Mitogenomic Alterations in Breast Cancer: Identification of Potential Biomarkers of Risk and Prognosis
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. Landscape of Mitochondrial DNA Germline Variants in Breast Tumors
2.3. High Mutational Rate of Mitochondrial Genome from Peripheral Blood of Patients with Breast Cancer
2.4. Low Mutational Rate of Mitochondrial Genome from Peripheral Blood of Healthy Women
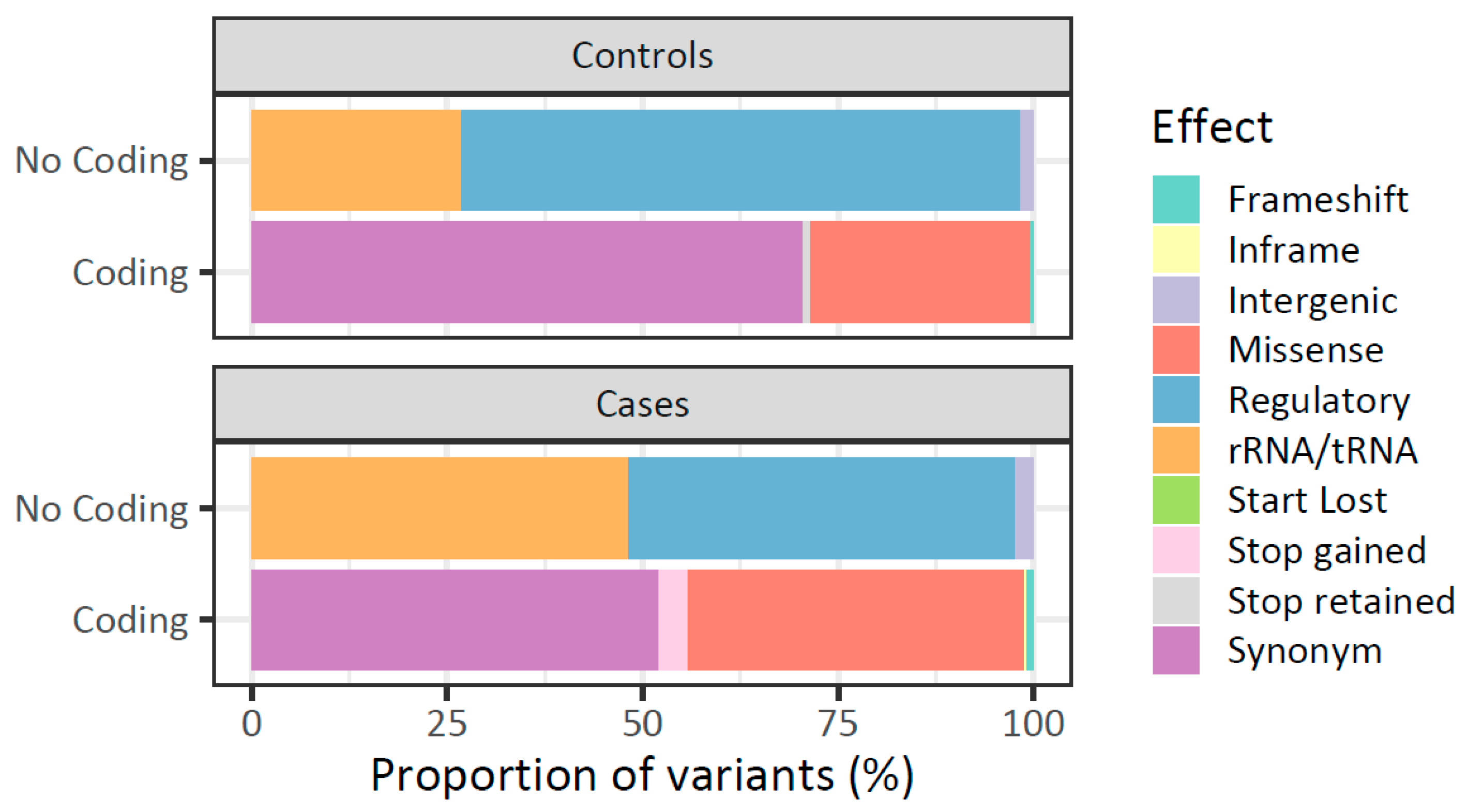
2.5. The Landscape of mtDNA Germline Variants Differs Between Patients with Breast Cancer and Healthy Women
2.6. Higher Heteroplasmy Variation in Peripheral Blood of Patients with Breast Cancer than Healthy Women
2.7. Association Analysis Between Mitochondrial DNA Variants and Breast Cancer
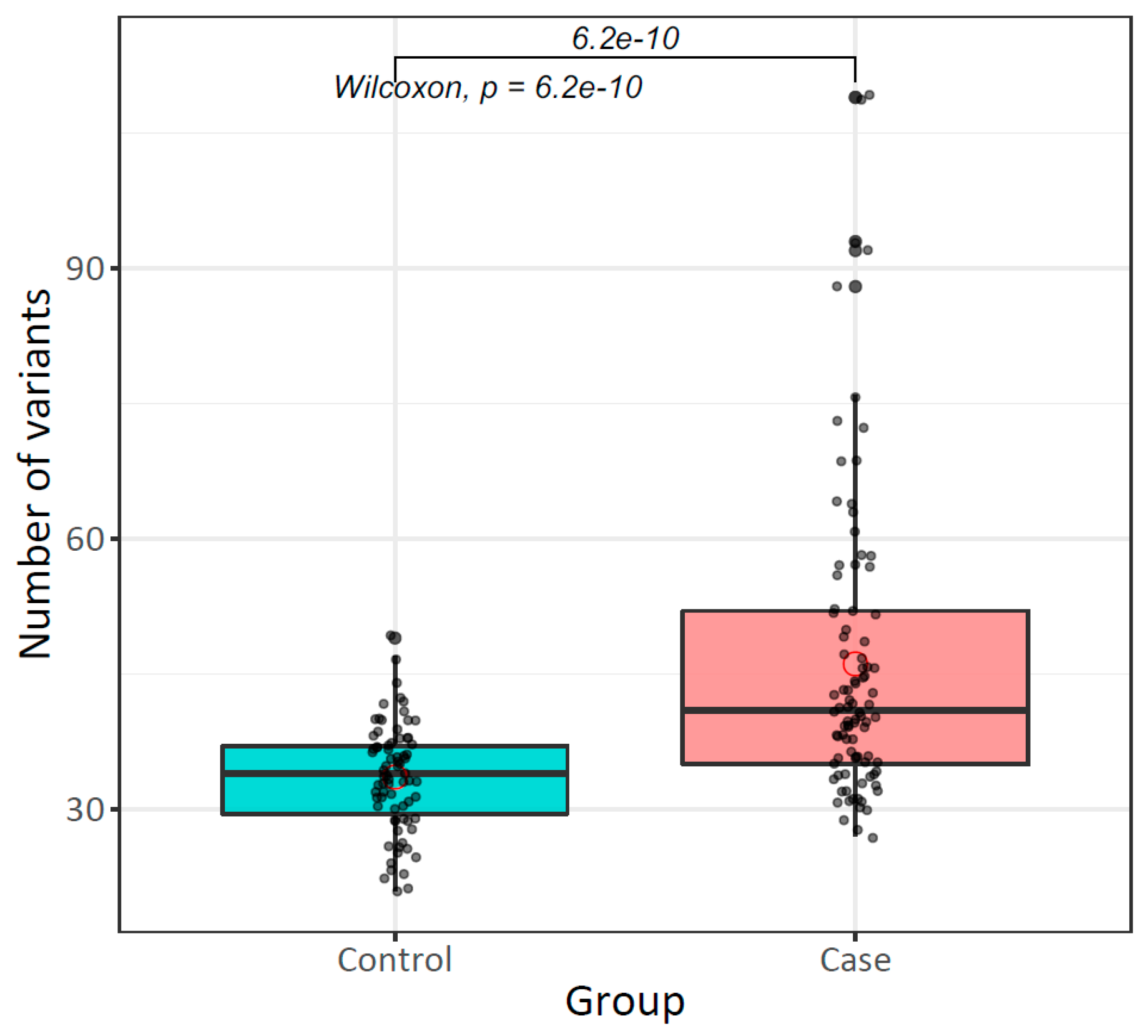
2.7.1. The Mitochondrial Mutational Burden in Peripheral Blood Increases Risk for Breast Cancer
2.7.2. Mitochondrial DNA Germline Variants Are Associated with Breast Cancer Development Risk
2.7.3. Higher Peripheral Blood Mitochondrial DNA Content in Patients with Breast Cancer than Healthy Women
