Comprehensive Characterisation of Extracellular Vesicle Preparations Using Multiparametric Size-Exclusion Chromatography
Abstract
1. Introduction
2. Results
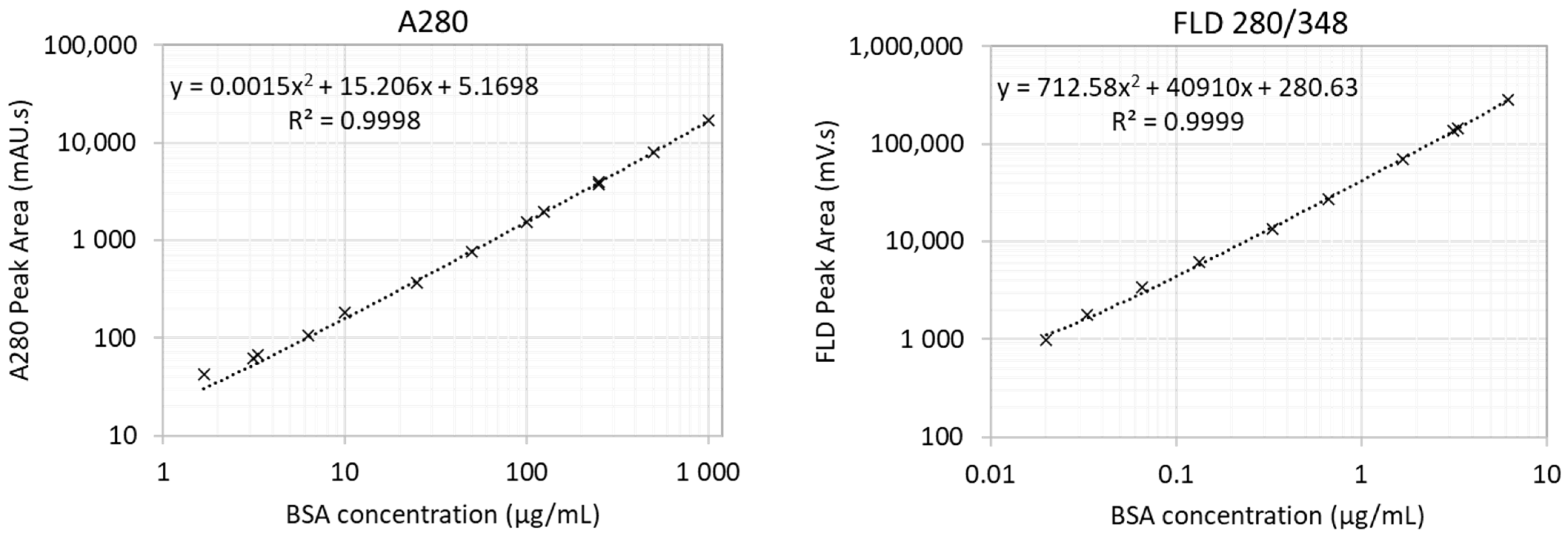
2.1. SEC Analysis and Characterisation Strategy
2.2. Robustness of Analysis
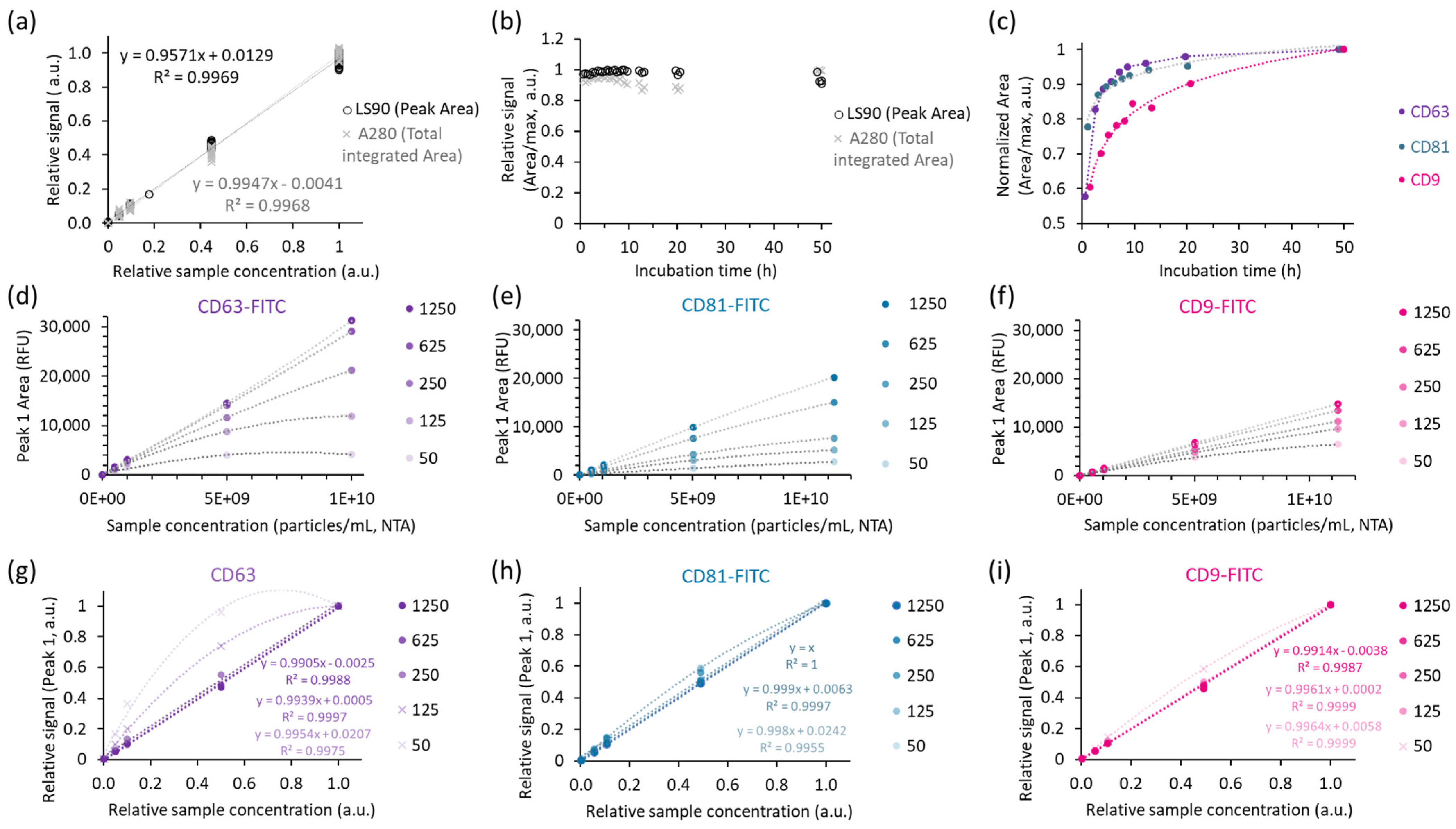
2.2.1. Assuring Reproducible Immunolabelling and Quantitation
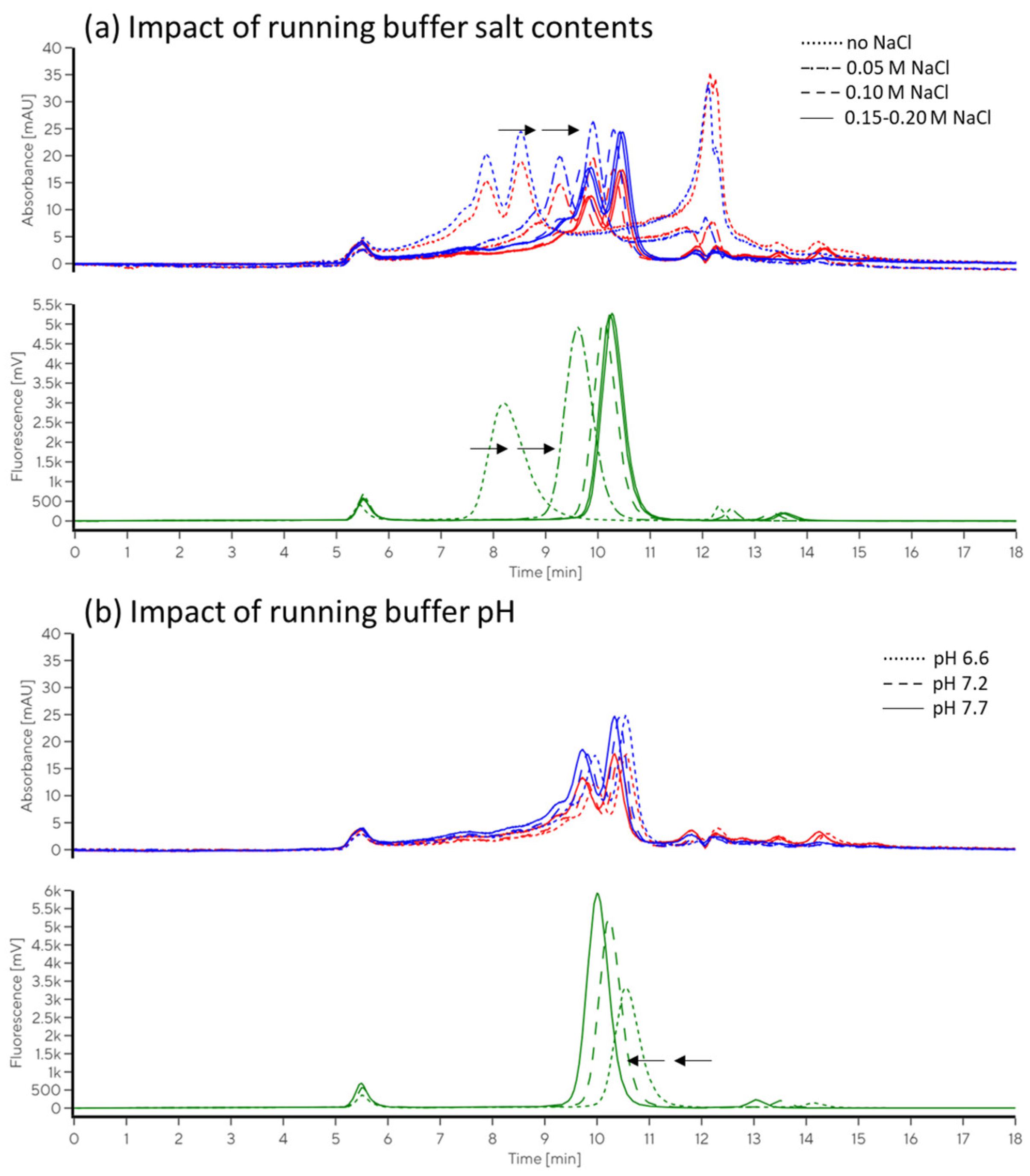
2.2.2. Impacts of Running Buffer Composition
2.2.3. Evaluation of Sample Changes After Exposure to Different Conditions and Immunolabelling Efficiency
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Chromatography
4.3. Nanoparticle Tracking Analysis
4.4. Protein Analysis
4.5. Design of Experiments for Method Optimisation and Identification of Critical Parameters for Repeatable Assay Readout
4.5.1. Sample and Antibody Titration
4.5.2. Evaluation of the Impact of Incubation Time
4.5.3. Evaluation of the Impact of Running Buffer Composition
4.5.4. Evaluation of the Impacts of Salt and pH on Immunolabelling Efficiency
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| A260 | Absorbance at a wavelength of 260 nm |
| A280 | Absorbance at a wavelength of 280 nm |
| BSA | Bovine serum albumin |
| CCM | Clarified conditioned medium |
| dPBS | Dulbecco’s phosphate-buffered saline |
| EV | Extracellular vesicle |
| FLD | Fluorescence |
| FLD 280/348 | Fluorescence signal using excitation at 280 nm and emission detection at 348 nm |
| LS | Light scattering |
| LS90 | Light scattering at a 90° angle |
| MALS | Multi-angle light scattering |
| MSC | Mesenchymal stem cell |
| NTA | Nanoparticle tracking analysis |
| RAU | Relative absorbance units |
| RET | Retentate |
| RFU | Relative fluorescence units |
| RSU | Relative scattering units |
| SEC | Size-exclusion chromatography |
| TFF | Tangential flow filtration |
| TRP | Tryptophan autofluorescence |
Appendix A
| A | B | C | D | |
|---|---|---|---|---|
| 1 | 50 mM HEPES pH 6.8 | 50 mM HEPES pH 6.8, 0.2% poloxamer | 600 mM NaCl | ddH2O |
| 2 | 50 mM HEPES pH 8, 0.2% poloxamer | 50 mM HEPES pH 8 | ||
| 3 | 50 mM MES pH 5.5 | 50 mM MES pH 5.5, 0.2% poloxamer | ||
| 4 | 50 mM MES pH 6.5, 0.2% poloxamer | 50 mM MES, pH 6.5 | ||
| 5 | 50 mM KP, pH 6.4 | 50 mM KP, pH 6.4, 0.2% poloxamer | ||
| 6 | 50 mM KP pH 7.8, 0.2% poloxamer | 50 mM KP, pH 7.8 | ||
| 7 | Same as B1 | Same as A6 | ||
| 8 | Same as B3 | dPBS |
| No. ID | Buffer | A | B | C | D | NaCl, mM | Poloxamer % | pH | Cond. | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1, 41 | dPBS | - | 0% | B8 | 100% | 0% | 0% | 150 | 0 | 7.4 | 15.4 |
| 2 | HEPES | A1 | 50% | - | 0% | 0% | 50% | 0 | 0 | 6.6 | 0.15 |
| 3 | HEPES | A2 | 50% | - | 0% | 50% | 0% | 300 | 0.1 | 7.8 | 29.3 |
| 4 | HEPES | A2 | 35% | B2 | 15% | 8% | 42% | 48 | 0.07 | 7.9 | 6.2 |
| 5 | HEPES | A2 | 16% | B2 | 34% | 27% | 23% | 162 | 0.03 | 7.8 | 17.2 |
| 6 | HEPES | - | 0% | B2 | 50% | 25% | 25% | 150 | 0 | 7.8 | 16.1 |
| 7 | HEPES | A1 | 25% | B2 | 25% | 50% | 0% | 300 | 0 | 6.8 | 26 |
| 8 | HEPES | A7 | 25% | B2 | 25% | 17% | 33% | 102 | 0.05 | 7.3 | 11.1 |
| 9 | HEPES | A7 | 30% | B2 | 20% | 28% | 22% | 168 | 0.06 | 7.2 | 17.5 |
| 10 | HEPES | A7 | 25% | B2 | 25% | 26% | 24% | 156 | 0.05 | 7.2 | 16.6 |
| 11 | HEPES | A7 | 25% | B2 | 25% | 32% | 18% | 192 | 0.05 | 7.2 | 20 |
| 12 | HEPES | A2 | 25% | B1 | 25% | 0% | 50% | 0 | 0.1 | 7.3 | 0.6 |
| 13 | HEPES | A2 | 25% | B1 | 25% | 22% | 28% | 132 | 0.1 | 7.3 | 13.8 |
