Radiation Modifies Let-7 miRNA Binding to AGO2 Independent of Changes in Transcription to Influence Tumor Cell Radiosensitivity
Abstract
1. Introduction
2. Results
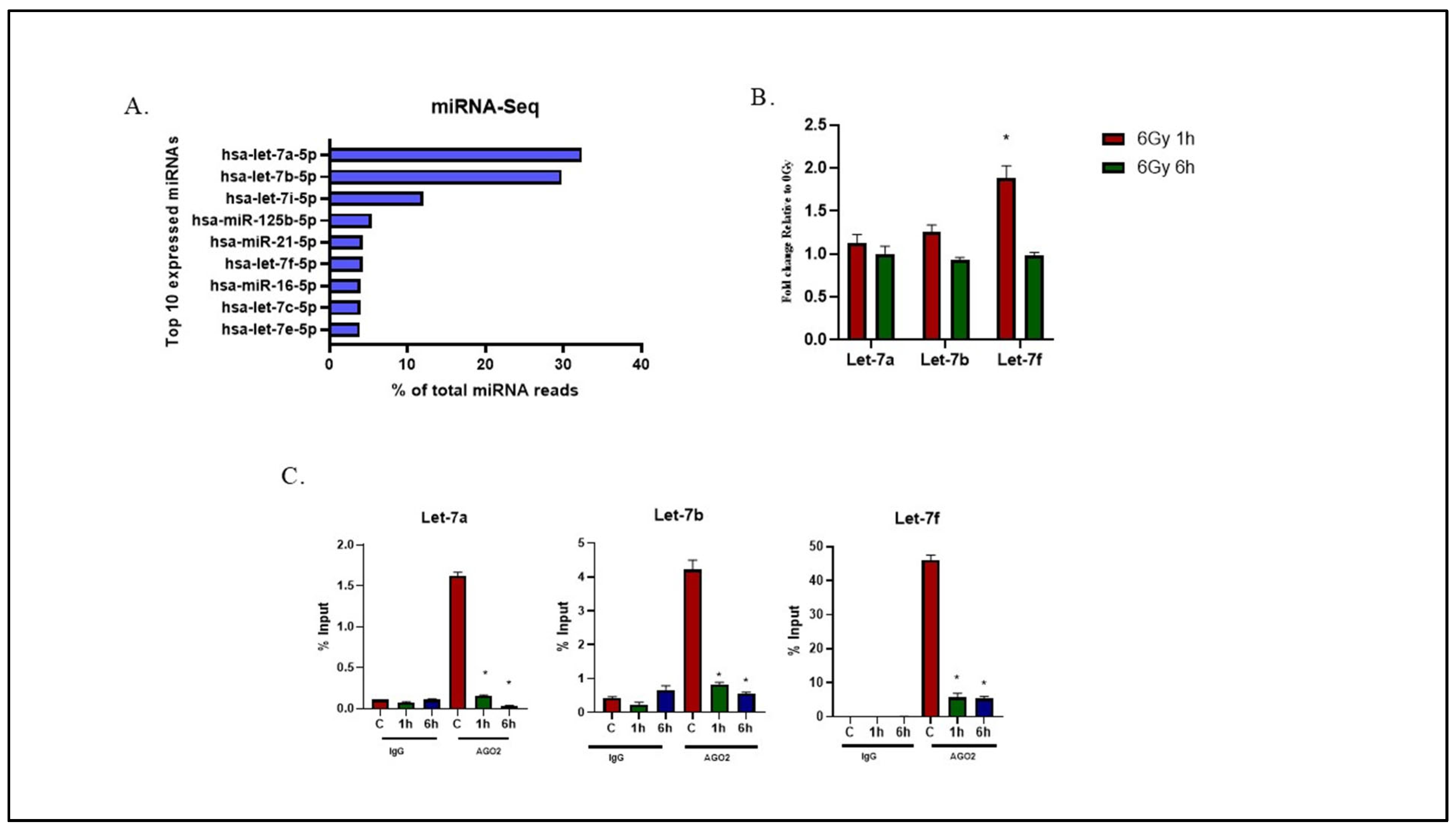
2.1. Effects of Radiation on Let-7 miRNA Expression and AGO2 Binding in U-251 Glioma Cells
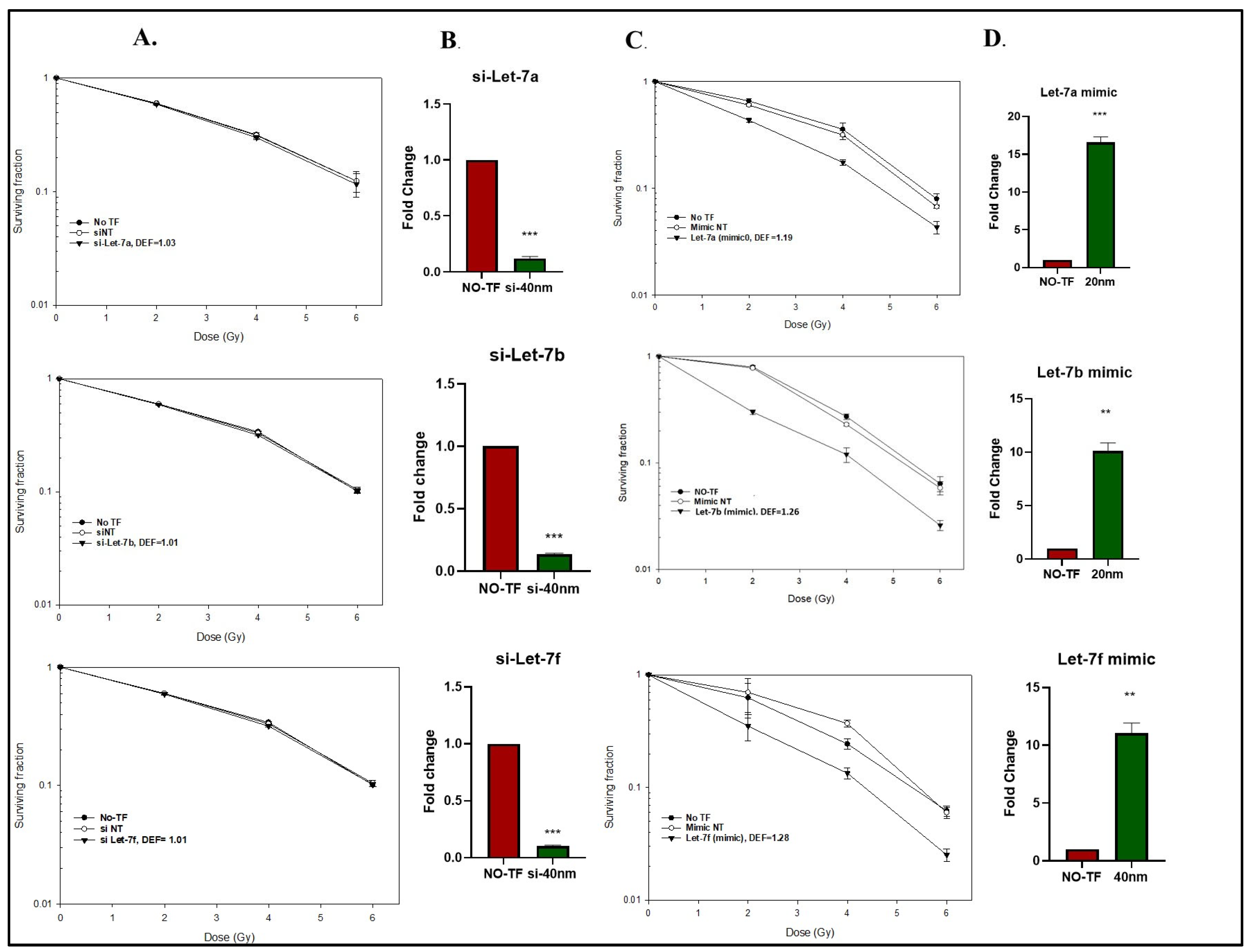
2.2. Modifying Let-7 Expression and U-251 Cell Radiosensitivity
2.3. Mechanisms Mediating Radio Sensitization Induced by Let-7a Mimic
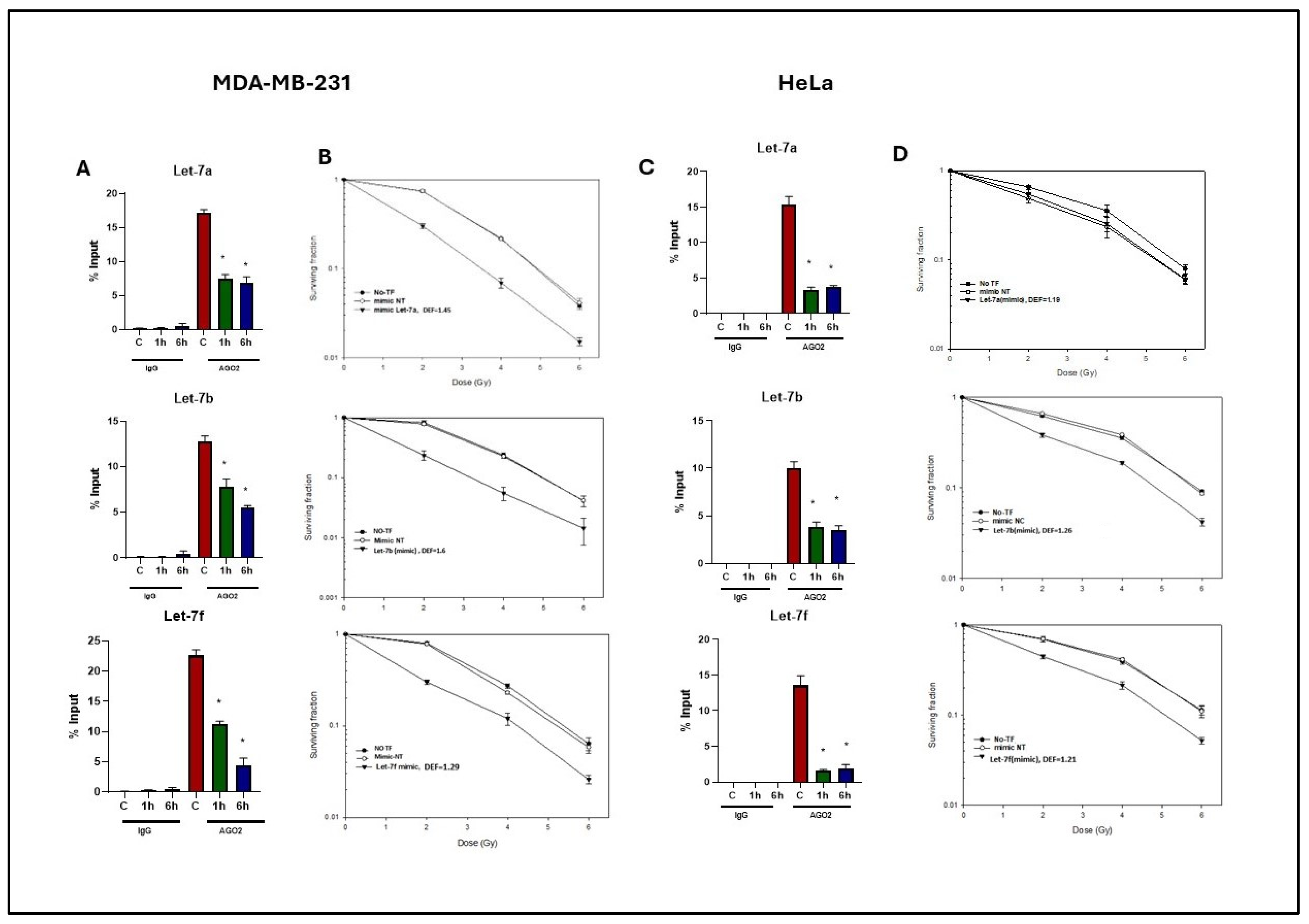
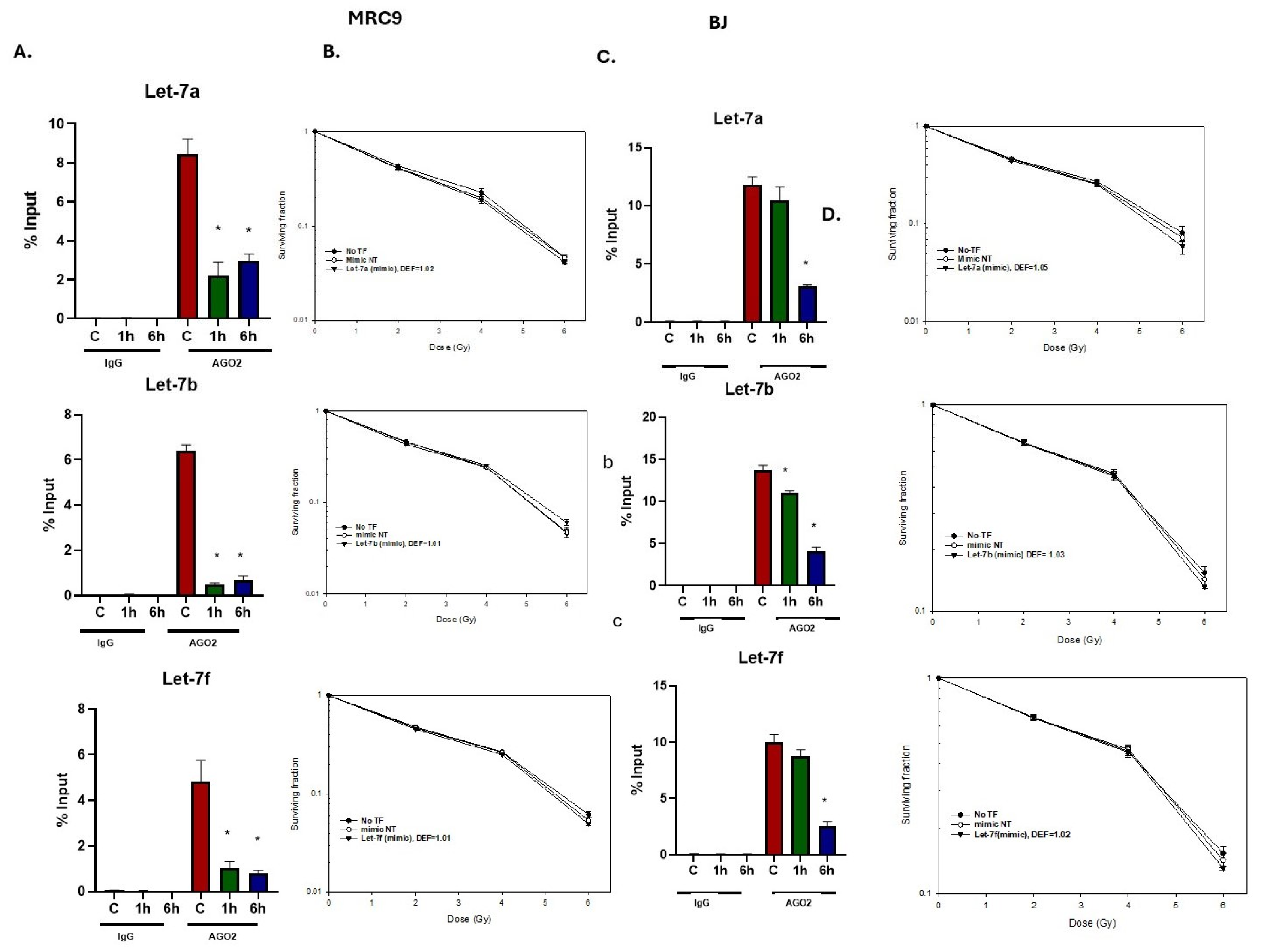
2.4. Effects of Radiation on Let-7-AGO2 Binding in Other Tumor and Normal Cell Lines
3. Discussion
4. Methods
4.1. Cell Culture
4.2. miRNA Sequencing
4.3. Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.4. siRNA and miRNA Mimic Transfection
4.5. AGO2-RNA Immunoprecipitation Assay (RIP)
4.6. Clonogenic Survival
4.7. γH2AX Foci
4.8. Flow Cytometry
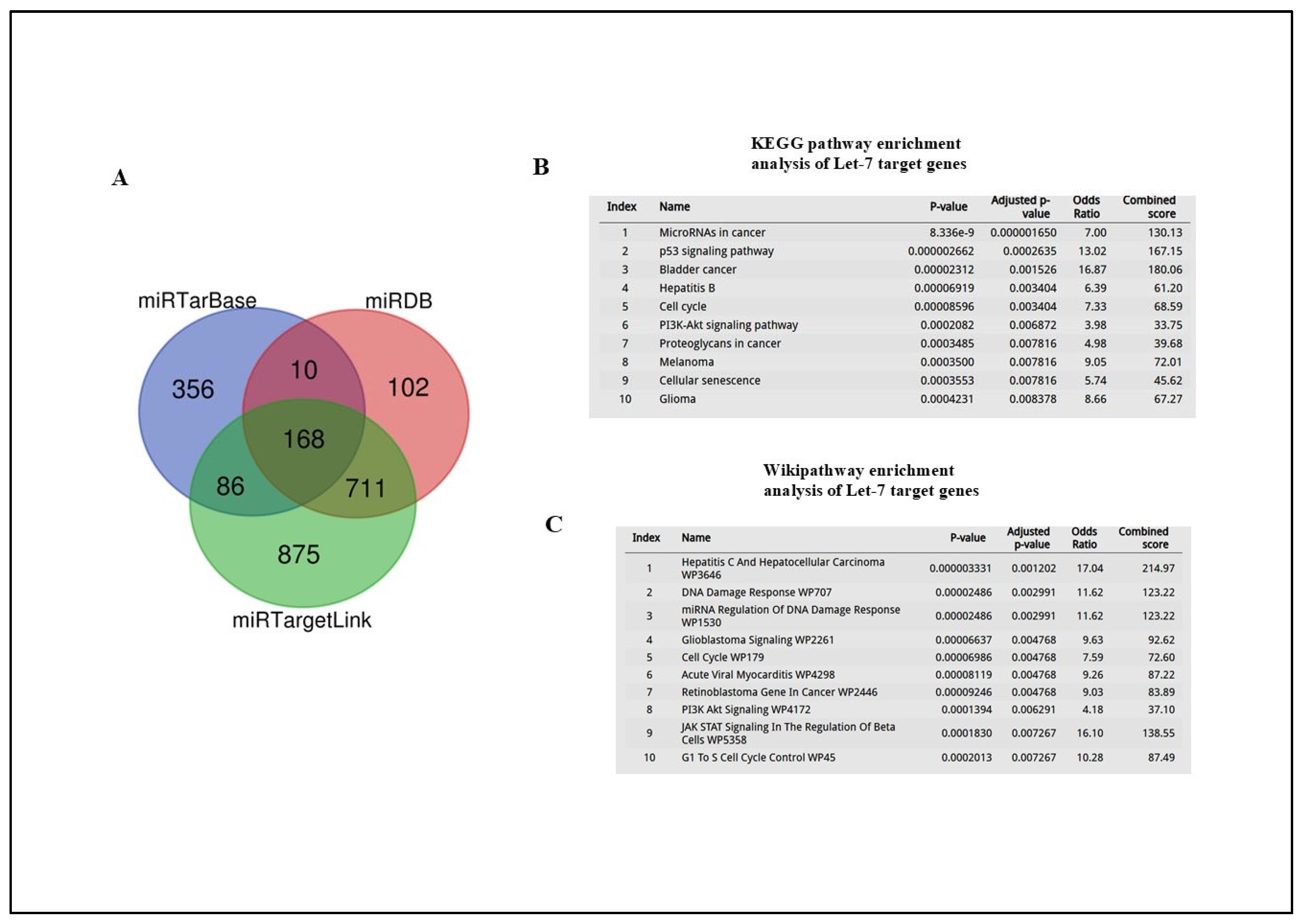