2.8. Native American Mitochondrial Haplogroups Were Enriched in Patients with Breast Cancer and Healthy Women
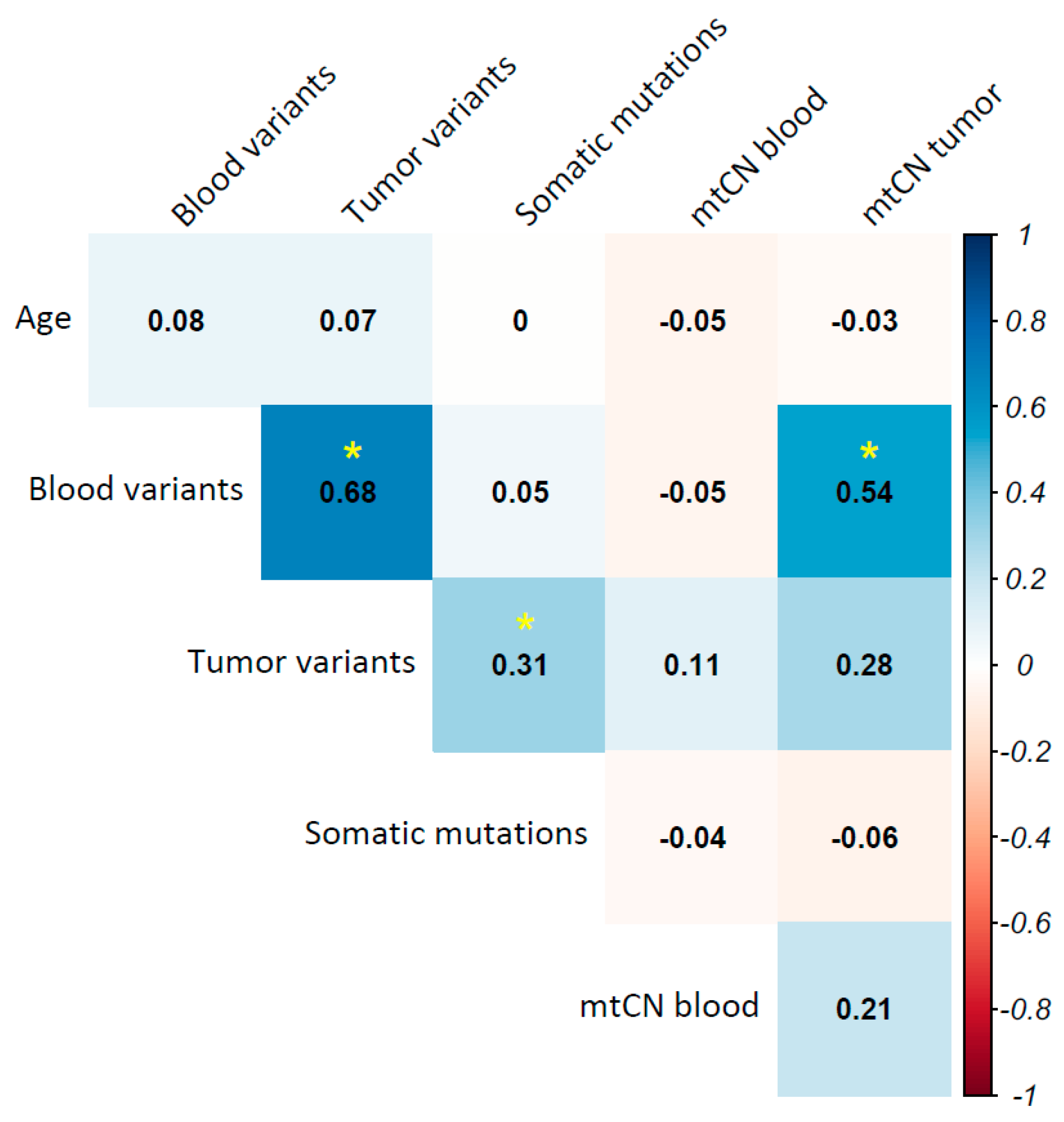
2.9. Tumor Mitochondrial DNA Mutational Burden Is Associated with Overall Survival in Breast Cancer
2.10. Mitochondrial Haplogroups Are Associated with Prognosis in Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Genomic DNA Extraction
4.3. Direct Mitochondrial DNA Sequencing
4.4. Mitochondrial DNA Content Quantification
4.5. Bioinformatic Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BWA | Burrows–Wheeler Aligner |
| CSB | Conserved sequence blocks |
| ER | Estrogen receptor |
| GATK | Genome Analysis Tool Kit |
| HBB | Hemoglobin Subunit Beta |
| H2 | HER2 positive breast cancer subtype |
| HER2 | Human epidermal growth factor receptor 2 |
| HR | Hazzard ratio |
| IHC | Immunohistochemistry |
| LA | Luminal A breast cancer subtype |
| LB | Luminal B breast cancer subtype |
| MAF | Mutant allele fraction |
| MT-ATP6 | Mitochondrial ATP Synthase Membrane Subunit 6 |