| 14 | HEPES | - | 0% | B1 | 50% | 25% | 25% | 150 | 0.1 | 6.6 | 15 |
| 15 | HEPES | A1 | 25% | B1 | 25% | 50% | 0% | 300 | 0.05 | 6.6 | 28.4 |
| 16 | HEPES | A1 | 50% | - | 0% | 2% | 48% | 12 | 0 | 6.7 | 2 |
| 17 | MES | A3 | 50% | - | 0% | 10% | 40% | 60 | 0 | 5.4 | 6.6 |
| 18 | MES | A3 | 25% | B3 | 25% | 50% | 0% | 300 | 0.05 | 5.4 | 28.4 |
| 19 | MES | - | 0% | B3 | 50% | 25% | 25% | 150 | 0.1 | 5.3 | 15.1 |
| 20 | MES | - | 0% | B3 | 50% | 42% | 8% | 252 | 0.1 | 5.3 | 24.5 |
| 21 | MES | A4 | 25% | B3 | 25% | 12% | 38% | 72 | 0.1 | 5.9 | 8.15 |
| 22 | MES | A8 | 15% | B4 | 35% | 21% | 29% | 126 | 0.03 | 6 | 13.4 |
| 23 | MES | A8 | 30% | B4 | 20% | 18% | 32% | 108 | 0.06 | 5.8 | 11.6 |
| 24 | MES | A3 | 25% | B4 | 25% | 35% | 15% | 210 | 0 | 5.9 | 21.4 |
| 25 | MES | A3 | 50% | - | 0% | 25% | 25% | 150 | 0 | 5.7 | 15.7 |
| 26 | MES | - | 0% | B4 | 50% | 23% | 27% | 138 | 0 | 6.3 | 15 |
| 27 | MES | A4 | 20% | B4 | 30% | 15% | 35% | 90 | 0 | 6.4 | 10.3 |
| 28 | MES | A4 | 50% | - | 0% | 5% | 45% | 30 | 0 | 6.4 | 4.5 |
| 29 | Phosphate | - | 0% | B5 | 50% | 21% | 29% | 126 | 0.1 | 6.6 | 14.7 |
| 30 | Phosphate | A5 | 35% | B5 | 15% | 4% | 46% | 24 | 0.03 | 6.5 | 5.5 |
| 31 | Phosphate | A5 | 35% | B5 | 15% | 45% | 5% | 270 | 0.03 | 6.2 | 28.2 |
| 32 | Phosphate | A5 | 50% | - | 0% | 25% | 25% | 150 | 0 | 6.2 | 17.8 |
| 33 | Phosphate | A5 | 25% | B6 | 25% | 5% | 45% | 30 | 0 | 6.9 | 7 |
| 34 | Phosphate | A5 | 25% | B7 | 25% | 32% | 18% | 192 | 0.05 | 6.7 | 22.3 |
| 35 | Phosphate | A6 | 25% | B5 | 25% | 20% | 30% | 120 | 0.1 | 7 | 15.15 |
| 36 | Phosphate | A6 | 50% | - | 0% | 35% | 15% | 210 | 0.1 | 7.3 | 24.2 |
| 37 | Phosphate | A6 | 50% | - | 0% | 50% | 0% | 300 | 0.1 | 7.2 | 32 |
| 38 | Phosphate | A6 | 10% | B6 | 40% | 12% | 38% | 72 | 0.02 | 7.44 | 11.6 |
| 39 | Phosphate | - | 0% | B6 | 50% | 28% | 22% | 168 | 0 | 7.3 | 21 |
| 40 | Phosphate | A5 | 20% | B6 | 30% | 40% | 10% | 240 | 0 | 6.8 | 26.8 |
References
- Couch, Y.; Buzàs, E.I.; Di Vizio, D.; Gho, Y.S.; Harrison, P.; Hill, A.F.; Lötvall, J.; Raposo, G.; Stahl, P.D.; Théry, C.; et al. A brief history of nearly EV-erything—The rise and rise of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12144. [Google Scholar] [CrossRef]
- Zocco, D. Extracellular vesicles: Setting your path to IND with advanced characterization package. Cell Gene Ther. Insights 2024, 10, 1273–1287. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’DRiscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404, Erratum in J. Extracell. Vesicles 2024, 13, e12451. https://doi.org/10.1002/jev2.12451. [Google Scholar] [CrossRef]
- Xu, R.; Fitts, A.; Li, X.; Fernandes, J.; Pochampally, R.; Mao, J.; Liu, Y.-M. Quantification of Small Extracellular Vesicles by Size Exclusion Chromatography with Fluorescence Detection. Anal. Chem. 2016, 88, 10390–10394. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; He, J. Characterization of Extracellular Vesicles by Size-Exclusion High-Performance Liquid Chromatography (HPLC). Methods Mol. Biol. 2017, 1660, 191–199. [Google Scholar] [CrossRef]
- Kitka, D.; Mihály, J.; Fraikin, J.-L.; Beke-Somfai, T.; Varga, Z. Detection and phenotyping of extracellular vesicles by size exclusion chromatography coupled with on-line fluorescence detection. Sci. Rep. 2019, 9, 19868. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jalaludin, I.; Hwang, H.; Ko, M.; Adelipour, M.; Hwan, M.; Cho, N.; Kim, K.K.; Lubman, D.M.; Kim, J. Size-exclusion chromatography for the characterization of urinary extracellular vesicles. J. Chromatogr. B 2023, 1228, 123828. [Google Scholar] [CrossRef]