4.9. miRNA Target Prediction Databases and Pathway Enrichment Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institution Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moding, E.J.; Kastan, M.B.; Kirsch, D.G. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat. Rev. Drug Discov. 2013, 12, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 26, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Gao, P.; Yang, H.; Hu, J.; Xiao, J.J.; Shi, M.; Zhao, L.N. Inhibition of cell proliferation and radioresistance by miR-383-5p through targeting RNA binding protein motif (RBM3) in nasopharyngeal carcinoma. Ann. Transl. Med. 2021, 9, 123. [Google Scholar] [CrossRef]
- Javanmardi, S.; Aghamaali, M.R.; Abolmaali, S.S.; Mohammadi, S.; Tamaddon, A.M. miR-21, An Oncogenic Target miRNA for Cancer Therapy: Molecular Mechanisms and Recent Advancements in Chemo and Radio-resistance. Curr. Gene Ther. 2017, 16, 375–389. [Google Scholar] [CrossRef]
- Aryankalayil, M.J.; Chopra, S.; Makinde, A.; Eke, I.; Levin, J.; Shankavaram, U.; MacMillan, L.; Vanpouille-Box, C.; Demaria, S.; Coleman, C.N. Microarray analysis of miRNA expression profiles following whole body irradiation in a mouse model. Biomarkers 2018, 23, 689–703. [Google Scholar] [CrossRef]