| MT-ATP8 | Mitochondrial ATP Synthase Membrane Subunit 8 |
| MT-CO (1-3) | Mitochondrial Cytochrome C Oxidase Subunit (1-3) |
| mtCN | Mitochondrial DNA copy number |
| MT-CYB | Mitochondrial Cytochrome B |
| mtDNA | Mitochondrial DNA |
| MT-ND (1-6) | Mitochondrial NADH Dehydrogenase Subunits (1-6) |
| MT-ND4L | Mitochondrial NADH Dehydrogenase Subunits 4L |
| mtTF1 | Mitochondrial transcription factor 1 |
| NUMTS | Nuclear mitochondrial segments |
| OHR | H-strand replication origin region |
| OR | Odds ratio |
| OXPHOS | Oxidative phosphorylation |
| PB | Peripheral blood |
| PCR | Polymerase Chain Reaction |
| PR | Progesterone receptor |
| ROS | Reactive oxygen species |
| rRNA | Ribosomal RNA |
| SNV | Single-nucleotide variant |
| TN | Triple negative |
| tRNA | Transfer RNA |
| VEP | Variant Effect Predictor |
References
- Breastcancer org. Available online: https://www.breastcancer.org/ (accessed on 1 January 2019).
- GLOBOCAN. The Global Cancer Observatory: Globocan 2020 (México). Available online: https://gco.iarc.fr/today/home (accessed on 5 October 2024).