- Normak, K.; Papp, M.; Ullmann, M.; Paganini, C.; Manno, M.; Bongiovanni, A.; Bergese, P.; Arosio, P. Multiparametric Orthogonal Characterization of Extracellular Vesicles by Liquid Chromatography Combined with In-Line Light Scattering and Fluorescence Detection. Anal. Chem. 2023, 95, 12443–12451. [Google Scholar] [CrossRef]
- Lai, J.J.; Hill, J.J.; Huang, C.Y.; Lee, G.C.; Mai, K.W.; Shen, M.Y.; Wang, S.K. Unveiling the Complex World of Extracellular Vesicles: Novel Characterization Techniques and Manufacturing Considerations. Chonnam Med. J. 2024, 60, 1–12. [Google Scholar] [CrossRef]
- Kolenc, A.; Maličev, E. Current Methods for Analysing Mesenchymal Stem Cell-Derived Extracellular Vesicles. Int. J. Mol. Sci. 2024, 25, 3439. [Google Scholar] [CrossRef]
- Takakura, Y.; Hanayama, R.; Akiyoshi, K.; Futaki, S.; Hida, K.; Ichiki, T.; Ishii-Watabe, A.; Kuroda, M.; Maki, K.; Miura, Y.; et al. Quality and Safety Considerations for Therapeutic Products Based on Extracellular Vesicles. Pharm. Res. 2024, 41, 1573–1594. [Google Scholar] [CrossRef]
- Aitken, A.; Learmonth, M.P. Protein Determination by UV Absorption. In The Protein Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks: Totowa, NJ, USA, 2009; pp. 3–6. [Google Scholar] [CrossRef]
- Compton, S.J.; Jones, C.G. Mechanism of dye response and interference in the Bradford protein assay. Anal. Biochem. 1985, 151, 369–374. [Google Scholar] [CrossRef]
- Kopaciewicz, W.; Regnier, F. Nonideal size-exclusion chromatography of proteins: Effects of pH at low ionic strength. Anal. Biochem. 1982, 126, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Fekete, S.; Beck, A.; Veuthey, J.-L.; Guillarme, D. Theory and practice of size exclusion chromatography for the analysis of protein aggregates. J. Pharm. Biomed. Anal. 2014, 101, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Mäntele, W.; Deniz, E. UV–VIS absorption spectroscopy: Lambert-Beer reloaded. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 173, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Eremin, Y.A. SCATTERING|Scattering Theory. In Encyclopedia of Modern Optics; Elsevier: Amsterdam, The Netherlands, 2005; pp. 326–330. [Google Scholar] [CrossRef]
- Lee, H.J.; McAuley, A.; Schilke, K.F.; McGuire, J. Molecular origins of surfactant-mediated stabilization of protein drugs. Adv. Drug Deliv. Rev. 2011, 63, 1160–1171. [Google Scholar] [CrossRef]
- Chang, D.; Fox, R.; Hicks, E.; Ferguson, R.; Chang, K.; Osborne, D.; Hu, W.; Velev, O.D. Investigation of interfacial properties of pure and mixed poloxamers for surfactant-mediated shear protection of mammalian cells. Colloids Surf. B Biointerfaces 2017, 156, 358–365. [Google Scholar] [CrossRef]
- Desai, R.; Jain, R.; Dandekar, P. Surfactants reduce aggregation of monoclonal antibodies in cell culture medium with improvement in performance of mammalian cell culture. Biotechnol. Prog. 2023, 39, e3370. [Google Scholar] [CrossRef]
- Trenkenschuh, E.; Richter, M.; Heinrich, E.; Koch, M.; Fuhrmann, G.; Friess, W. Enhancing the Stabilization Potential of Lyophilization for Extracellular Vesicles. Adv. Healthc. Mater. 2021, 11, 2100538. [Google Scholar] [CrossRef]