- Wang, X.-C.; Wang, W.; Zhang, Z.-B.; Zhao, J.; Tan, X.-G.; Luo, J.-C. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat. Oncol. 2013, 8, 146. [Google Scholar]
- Pogosova-Agadjanyan, E.L.; Fan, W.; Georges, G.E.; Schwartz, J.L.; Kepler, C.M.; Lee, H.; Suchanek, A.; Cronk, M.R.; Brumbaugh, A.; Engel, J.H.; et al. Identification of radiation-induced expression changes in nonimmortalized human T cells. Radiat. Res. 2011, 175, 72–84. [Google Scholar] [CrossRef]
- Becker, W.R.; Ober-Reynolds, B.; Jouravleva, K.; Jolly, S.M.; Zamore, P.D.; Greenleaf, W.J. High-Throughput Analysis Reveals Rules for Target RNA Binding and Cleavage by AGO2. Mol. Cell 2019, 75, 741–755. [Google Scholar] [CrossRef]
- Tomasetti, M.; Monaco, F.; Rubini, C.; Rossato, M.; De Quattro, C.; Beltrami, C.; Sollini, G.; Pasquini, E.; Amati, M.; Goteri, G.; et al. AGO2-RIP-Seq reveals miR-34/miR-449 cluster targetome in sinonasal cancers. PLoS ONE 2024, 19, e0295997. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, X.; Pestell, T.G.; Fan, C.; Qin, S.; Mirabelli, E.; Ren, H.; Pestell, R.G. MicroRNAs and cancer stem cells: The sword and the shield. Oncogene 2014, 33, 4967–4977. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Weinstein, E.G.; Abdelhakim, A.; Yekta, S.; Rhoades, M.W.; Burge, C.B.; Bartel, D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003, 17, 991–1008. [Google Scholar] [CrossRef]
- Büssing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Slack, F.J. Potential microRNA therapies targeting Ras, N. FkappaB and p53 signaling. Curr. Opin. Mol. Ther. 2010, 12, 145–157. [Google Scholar]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Szkanderova, S.; Port, M.; Stulik, J.; Hernychova, L.; Kasalova, I.; Van Beuningen, D.; Abend, M. Comparison of the abundance of 10 radiation-induced proteins with their differential gene expression in L929 cells. Int. J. Radiat. Biol. 2003, 79, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Lü, X.; de la Pena, L.; Barker, C.; Camphausen, K.; Tofilon, P.J. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006, 66, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Wee, L.M.; Flores-Jasso, C.F.; Salomon, W.E.; Zamore, P.D. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 2012, 151, 1055–1067. [Google Scholar] [CrossRef]
- Gandellini, P.; Rancati, T.; Valdagni, R.; Zaffaroni, N. MiRNAs in Tumor Radiation Response: Bystanders or Participants? Trends Mol. Med. 2014, 20, 529–539. [Google Scholar] [CrossRef]
- Kura, B.; Pavelkova, P.; Kalocayova, B.; Pobijakova, M.; Slezak, J. MicroRNAs as Regulators of Radiation-Induced Oxidative Stress. Curr. Issues Mol. Biol. 2024, 46, 7097–7113. [Google Scholar] [CrossRef]
- Karginov, F.V.; Hannon, G.J. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013, 27, 1624–1632. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 302. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Patel, V.L.; Mitra, S.; Harris, R.; Buxbaum, A.R.; Lionnet, T.; Brenowitz, M.; Girvin, M.; Levy, M.; Almo, S.C.; Singer, R.H.; et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012, 26, 43–53. [Google Scholar] [CrossRef]