- Consenso Mexicano sobre el Diagnóstico y Tratamiento del Cáncer Mamario. Décima Revisión del Consenso Mexicano sobre el Diagnóstico y Tratamiento del Cáncer Mamario; Consenso Mexicano sobre el Diagnóstico y Tratamiento del Cáncer Mamario: Colima, México, 2023; p. 119. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Provenzano, E.; Ulaner, G.A.; Chin, S.F. Molecular Classification of Breast Cancer. PET Clin. 2018, 13, 325–338. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Parker, J.S.; Leung, S.; Voduc, D.; Ebbert, M.; Vickery, T.; Davies, S.R.; Snider, J.; Stijleman, I.J.; Reed, J.; et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 2010, 16, 5222–5232. [Google Scholar] [CrossRef]
- Jimenez-Morales, S.; Perez-Amado, C.J.; Langley, E.; Hidalgo-Miranda, A. Overview of mitochondrial germline variants and mutations in human disease: Focus on breast cancer (Review). Int. J. Oncol. 2018, 53, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Dominguez-de-la-Cruz, E.; Munoz, M.L.; Perez-Munoz, A.; Garcia-Hernandez, N.; Moctezuma-Meza, C.; Hinojosa-Cruz, J.C. Reduced mitochondrial DNA copy number is associated with the haplogroup, and some clinical features of breast cancer in Mexican patients. Gene 2020, 761, 145047. [Google Scholar] [CrossRef]
- Guo, W.; Liu, Y.; Ji, X.; Guo, S.; Xie, F.; Chen, Y.; Zhou, K.; Zhang, H.; Peng, F.; Wu, D.; et al. Mutational signature of mtDNA confers mechanistic insight into oxidative metabolism remodeling in colorectal cancer. Theranostics 2023, 13, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.S.; Wang, H.S.; Mugaka, B.P.; Yang, G.J.; Ding, Y. Mitochondria: Promising organelle targets for cancer diagnosis and treatment. Biomater. Sci. 2018, 6, 2786–2797. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Bioenergetic origins of complexity and disease. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 1–16. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Gorelick, A.N.; Kim, M.; Chatila, W.K.; La, K.; Hakimi, A.A.; Berger, M.F.; Taylor, B.S.; Gammage, P.A.; Reznik, E. Respiratory complex and tissue lineage drive recurrent mutations in tumour mtDNA. Nat. Metab. 2021, 3, 558–570. [Google Scholar] [CrossRef]
- Grandhi, S.; Bosworth, C.; Maddox, W.; Sensiba, C.; Akhavanfard, S.; Ni, Y.; LaFramboise, T. Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum. Mol. Genet. 2017, 26, 2912–2922. [Google Scholar] [CrossRef]
- Ju, Y.S.; Alexandrov, L.B.; Gerstung, M.; Martincorena, I.; Nik-Zainal, S.; Ramakrishna, M.; Davies, H.R.; Papaemmanuil, E.; Gundem, G.; Shlien, A.; et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 2014, 3, e02935. [Google Scholar] [CrossRef]
- Perez-Amado, C.J.; Tovar, H.; Gomez-Romero, L.; Beltran-Anaya, F.O.; Bautista-Pina, V.; Dominguez-Reyes, C.; Villegas-Carlos, F.; Tenorio-Torres, A.; Alfaro-Ruiz, L.A.; Hidalgo-Miranda, A.; et al. Mitochondrial DNA Mutation Analysis in Breast Cancer: Shifting From Germline Heteroplasmy Toward Homoplasmy in Tumors. Front. Oncol. 2020, 10, 572954. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Canter, J.A.; Kallianpur, A.R.; Parl, F.F.; Millikan, R.C. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005, 65, 8028–8033. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, K.; Sharma, S.; Bhat, A.K.; Rai, E.; Bamezai, R.N. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007, 249, 249–255. [Google Scholar] [CrossRef]
- McMahon, S.; LaFramboise, T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis 2014, 35, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Reznik, E.; Miller, M.L.; Senbabaoglu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA copy number variation across human cancers. eLife 2016, 5, e10769. [Google Scholar] [CrossRef]
- Perez-Amado, C.J.; Bazan-Cordoba, A.; Hidalgo-Miranda, A.; Jimenez-Morales, S. Mitochondrial Heteroplasmy Shifting as a Potential Biomarker of Cancer Progression. Int. J. Mol. Sci. 2021, 22, 7369. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Chinnery, P.F. The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat. Rev. Genet. 2015, 16, 530–542. [Google Scholar] [CrossRef]
- Yin, C.; Li, D.Y.; Guo, X.; Cao, H.Y.; Chen, Y.B.; Zhou, F.; Ge, N.J.; Liu, Y.; Guo, S.S.; Zhao, Z.; et al. NGS-based profiling reveals a critical contributing role of somatic D-loop mtDNA mutations in HBV-related hepatocarcinogenesis. Ann. Oncol. 2019, 30, 953–962. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.D.; Sun, Y.B.; Li, E.M.; Xu, L.Y.; Zhang, Y.P.; Yao, Y.G.; Kong, Q.P. Deciphering the signature of selective constraints on cancerous mitochondrial genome. Mol. Biol. Evol. 2012, 29, 1255–1261. [Google Scholar] [CrossRef]