- Shahsavari, M.; Nieuwland, R.; van Leeuwen, T.G.; van der Pol, E.; Singh, A.K. Poloxamer-188 as a wetting agent for microfluidic resistive pulse sensing measurements of extracellular vesicles. PLoS ONE 2024, 19, e0295849. [Google Scholar] [CrossRef]
- Fekete, S.; Kizekai, L.; Sarisozen, Y.T.; Lawrence, N.; Shiner, S.; Lauber, M. Investigating the secondary interactions of packing materials for size-exclusion chromatography of therapeutic proteins. J. Chromatogr. A 2022, 1676, 463262. [Google Scholar] [CrossRef]
- Božič, D.; Hočevar, M.; Kisovec, M.; Pajnič, M.; Pađen, L.; Jeran, M.; Zavec, A.B.; Podobnik, M.; Kogej, K.; Iglič, A.; et al. Stability of Erythrocyte-Derived Nanovesicles Assessed by Light Scattering and Electron Microscopy. Int. J. Mol. Sci. 2021, 22, 12772. [Google Scholar] [CrossRef]
- Le Guern, F.; Mussard, V.; Gaucher, A.; Rottman, M.; Prim, D. Fluorescein Derivatives as Fluorescent Probes for pH Monitoring along Recent Biological Applications. Int. J. Mol. Sci. 2020, 21, 9217. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Gobalasingham, N.; Purchel, A.; Lin, J. The Power of Field-Flow Fractionation in Characterization of Nanoparticles in Drug Delivery. Molecules 2023, 28, 4169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chen, X.; Wang, J.; Qing, X.; Wang, Z.; Ding, X.; Xie, Z.; Niu, L.; Guo, X.; Cai, T.; et al. Separation and characterization of extracellular vesicles from human plasma by asymmetrical flow field-flow fractionation. Anal. Chim. Acta 2020, 1127, 234–245. [Google Scholar] [CrossRef]
- Sitar, S.; Kejžar, A.; Pahovnik, D.; Kogej, K.; Tušek-Žnidarič, M.; Lenassi, M.; Žagar, E. Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation. Anal. Chem. 2015, 87, 9225–9233. [Google Scholar] [CrossRef]
- Oeyen, E.; Van Mol, K.; Baggerman, G.; Willems, H.; Boonen, K.; Rolfo, C.; Pauwels, P.; Jacobs, A.; Schildermans, K.; Cho, W.C.; et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J. Extracell. Vesicles 2018, 7, 1490143. [Google Scholar] [CrossRef]
- Vogel, R.; Savage, J.; Muzard, J.; Della Camera, G.; Vella, G.; Law, A.; Marchioni, M.; Mehn, D.; Geiss, O.; Peacock, B.; et al. Measuring particle concentration of multimodal synthetic reference materials and extracellular vesicles with orthogonal techniques: Who is up to the challenge? J. Extracell. Vesicles 2021, 10, e12052. [Google Scholar] [CrossRef]
- Maas, S.L.; de Vrij, J.; van der Vlist, E.J.; Geragousian, B.; van Bloois, L.; Mastrobattista, E.; Schiffelers, R.M.; Wauben, M.H.M.; Broekman, M.L.D.; Hoen, E.N.N. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J. Control. Release 2015, 200, 87–96. [Google Scholar] [CrossRef]
- Enjeti, A.K.; Ariyarajah, A.; Warwick, E.; Seldon, M.; Lincz, L.F. Challenges in Analysis of Circulating Extracellular Vesicles in Human Plasma Using Na-notracking and Tunable Resistive Pulse Sensing. J. Nanomed. Nanotechnol 2017, 8, 468. [Google Scholar] [CrossRef]
- Fortunato, D.; Mladenović, D.; Criscuoli, M.; Loria, F.; Veiman, K.-L.; Zocco, D.; Koort, K.; Zarovni, N. Opportunities and Pitfalls of Fluorescent Labeling Methodologies for Extracellular Vesicle Profiling on High-Resolution Single-Particle Platforms. Int. J. Mol. Sci. 2021, 22, 10510. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Arkesteijn, G.J.A.; Bremer, M.; Cimorelli, M.; Dignat-George, F.; Giebel, B.; Görgens, A.; Hendrix, A.; Kuiper, M.; Lacroix, R.; et al. A compendium of single extracellular vesicle flow cytometry. J. Extracell. Vesicles 2023, 12, e12299. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Xu, S.; Jung, S.; Nguyen, A.; Cheng, Y.; Zhao, M.; Fujimoto, B.S.; Nelson, W.; Schiro, P.; Franklin, J.L.; et al. Comparison of EV characterization by commercial high-sensitivity flow cytometers and a custom single-molecule flow cytometer. J. Extracell. Vesicles 2024, 13, e12498. [Google Scholar] [CrossRef]