- Zhuo, Z.; Wan, Y.; Guan, D.; Ni, S.; Wang, L.; Zhang, Z.; Liu, J.; Liang, C.; Yu, Y.; Lu, A.; et al. A Loop-Based and AGO-Incorporated Virtual Screening Model Targeting AGO-Mediated miRNA-mRNA Interactions for Drug Discovery to Rescue Bone Phenotype in Genetically Modified Mice. Adv. Sci. 2020, 7, 1903451. [Google Scholar] [CrossRef]
- Fuentes-Iglesias, A.; Garcia-Outeiral, V.; Pardavila, J.A.; Wang, J.; Fidalgo, M.; Guallar, D. An Optimized Immunoprecipitation Protocol for Assessing Protein-RNA Interactions In Vitro. STAR Protoc. 2020, 1, 100093. [Google Scholar] [CrossRef]
- Wang, X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. Rna 2008, 14, 1012–1017. [Google Scholar] [CrossRef]
- Cui, S.; Yu, S.; Huang, H.Y.; Lin, Y.C.; Huang, Y.; Zhang, B.; Xiao, J.; Zuo, H.; Wang, J.; Li, Z.; et al. miRTarBase 2025: Updates to the collection of experimentally validated microRNA-target interactions. Nucleic Acids Res. 2025, 53, 147–156. [Google Scholar] [CrossRef]
- Hamberg, M.; Backes, C.; Fehlmann, T.; Hart, M.; Meder, B.; Meese, E.; Keller, A. MiRTargetLink—miRNAs, Genes and Interaction Networks. Int. J. Mol. Sci. 2016, 17, 564. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, 90–97. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, T.; Degorre, C.; Tofilon, P.J. Radiation Modifies Let-7 miRNA Binding to AGO2 Independent of Changes in Transcription to Influence Tumor Cell Radiosensitivity. Int. J. Mol. Sci. 2025, 26, 8483. https://doi.org/10.3390/ijms26178483
Ali T, Degorre C, Tofilon PJ. Radiation Modifies Let-7 miRNA Binding to AGO2 Independent of Changes in Transcription to Influence Tumor Cell Radiosensitivity. International Journal of Molecular Sciences. 2025; 26(17):8483. https://doi.org/10.3390/ijms26178483
Chicago/Turabian StyleAli, Taqveema, Charlotte Degorre, and Philip J. Tofilon. 2025. "Radiation Modifies Let-7 miRNA Binding to AGO2 Independent of Changes in Transcription to Influence Tumor Cell Radiosensitivity" International Journal of Molecular Sciences 26, no. 17: 8483. https://doi.org/10.3390/ijms26178483
APA StyleAli, T., Degorre, C., & Tofilon, P. J. (2025). Radiation Modifies Let-7 miRNA Binding to AGO2 Independent of Changes in Transcription to Influence Tumor Cell Radiosensitivity. International Journal of Molecular Sciences, 26(17), 8483. https://doi.org/10.3390/ijms26178483