- Larman, T.C.; DePalma, S.R.; Hadjipanayis, A.G.; Cancer Genome Atlas Research, N.; Protopopov, A.; Zhang, J.; Gabriel, S.B.; Chin, L.; Seidman, C.E.; Kucherlapati, R.; et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl. Acad. Sci. USA 2012, 109, 14087–14091. [Google Scholar] [CrossRef]
- Li, Y.; Sundquist, K.; Vats, S.; Hong, M.G.; Wang, X.; Chen, Y.; Hedelius, A.; Saal, L.H.; Sundquist, J.; Memon, A.A. Mitochondrial heteroplasmic shifts reveal a positive selection of breast cancer. J. Transl. Med. 2023, 21, 696. [Google Scholar] [CrossRef] [PubMed]
- Tseng, L.M.; Yin, P.H.; Yang, C.W.; Tsai, Y.F.; Hsu, C.Y.; Chi, C.W.; Lee, H.C. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer 2011, 50, 800–811. [Google Scholar] [CrossRef]
- Weerts, M.J.A.; Sleijfer, S.; Martens, J.W.M. The role of mitochondrial DNA in breast tumors. Drug Discov. Today 2019, 24, 1202–1208. [Google Scholar] [CrossRef]
- Jayasekera, L.P.; Ranasinghe, R.; Senathilake, K.S.; Kotelawala, J.T.; de Silva, K.; Abeygunasekara, P.H.; Goonesinghe, R.; Tennekoon, K.H. Mitochondrial genome in sporadic breast cancer: A case control study and a proteomic analysis in a Sinhalese cohort from Sri Lanka. PLoS ONE 2023, 18, e0281620. [Google Scholar] [CrossRef]
- Kim, M.Y.; Kim, H.; Sung, J.A.; Koh, J.; Cho, S.; Chung, D.H.; Jeon, Y.K.; Lee, S.D. Whole Mitochondrial Genome Analysis in Non-Small Cell Lung Carcinoma Reveals Unique Tumor-Specific Somatic Mutations. Arch. Pathol. Lab. Med. 2023, 147, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, M.; Liu, E.M.; Shergold, A.L.; Tolla, E.; Tait-Mulder, J.; Huerta Uribe, A.; Shokry, E.; Young, A.L.; Lilla, S.; Kim, M.; et al. Tumour mitochondrial DNA mutations drive aerobic glycolysis to enhance checkpoint blockade. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kauppila, J.H.K.; Baines, H.L.; Bratic, A.; Simard, M.L.; Freyer, C.; Mourier, A.; Stamp, C.; Filograna, R.; Larsson, N.G.; Greaves, L.C.; et al. A Phenotype-Driven Approach to Generate Mouse Models with Pathogenic mtDNA Mutations Causing Mitochondrial Disease. Cell Rep. 2016, 16, 2980–2990. [Google Scholar] [CrossRef]
- Pereira, C.V.; Gitschlag, B.L.; Patel, M.R. Cellular mechanisms of mtDNA heteroplasmy dynamics. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 510–525. [Google Scholar] [CrossRef]
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Wredenberg, A.; Cansu, Z.; Trifunovic, A.; Larsson, N.G. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008, 6, e10. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.J.; Zhao, Y.P.; Jiang, Z.C.; Zhou, D.T.; Zhu, R. Analysis of Mitochondrial Transfer RNA Mutations in Breast Cancer. Balk. J. Med. Genet. 2023, 25, 15–22. [Google Scholar] [CrossRef]
- Stefano, G.B.; Kream, R.M. Mitochondrial DNA heteroplasmy in human health and disease. Biomed. Rep. 2016, 4, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Cararo-Lopes, E.; Sawant, A.; Moore, D.; Ke, H.; Shi, F.; Laddha, S.; Chen, Y.; Sharma, A.; Naumann, J.; Guo, J.Y.; et al. Integrated metabolic and genetic analysis reveals distinct features of primary differentiated thyroid cancer and its metastatic potential in humans. medRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Guo, W.; Gu, X.; Guo, S.; Zhou, K.; Su, L.; Yuan, Q.; Liu, Y.; Guo, X.; Huang, Q.; et al. Mutational profiling of mtDNA control region reveals tumor-specific evolutionary selection involved in mitochondrial dysfunction. EBioMedicine 2022, 80, 104058. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, Y.; Wang, Y.; Su, J.; Chan, T.; Zhou, J.; Gong, Y.; Wang, K.; Gu, Y.; Zhang, C.; et al. Molecular fingerprints of nuclear genome and mitochondrial genome for early diagnosis of lung adenocarcinoma. J. Transl. Med. 2023, 21, 250. [Google Scholar] [CrossRef]
- Tipirisetti, N.R.; Lakshmi, R.K.; Govatati, S.; Govatati, S.; Vuree, S.; Singh, L.; Raghunadha Rao, D.; Bhanoori, M.; Vishnupriya, S. Mitochondrial genome variations in advanced stage breast cancer: A case-control study. Mitochondrion 2013, 13, 372–378. [Google Scholar] [CrossRef]
- Hofmann, J.N.; Hosgood, H.D., 3rd; Liu, C.S.; Chow, W.H.; Shuch, B.; Cheng, W.L.; Lin, T.T.; Moore, L.E.; Lan, Q.; Rothman, N.; et al. A nested case-control study of leukocyte mitochondrial DNA copy number and renal cell carcinoma in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Carcinogenesis 2014, 35, 1028–1031. [Google Scholar] [CrossRef]
- Yang, K.; Li, X.; Forman, M.R.; Monahan, P.O.; Graham, B.H.; Joshi, A.; Song, M.; Hang, D.; Ogino, S.; Giovannucci, E.L.; et al. Pre-diagnostic leukocyte mitochondrial DNA copy number and colorectal cancer risk. Carcinogenesis 2019, 40, 1462–1468. [Google Scholar] [CrossRef]