- van der Pol, E.; de Rond, L.; Coumans, F.A.; Gool, E.L.; Böing, A.N.; Sturk, A.; Nieuwland, R.; van Leeuwen, T.G. Absolute sizing and label-free identification of extracellular vesicles by flow cytometry. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 801–810. [Google Scholar] [CrossRef]
- Breitwieser, K.; Koch, L.F.; Tertel, T.; Proestler, E.; Burgers, L.D.; Lipps, C.; Adjaye, J.; Fürst, R.; Giebel, B.; Saul, M.J. Detailed Characterization of Small Extracellular Vesicles from Different Cell Types Based on Tetraspanin Composition by ExoView R100 Platform. Int. J. Mol. Sci. 2022, 23, 8544. [Google Scholar] [CrossRef]
- Andersson, K.; Björkelund, H.; Malmqvist, M. Antibody-antigen interactions: What is the required time to equilibrium? Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Tertel, T.; Bremer, M.; Maire, C.; Lamszus, K.; Peine, S.; Jawad, R.; Andaloussi, S.E.; Giebel, B.; Ricklefs, F.L.; Görgens, A. High-Resolution Imaging Flow Cytometry Reveals Impact of Incubation Temperature on Labeling of Extracellular Vesicles with Antibodies. Cytom. Part A 2020, 97, 602–609. [Google Scholar] [CrossRef]
- Bonilla, D.L.; Paul, A.; Gil-Pulido, J.; Park, L.M.; Jaimes, M.C. The Power of Reagent Titration in Flow Cytometry. Cells 2024, 13, 1677. [Google Scholar] [CrossRef]
- Sivanantham, A.; Jin, Y. Impact of Storage Conditions on EV Integrity/Surface Markers and Cargos. Life 2022, 12, 697. [Google Scholar] [CrossRef]


| V [mL] | NTA Particle Conc. [Particles/mL] | LS90 [RSU/mL] | NTA Sizing Mode, Mean (D10-D90) [nm] | MALS Sizing at Peak Apex (Range a) [nm] | Protein Conc. Bradford Assay [µg/mL] | Protein Conc. Estim. b from SEC A280 [µg/mL] | Protein Depletion Bradford Assay | Protein Depletion Estim. b (SEC A280) | CD9 [RFU/mL] | CD63 [RFU/mL] | CD81 [RFU/mL] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Process 1 | CCM | 200 | 2.30 × 109 | 15 ± 1 | 145 ± 7, 170 ± 1 (112–236) | 169 ± 12 (126–252) | 527 ± 20 | 523 ± 36 | n.a. | n.a. | 11.6 | 24.6 | 13.5 |
| PER1 | 180 | n.a. | n.a. | n.a. | n.a. | 213 ± 5 | 252 ± 13 | n.a. | n.a. | bld | bld | bld | |
| RET+Flush | 20 | 1.20 × 1010 | 67 ± 3 | 140 ± 7, 172 ± 1 (119–241) | 171 ± 7 (120–242) | 251 ± 8 | 201 ± 22 | −95% | −96% | 132.5 | 283.3 | 137.6 | |
| Enrichment factor | 5 | 4 | Enrichment factor | 11.4 | 11.5 | 10.2 | |||||||
| Process 2 | CCM | 400 | 1.16 × 109 | 13 ± 2 | 130 ± 2, 192 ± 2 (122–292) | 194 ± 12 (152–284) | 479 ± 55 | 504 ± 2 | n.a. | n.a. | 20.00 | 34.9 | 18.3 |
| PER1 | 320 | n.a. | n.a. | n.a. | n.a. | 258 ± 52 | 267 ± 8 | n.a. | n.a. | bld | bld | bld | |
| PER2 | 620 | n.a. | n.a. | n.a. | n.a. | 191 ± 47 | 84 ± 10 | n.a. | n.a. | bld | bld | bld | |
| RET | 12 | 2.99 × 1010 | 271 ± 40 | 161 ± 8, 215 ± 3 (136–327) | 183 ± 9 (165–248) | 3405 ± 73 | 3019 ± 119 | −79% | −82% | 486.8 | 924.2 | 536.3 | |
| Flush | 15 | 5.17 × 109 | 35 ± 1 | 157 ± 6, 211 ± 5 (132–312) | 188 ± 2 (151–287) | 309 ± 38 | 359 ± 4 | n.a. | n.a. | 48.3 | 97.7 | 62.5 | |
| Enrichment factor | 26 | 21 | Enrichment factor | 24.3 | 26.5 | 29.2 | |||||||
