G/C | 0.288 | 0.2623651 | 2.88412 × 10−8 | 0.008916544 | RAB6A | ENSGALG00000017320 | 4.86582 |
| 89–113 FCR | 1 | 170070606 | G/C | 0.162 | 0.3721619 | 2.05114 × 10−10 | 0.011752907 | RB1 | ENSGALG00000016997 | 2.29389 |
| 89–113 FCR | 25 | 2987929 | A/G | 0.519 | 0.4489232 | 2.64559 × 10−39 | 0.047156239 | SHC1 | ENSGALG00000039775 | 2.42158 |
| 89–113 FCR | 14 | 750730 | T/C | 0.388 | −0.4576204 | 3.2049 × 10−46 | 0.072646283 | SHISA9 | ENSGALG00000035799 | 2.72137 |
| 89–113 FCR | 5 | 30932965 | C/T | 0.271 | 0.2923482 | 4.41261 × 10−9 | 0.009977342 | SPRED1 | ENSGALG00000028203 | 0.64103 |
| 89–113 FCR | 1 | 46624790 | A/G | 0.353 | 0.2596588 | 1.599 × 10−9 | 0.010485056 | TMPO | ENSGALG00000011504 | 6.83232 |
| 89–113 FCR | 3 | 52101965 | G/A | 0.066 | 0.4624581 | 1.73306 × 10−7 | 0.00804205 | TULP4 | ENSGALG00000037377 | 1.36685 |
| 89–113 FCR | 2 | 1951183 | G/C | 0.161 | 0.3088039 | 1.62009 × 10−7 | 0.008010282 | VIPR1 | ENSGALG00000005259 | 7.10688 |
| 89–113 FCR | 2 | 10466524 | A/C | 0.111 | 0.3675874 | 7.28495 × 10−8 | 0.008488573 | WDR37 | ENSGALG00000006749 | 0.2238 |
| 89–113 FCR | 5 | 5671784 | A/G | 0.396 | −0.4519878 | 1.24313 × 10−44 | 0.069699203 | WT1 | ENSGALG00000012115 | 6.0281 |
| 113–120 FCR | 14 | 8104013 | G/A | 0.411 | 0.2407186 | 6.91816 × 10−6 | 0.014425553 | ABCC6 | ENSGALG00000038152 | 1.79817 |
| 113–120 FCR | 1 | 106979821 | C/T | 0.297 | −0.2535606 | 1.25899 × 10−5 | 0.015346278 | MRPS6 | ENSGALG00000027579 | 0.25737 |
| 81–89 RFI | 18 | 2827691 | T/C | 0.101 | −2.33695 | 2.09453 × 10−5 | 0.013461153 | HS3ST3B1 | ENSGALG00000001371 | 2.81062 |
| 81–89 RFI | 5 | 52305921 | A/T | 0.397 | −1.471009 | 1.36042 × 10−5 | 0.014130899 | JAG2 | ENSGALG00000011696 | 3.88963 |
| 81–89 RFI | 2 | 110316845 | T/C | 0.307 | −1.470263 | 3.34978 × 10−5 | 0.01283052 | RGS20 | ENSGALG00000025941 | 0.47986 |
| 113–120 RFI | 2 | 123744069 | G/A | 0.375 | −2.928533 | 2.74233 × 10−5 | 0.013398714 | MMP16 | ENSGALG00000032031 | 1.16648 |
| 113–120 RFI | 1 | 106979821 | C/T | 0.297 | −0.2535606 | 5.87375 × 10−6 | 0.015654765 | MRPS6 | ENSGALG00000027579 | 0.25737 |
| 113–120 RFI | 1 | 107037036 | G/A | 0.251 | −3.274938 | 3.72438 × 10−5 | 0.01294475 | MRPS6 | ENSGALG00000027579 | 1.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, N.; Liu, P.; Wei, L.; Wen, J.; Zhao, G.; An, B. Identifying New Loci and Genes Associated with Feed Efficiency in Broilers. Int. J. Mol. Sci. 2025, 26, 8492. https://doi.org/10.3390/ijms26178492
Luo N, Liu P, Wei L, Wen J, Zhao G, An B. Identifying New Loci and Genes Associated with Feed Efficiency in Broilers. International Journal of Molecular Sciences. 2025; 26(17):8492. https://doi.org/10.3390/ijms26178492
Chicago/Turabian StyleLuo, Na, Peihao Liu, Limin Wei, Jie Wen, Guiping Zhao, and Bingxing An. 2025. "Identifying New Loci and Genes Associated with Feed Efficiency in Broilers" International Journal of Molecular Sciences 26, no. 17: 8492. https://doi.org/10.3390/ijms26178492
APA StyleLuo, N., Liu, P., Wei, L., Wen, J., Zhao, G., & An, B. (2025). Identifying New Loci and Genes Associated with Feed Efficiency in Broilers. International Journal of Molecular Sciences, 26(17), 8492. https://doi.org/10.3390/ijms26178492