- Li, L.; Chen, L.; Li, J.; Zhang, W.; Liao, Y.; Chen, J.; Sun, Z. Correlational study on mitochondrial DNA mutations as potential risk factors in breast cancer. Oncotarget 2016, 7, 31270–31283. [Google Scholar] [CrossRef] [PubMed]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1.23.1–1.23.26. [Google Scholar] [CrossRef]
- Bai, R.K.; Leal, S.M.; Covarrubias, D.; Liu, A.; Wong, L.J. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007, 67, 4687–4694. [Google Scholar] [CrossRef]
- Covarrubias, D.; Bai, R.K.; Wong, L.J.; Leal, S.M. Mitochondrial DNA variant interactions modify breast cancer risk. J. Hum. Genet. 2008, 53, 924–928. [Google Scholar] [CrossRef]
- Tengku Baharudin, N.; Jaafar, H.; Zainuddin, Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. Malays. J. Med. Sci. 2012, 19, 36–42. [Google Scholar] [PubMed]
- Czarnecka, A.M.; Krawczyk, T.; Zdrozny, M.; Lubinski, J.; Arnold, R.S.; Kukwa, W.; Scinska, A.; Golik, P.; Bartnik, E.; Petros, J.A. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res. Treat. 2010, 121, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Jahani, M.M.; Azimi Meibody, A.; Karimi, T.; Banoei, M.M.; Houshmand, M. An A10398G mitochondrial DNA alteration is related to increased risk of breast cancer, and associates with Her2 positive receptor. Mitochondrial DNA. Part A DNA Mapp. Seq. Anal. 2020, 31, 11–16. [Google Scholar] [CrossRef]
- Setiawan, V.W.; Chu, L.H.; John, E.M.; Ding, Y.C.; Ingles, S.A.; Bernstein, L.; Press, M.F.; Ursin, G.; Haiman, C.A.; Neuhausen, S.L. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet. Cytogenet. 2008, 181, 16–19. [Google Scholar] [CrossRef][Green Version]
- Gonzalez, M.D.M.; Santos, C.; Alarcon, C.; Ramos, A.; Cos, M.; Catalano, G.; Acebes, J.J.; Aluja, M.P. Mitochondrial DNA haplogroups J and T increase the risk of glioma. Mitochondrion 2021, 58, 95–101. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Jenkins, E.C.; Lechuga-Vieco, A.V.; Nie, K.; Fiel, M.I.; Rialdi, A.; Guccione, E.; Enriquez, J.A.; Sia, D.; Lujambio, A.; et al. The portrait of liver cancer is shaped by mitochondrial genetics. Cell Rep. 2022, 38, 110254. [Google Scholar] [CrossRef]
- Brinker, A.E.; Vivian, C.J.; Beadnell, T.C.; Koestler, D.C.; Teoh, S.T.; Lunt, S.Y.; Welch, D.R. Mitochondrial Haplotype of the Host Stromal Microenvironment Alters Metastasis in a Non-cell Autonomous Manner. Cancer Res. 2020, 80, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Brinker, A.E.; Vivian, C.J.; Koestler, D.C.; Tsue, T.T.; Jensen, R.A.; Welch, D.R. Mitochondrial Haplotype Alters Mammary Cancer Tumorigenicity and Metastasis in an Oncogenic Driver-Dependent Manner. Cancer Res. 2017, 77, 6941–6949. [Google Scholar] [CrossRef]
- Gaude, E.; Schmidt, C.; Gammage, P.A.; Dugourd, A.; Blacker, T.; Chew, S.P.; Saez-Rodriguez, J.; O’Neill, J.S.; Szabadkai, G.; Minczuk, M.; et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol. Cell 2018, 69, 581–593.e587. [Google Scholar] [CrossRef]
- Tasdogan, A.; McFadden, D.G.; Mishra, P. Mitochondrial DNA Haplotypes as Genetic Modifiers of Cancer. Trends Cancer 2020, 6, 1044–1058. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, G.C.; Marinho, A.N.R.; Anaissi, A.K.; Vinasco-Sandoval, T.; Ribeiro-Dos-Santos, A.; Vidal, A.F.; de Araujo, G.S.; Demachki, S.; Ribeiro-Dos-Santos, A. Whole mitochondrial genome sequencing highlights mitochondrial impact in gastric cancer. Sci. Rep. 2019, 9, 15716. [Google Scholar] [CrossRef] [PubMed]
- Guardado-Estrada, M.; Juarez-Torres, E.; Medina-Martinez, I.; Wegier, A.; Macias, A.; Gomez, G.; Cruz-Talonia, F.; Roman-Bassaure, E.; Pinero, D.; Kofman-Alfaro, S.; et al. A great diversity of Amerindian mitochondrial DNA ancestry is present in the Mexican mestizo population. J. Hum. Genet. 2009, 54, 695–705. [Google Scholar] [CrossRef]
- Kenney, M.C.; Chwa, M.; Atilano, S.R.; Falatoonzadeh, P.; Ramirez, C.; Malik, D.; Tarek, M.; Del Carpio, J.C.; Nesburn, A.B.; Boyer, D.S.; et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: Implications for population susceptibility to diseases. Biochim. Biophys. Acta 2014, 1842, 208–219. [Google Scholar] [CrossRef]
- Bi, C.; Wang, L.; Fan, Y.; Yuan, B.; Ramos-Mandujano, G.; Zhang, Y.; Alsolami, S.; Zhou, X.; Wang, J.; Shao, Y.; et al. Single-cell individual full-length mtDNA sequencing by iMiGseq uncovers unexpected heteroplasmy shifts in mtDNA editing. Nucleic Acids Res. 2023, 51, e48. [Google Scholar] [CrossRef]
- Bi, C.; Wang, L.; Fan, Y.; Yuan, B.; Alsolami, S.; Zhang, Y.; Zhang, P.Y.; Huang, Y.; Yu, Y.; Izpisua Belmonte, J.C.; et al. Quantitative haplotype-resolved analysis of mitochondrial DNA heteroplasmy in Human single oocytes, blastoids, and pluripotent stem cells. Nucleic Acids Res. 2023, 51, 3793–3805. [Google Scholar] [CrossRef]