| Factors | Responses | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buf(B) | Buf(C) | pH | c NaCl | Cond. | % p188 | RT Pk1 | Res 2–3 | ||
| Factors | Buf(B) | 1 | 0.41 | 0.28 | −0.09 | −0.19 | 0.08 | −0.34 | −0.25 |
| Buf(C) | 0.41 | 1 | −0.59 | −0.09 | −0.19 | 0.07 | 0.17 | 0.22 | |
| pH | 0.28 | −0.59 | 1 | 0.00 | 0.05 | −0.02 | −0.30 | −0.39 | |
| c NaCl | −0.09 | −0.09 | 0.00 | 1 | 0.99 | 0.30 | 0.51 | 0.70 | |
| Cond. | −0.19 | −0.19 | 0.05 | 0.99 | 1 | 0.28 | 0.54 | 0.69 | |
| % p188 | 0.08 | 0.07 | −0.02 | 0.30 | 0.28 | 1 | 0.17 | 0.20 | |
| Resp. | RT Pk 1 | −0.34 | 0.17 | −0.30 | 0.51 | 0.54 | 0.17 | 1 | 0.75 |
| Res 2–3 | −0.25 | 0.22 | −0.39 | 0.70 | 0.69 | 0.20 | 0.75 | 1 | |
| Factors | Responses | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | c NaCl | % p188 | FLD RT Pk1 | FLD Area Total | FLD Res 1–2 | FLD Res 2–3 | FLD S/T Pk1 | FLD S/T Pk2 | FLD S/T Pk3 | LS90 Pk1 RT | Rel. LS90 Pk1 Area | Rel. LS Pk1 Height | LS90 S/T | ||
| Factors | pH | 1 | −0.07 | −0.08 | −0.16 | 0.98 | 0.35 | 0.81 | −0.20 | −0.81 | −0.25 | −0.16 | 0.41 | 0.44 | −0.53 |
| c NaCl | −0.07 | 1 | 0.30 | 0.36 | 0.02 | 0.32 | −0.01 | −0.46 | −0.05 | −0.29 | −0.16 | 0.34 | 0.52 | 0.25 | |
| % p188 | −0.08 | 0.30 | 1 | 0.36 | −0.02 | −0.04 | −0.11 | −0.18 | 0.23 | −0.15 | −0.07 | 0.12 | 0.17 | 0.19 | |
| Responses | FLD RT Pk1 | −0.16 | 0.36 | 0.36 | 1 | −0.08 | 0.41 | −0.29 | −0.07 | 0.02 | −0.03 | 0.34 | −0.20 | 0.01 | 0.27 |
| FLD RT Pk2 | −0.60 | 0.41 | 0.16 | 0.69 | −0.51 | 0.41 | −0.64 | −0.08 | 0.26 | 0.29 | 0.39 | −0.44 | −0.30 | 0.59 | |
| FLD RT Pk3 | −0.81 | 0.38 | 0.15 | 0.52 | −0.73 | 0.01 | −0.65 | −0.03 | 0.62 | 0.06 | 0.35 | −0.39 | −0.36 | 0.60 | |
| FLD W05 Pk1 | −0.57 | 0.26 | −0.12 | 0.00 | −0.55 | −0.11 | −0.52 | −0.23 | 0.46 | 0.12 | −0.04 | −0.23 | −0.36 | 0.43 | |
| FLD W05 Pk2 | −0.80 | −0.10 | 0.17 | 0.00 | −0.84 | −0.79 | −0.77 | 0.17 | 0.95 | −0.11 | −0.01 | −0.13 | −0.27 | 0.23 | |
| FLD W05 Pk3 | −0.74 | 0.28 | 0.04 | 0.45 | −0.73 | −0.03 | −0.77 | −0.05 | 0.57 | 0.07 | 0.31 | −0.35 | −0.37 | 0.53 | |
| FLD Area Total | 0.98 | 0.02 | −0.02 | −0.08 | 1 | 0.44 | 0.84 | −0.25 | −0.82 | −0.26 | −0.14 | 0.40 | 0.46 | −0.49 | |
| FLD % Area Pk1 | 0.23 | 0.44 | 0.01 | 0.08 | 0.21 | 0.29 | 0.04 | −0.67 | −0.20 | −0.18 | −0.31 | 0.12 | 0.14 | 0.21 | |
| FLD % Area Pk2 | 0.26 | −0.23 | 0.07 | 0.01 | 0.29 | −0.26 | 0.43 | 0.36 | 0.01 | −0.47 | 0.30 | 0.14 | 0.14 | −0.43 | |
| FLD % Area Pk3 | −0.57 | −0.19 | −0.11 | −0.10 | −0.59 | 0.02 | −0.57 | 0.28 | 0.20 | 0.78 | −0.03 | −0.30 | −0.32 | 0.31 | |
| FLD Res 1–2 | 0.35 | 0.32 | −0.04 | 0.41 | 0.44 | 1 | 0.30 | −0.19 | −0.73 | 0.35 | 0.21 | −0.13 | 0.10 | 0.15 | |
| FLD Res 2–3 | 0.81 | −0.01 | −0.11 | −0.29 | 0.84 | 0.30 | 1 | −0.14 | −0.68 | −0.25 | −0.15 | 0.34 | 0.36 | −0.40 | |
| FLD S/T Pk1 | −0.20 | −0.46 | −0.18 | −0.07 | −0.25 | −0.19 | −0.14 | 1 | 0.09 | 0.27 | 0.36 | −0.48 | −0.42 | −0.03 | |
| FLD S/T Pk2 | −0.81 | −0.05 | 0.23 | 0.02 | −0.82 | −0.73 | −0.68 | 0.09 | 1 | −0.16 | 0.04 | −0.16 | −0.31 | 0.29 | |
| FLD S/T Pk3 | −0.25 | −0.29 | −0.15 | −0.03 | −0.26 | 0.35 | −0.25 | 0.27 | −0.16 | 1 | 0.18 | −0.43 | −0.42 | 0.24 | |
| LS90 RT | −0.16 | −0.16 | −0.07 | 0.34 | −0.14 | 0.21 | −0.15 | 0.36 | 0.04 | 0.18 | 1 | −0.49 | −0.49 | 0.22 | |
| Rel. LS90 Pk1 Area | 0.41 | 0.34 | 0.12 | −0.20 | 0.40 | −0.13 | 0.34 | −0.48 | −0.16 | −0.43 | −0.49 | 1 | 0.90 | −0.54 | |
| Rel. LS Pk1 Height | 0.44 | 0.52 | 0.17 | 0.01 | 0.46 | 0.10 | 0.36 | −0.42 | −0.31 | −0.42 | −0.49 | 0.90 | 1 | −0.49 | |
| LS90 W05 | −0.36 | −0.25 | −0.16 | −0.22 | −0.38 | −0.13 | −0.28 | 0.08 | 0.31 | 0.27 | 0.40 | −0.52 | −0.75 | 0.44 | |
| LS90 S/T | −0.53 | 0.25 | 0.19 | 0.27 | −0.49 | 0.15 | −0.40 | −0.03 | 0.29 | 0.24 | 0.22 | −0.54 | −0.49 | 1 | |
| A280 [RAU/mL] | rΔA280 % | TRP (Peak 1) [RFUs/mL] | rΔTRP (Peak 1) % | TRP (Total) [RFUs/mL] | rΔTRP % | LS90 [RSUs/mL] | rsd LS90 % | rΔLS90 % | MALS Size [nm] | rsd Size % | rΔSize % | ||||||||
| 50 mM NaCl, pH 6.5 | 508 | −3% | 14,346 | +1% | 278,361 | +1% | 474 | 3% | −4% | 105 | 4% | +2% | |||||||
| 50 mM NaCl, pH 7.2 | 528 | 0% | 14,418 | +2% | 283,596 | +3% | 515 | 2% | +4% | 101 | 3% | −2% | |||||||
| 50 mM NaCl, pH 8.0 | 534 | +2% | 15,096 | +7% | 288,651 | +5% | 515 | 3% | +4% | 97 | 2% | −5% | |||||||
| 1 M NaCl, pH 6.5 | 530 | +1% | 14,576 | +3% | 300,365 | +9% | 641 | 20% | +30% | 64 | 5% | −38% | |||||||
| 1 M NaCl, pH 7.2 | 534 | +1% | 14,484 | +2% | 299,127 | +9% | 592 | 15% | +20% | 66 | 4% | −36% | |||||||
| 1 M NaCl, pH 8.0 | 543 | +3% | 14,508 | +3% | 300,481 | +9% | 595 | 15% | +20% | 64 | 6% | −38% | |||||||
| 137 mM NaCl, pH 7.4 (dPBS reference) | 526 | n.a. | 14,134 | n.a. | 275,244 | n.a. | 494 | 8% | n.a. | 103 | 4% | n.a. | |||||||
| CD9 (Peak 1) [RFUs/mL] | rΔCD9 % | CD9 (Total) [RFUs/mL] | CD63 (Peak 1) [RFUs/mL] | rΔCD63 % | CD63 (Total) [RFUs/mL] | CD81 (Peak 1) [RFUs/mL] | rΔCD81 % | CD81 (Total) [RFUs/mL] | |||||||||||
| 50 mM NaCl, pH 6.5 | 513 | +2% | 13,351 | 910 | ±0% | 10,141 | 576 | +1% | 14,222 | ||||||||||
| 50 mM NaCl, pH 7.2 | 487 | −3% | 13,775 | 928 | +2% | 10,289 | 611 | +7% | 14,905 | ||||||||||
| 50 mM NaCl, pH 8.0 | 506 | +1% | 13,820 | 926 | +2% | 10,234 | 602 | +6% | 14,786 | ||||||||||
| 1 M NaCl, pH 6.5 | 460 | −8% | 13,671 | 877 | −3% | 10,127 | 379 | −33% | 14,454 | ||||||||||
| 1 M NaCl, pH 7.2 | 451 | −10% | 13,603 | 868 | −4% | 10,230 | 390 | −31% | 14,836 | ||||||||||
| 1 M NaCl, pH 8.0 | 451 | −10% | 13,675 | 860 | −5% | 10,143 | 363 | −36% | 14,497 | ||||||||||
| 137 mM NaCl, pH 7.4 (dPBS reference) | 502 | (±3%) | 13,283 | 907 | (±6%) | 9731 | 569 | (±1%) | 14,363 | ||||||||||
| Technique | Output Information | Benefits | Limitations | Ref. | |||
|---|---|---|---|---|---|---|---|
| Particle Concentration | Particle Size/Distribution | Marker Profile | Impurity Profile | ||||
| TRPS | + | + | − | − | Simple | Limited throughput; might not be suitable for complex samples; requires prior estimation of particle concentration | [31,32,33] |
| NTA | + | + | −/+ a | − | Simple | Presence of larger particles may impair analysis; fluorescence detection is prone to bleaching effects; evaluation of fluorescent particles is limited by the number of marker molecules per particle and fluorochrome brightness; fluorescent labelling requires extensive optimisation, often including additional sample pretreatment to remove excess fluorescent probe; requires prior estimation of particle concentration | [32,34] |
| FCM | + | −/+ b | + | − | Applicable to samples with low particle concentration and highly customisable | Hardly encounters the complete population of EVs/nanoparticles due to the limited sensitivity of scattering and fluorescence detectors; small changes in the lower limit of detection can lead to great differences in measured EV concentrations; fluorescent labelling requires extensive optimisation, often including additional sample pretreatment to remove excess fluorescent probe; requires prior estimation of particle concentration | [34,35,36,37] |