- Duan, M.; Tu, J.; Lu, Z. Recent Advances in Detecting Mitochondrial DNA Heteroplasmic Variations. Molecules 2018, 23, 323. [Google Scholar] [CrossRef]
- Roberts, N.D.; Kortschak, R.D.; Parker, W.T.; Schreiber, A.W.; Branford, S.; Scott, H.S.; Glonek, G.; Adelson, D.L. A comparative analysis of algorithms for somatic SNV detection in cancer. Bioinformatics 2013, 29, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.D.M.; Ramos, A.; Aluja, M.P.; Santos, C. Sensitivity of mitochondrial DNA heteroplasmy detection using Next Generation Sequencing. Mitochondrion 2020, 50, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, X.; Li, D.; Du, X.; Yin, C.; Chen, C.; Fang, W.; Bian, Z.; Zhang, J.; Li, B.; et al. Multi-regional sequencing reveals intratumor heterogeneity and positive selection of somatic mtDNA mutations in hepatocellular carcinoma and colorectal cancer. Int. J. Cancer 2018, 143, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Perez-Figueroa, A.; Alves, J.M.; Valecha, M.; Prado-Lopez, S.; Alvarino, P.; Cameselle-Teijeiro, J.M.; Chantada, D.; Fonseca, M.M.; Posada, D. Single-cell mtDNA heteroplasmy in colorectal cancer. Genomics 2022, 114, 110315. [Google Scholar] [CrossRef]
- Ludwig, L.S.; Lareau, C.A.; Ulirsch, J.C.; Christian, E.; Muus, C.; Li, L.H.; Pelka, K.; Ge, W.; Oren, Y.; Brack, A.; et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell 2019, 176, 1325–1339.E22. [Google Scholar] [CrossRef]
- Mori, K.M.; McElroy, J.P.; Weng, D.Y.; Chung, S.; Fadda, P.; Reisinger, S.A.; Ying, K.L.; Brasky, T.M.; Wewers, M.D.; Freudenheim, J.L.; et al. Lung mitochondrial DNA copy number, inflammatory biomarkers, gene transcription and gene methylation in vapers and smokers. eBioMedicine 2022, 85, 104301. [Google Scholar] [CrossRef]
- Wei, W.; Tuna, S.; Keogh, M.J.; Smith, K.R.; Aitman, T.J.; Beales, P.L.; Bennett, D.L.; Gale, D.P.; Bitner-Glindzicz, M.A.K.; Black, G.C.; et al. Germline selection shapes human mitochondrial DNA diversity. Science 2019, 364, eaau6520. [Google Scholar] [CrossRef]
- Kopinski, P.K.; Janssen, K.A.; Schaefer, P.M.; Trefely, S.; Perry, C.E.; Potluri, P.; Tintos-Hernandez, J.A.; Singh, L.N.; Karch, K.R.; Campbell, S.L.; et al. Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proc. Natl. Acad. Sci. USA 2019, 116, 16028–16035. [Google Scholar] [CrossRef]
- McMillan, R.P.; Stewart, S.; Budnick, J.A.; Caswell, C.C.; Hulver, M.W.; Mukherjee, K.; Srivastava, S. Quantitative Variation in m.3243A > G Mutation Produce Discrete Changes in Energy Metabolism. Sci. Rep. 2019, 9, 5752. [Google Scholar] [CrossRef] [PubMed]
- Vikramdeo, K.S.; Anand, S.; Khan, M.A.; Khushman, M.; Heslin, M.J.; Singh, S.; Singh, A.P.; Dasgupta, S. Detection of mitochondrial DNA mutations in circulating mitochondria-originated extracellular vesicles for potential diagnostic applications in pancreatic adenocarcinoma. Sci. Rep. 2022, 12, 18455. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, J.; Yuan, Q.; Su, J.; Li, Q.; Lu, X.; Zhang, L.; Cai, Z.; Han, J. Quantitative detection of circulating MT-ND1 as a potential biomarker for colorectal cancer. Bosn. J. Basic Med. Sci. 2021, 21, 577–586. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, S.; Lu, Y.; Wan, M.; Cheng, J.; Liu, J. MitoEVs: A new player in multiple disease pathology and treatment. J. Extracell. Vesicles 2023, 12, e12320. [Google Scholar] [CrossRef] [PubMed]
- Malkin, E.Z.; De Michino, S.; Lambie, M.; Gill, R.; Zhao, Z.; Rostami, A.; Arruda, A.; Minden, M.D.; Bratman, S.V. Cell-free DNA topology depends on its subcellular and cellular origins in cancer. JCI Insight 2022, 7, e159590. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; Sachidanandam, R.; Hales, D.B.; Brard, L.; Robinson, K.; Rahman, M.M.; Khadka, P.; Groesch, K.; Young, C.K.J. Identification of Somatic Mitochondrial DNA Mutations, Heteroplasmy, and Increased Levels of Catenanes in Tumor Specimens Obtained from Three Endometrial Cancer Patients. Life 2022, 12, 562. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 January 2018).
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstatter, A.; Forer, L.; Specht, G.; Bandelt, H.J.; Kronenberg, F.; Salas, A.; Schonherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Kallberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 1 August 2018).

 ) indicate the event (death, metastasis, death and/or metastasis, as appropriate), while crosses (+) indicate patients with censored follow-up.
) indicate the event (death, metastasis, death and/or metastasis, as appropriate), while crosses (+) indicate patients with censored follow-up.
 ) indicate the event (death, metastasis, death and/or metastasis, as appropriate), while crosses (+) indicate patients with censored follow-up.