| Commercial single particle imaging platforms (e.g., ExoView) | + c | + c | + | − | Easy to perform, automated, and applicable to complex samples | Only particles carrying specific markers are encountered in evaluation; requires prior estimation of particle concentration | [38] |
| AF4 (suppl. with UV, LS, and FLD detectors) | Relative (LS-intensity-based) | + | + | + | High resolution, gentle to fragile particles, highly customisable, and applicable to complex samples | Expensive; requires a rather concentrated sample; method development and optimisation require special expertise; LS-intensity-weighted size distribution | [26,27,28,29] |
| Analytical SEC (suppl. with UV, LS, and FLD detectors) | Relative (LS-intensity-based) | + | + | + | Applicable to complex samples; customisable; the sensitivity for marker detection increased due to the cumulative signal across the population | Does not provide number-based particle concentrations; sensitivity for nanoparticles at the expense of limited resolution; LS-intensity-weighted size distribution | |
| Antibody Concentration (ng/mL) | |||||||
|---|---|---|---|---|---|---|---|
| 1250 | 625 | 250 | 125 | 50 | 0 (No Add. Control) | ||
| Sample conc. (particles/mL) | 1 × 1010 | A1 | A2 | A3 | A4 | A5 | A6 |
| 5 × 109 | B1 | B2 | B3 | B4 | B5 | B6 | |
| 1 × 109 | C1 | C2 | C3 | C4 | C5 | C6 | |
| 5 × 108 | D1 | D2 | D3 | D4 | D5 | D6 | |
| 0 (Antibody Control) | E1 | E2 | E3 | E4 | E5 | Blank | |
| Condition Description | Buffer Type | Buffer | NaCl Concentration | pH |
|---|---|---|---|---|
| Physiological | dPBS | 9 mM phosphate | 137 mM | 7.4 |
| Low salt, low pH | MES | 50 mM MES | 50 mM | 6.5 |
| Low salt, mid pH | HEPES | 50 mM HEPES | 50 mM | 7.2 |
| Low salt, high pH | HEPES | 50 mM HEPES | 50 mM | 8.0 |
| High salt, low pH | MES | 50 mM MES | 1 M | 6.5 |
| High salt, mid pH | HEPES | 50 mM HEPES | 1 M | 7.2 |
| High salt, high pH | HEPES | 50 mM HEPES | 1 M | 8.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Božič, D.; Vrabec, K.; Železnik, A.; Raspor, A.; Novak, V.; Koshmak, I.P.; Leskovec, M.; Štrancar, A. Comprehensive Characterisation of Extracellular Vesicle Preparations Using Multiparametric Size-Exclusion Chromatography. Int. J. Mol. Sci. 2025, 26, 8477. https://doi.org/10.3390/ijms26178477
Božič D, Vrabec K, Železnik A, Raspor A, Novak V, Koshmak IP, Leskovec M, Štrancar A. Comprehensive Characterisation of Extracellular Vesicle Preparations Using Multiparametric Size-Exclusion Chromatography. International Journal of Molecular Sciences. 2025; 26(17):8477. https://doi.org/10.3390/ijms26178477
Chicago/Turabian StyleBožič, Darja, Katja Vrabec, Ana Železnik, Andrej Raspor, Valentina Novak, Ivana Petrović Koshmak, Maja Leskovec, and Aleš Štrancar. 2025. "Comprehensive Characterisation of Extracellular Vesicle Preparations Using Multiparametric Size-Exclusion Chromatography" International Journal of Molecular Sciences 26, no. 17: 8477. https://doi.org/10.3390/ijms26178477
APA StyleBožič, D., Vrabec, K., Železnik, A., Raspor, A., Novak, V., Koshmak, I. P., Leskovec, M., & Štrancar, A. (2025). Comprehensive Characterisation of Extracellular Vesicle Preparations Using Multiparametric Size-Exclusion Chromatography. International Journal of Molecular Sciences, 26(17), 8477. https://doi.org/10.3390/ijms26178477