) indicate the event (death, metastasis, death and/or metastasis, as appropriate), while crosses (+) indicate patients with censored follow-up.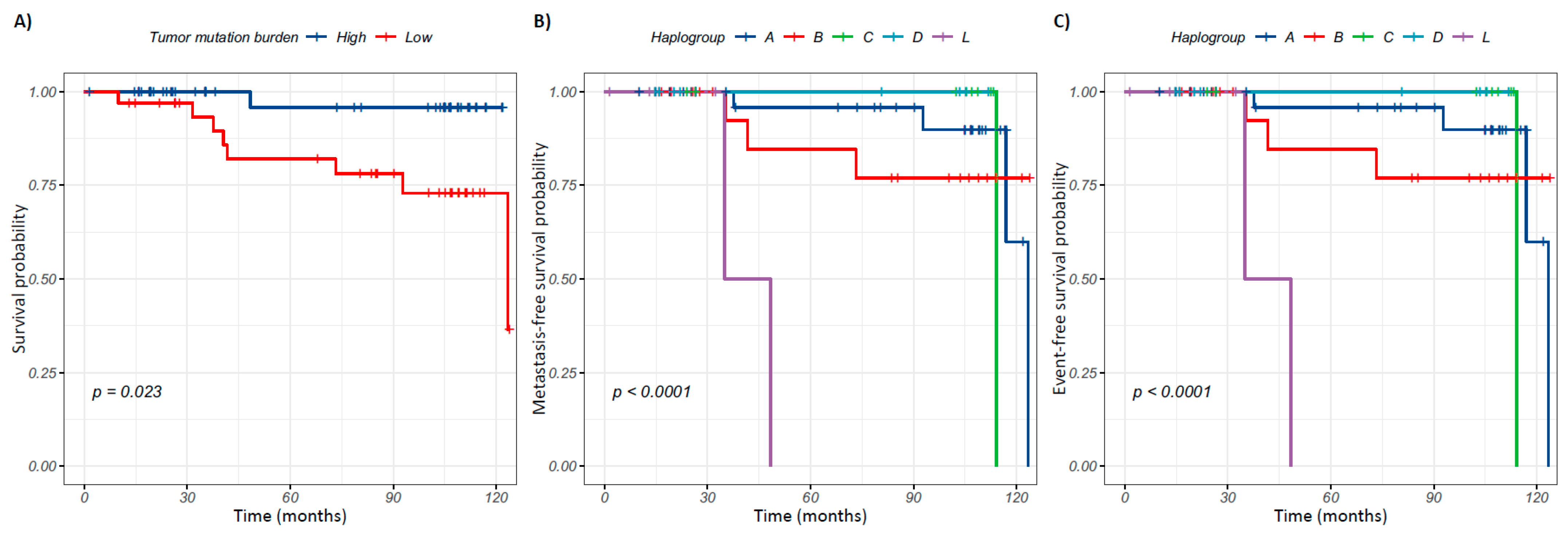
| Clinical Variable | Frequency n (%) | |
|---|---|---|
| Histological diagnosis | Invasive Ductal Carcinoma | 76 (82.6) |
| Ductal Carcinoma In Situ | 1 (1.51) | |
| Invasive Lobular Carcinoma | 11 (11.9) | |
| Mixed Carcinoma | 3 (3.3) | |
| NA | 1 (1.1) | |
| Clinical Stage | I | 8 (8.7) |
| IIA | 43 (46.7) | |
| IIB | 24 (26.1) | |
| IIIA | 7 (7.6) | |
| IIIB | 5 (5.4) | |
| IIIC | 3 (3.3) | |
| NA | 2 (2.2) | |
| IHC classification | Luminal A | 57 (61.9) |
| Luminal B | 22 (24.0) | |
| HER2 | 5 (5.4) | |
| Triple Negative | 6 (6.5) | |
| NA | 2 (2.2) | |
| Metastasis | Positive | 11 (11.9) |
| Negative | 65 (70.7) | |
| NA | 16 (17.4) | |
| Death | Positive | 9 (9.8) |
| Negative | 67 (72.8) | |
| NA | 16 (17.4) | |
| Controls (N = 75) n (%) | Cases (N = 92) n (%) | p-Value | |
|---|---|---|---|
| Total variants | 401 | 866 | - |
| Mutational rate (mut/kb) | 24.2 | 52.3 | - |
| SNVs | 378 (94.3) | 831(95.9) | 0.23 a |
| Insertions | 11 (2.7) | 16 (1.9) | 0.41 a |
| Deletions | 12 (3.0) | 19 (2.2) | 0.51 a |
| Variants per individual ( ± SE) | 33.6 ± 5.9 | 46.1 ± 16.8 | 6.225 × 10−10 *b |
| Range of variants per individual | 21–49 | 27–109 | - |
| Controls | Cases | ||
|---|---|---|---|
| Gen/Region | Total Variants a N = 402 (100%) | Total Variants a N = 863 (100%) | p-Value c |
| D-Loop | 119 (29.6) | 164 (19.0) | 2.7 × 10−5 * |
| MT-ND5 | 25 (6.2) | 94 (10.9) | 0.011 * |
| tRNAs | 13 (3.2) | 57 (6.6) | 0.022 * |
| rRNAs | 32 (8.0) | 103 (11.9) | 0.045 * |
| MT-CO1 | 32 (8.0) | 54 (6.3) | 0.179 |
| MT-ND6 | 6 (1.5) | 24 (2.8) | 0.234 |
| MT-CO2 | 14 (3.5) | 22 (2.5) | 0.443 |
| MT-ND4 | 26 (6.5) | 66 (7.6) | 0.542 |
| MT-ND4L | 4 (1.0) | 13 (1.5) | 0.643 |
| MT-ATP6 b | 26 (6.5) | 49 (5.7) | 0.651 |
| MT-CO3 | 17 (4.2) | 32 (3.7) | 0.756 |
| MT-ATP8 b | 8 (2.0) | 14 (1.6) | 0.803 |
| MT-ND2 | 20 (5.0) | 46 (5.3) | 0.915 |
| MT-CYB | 28 (7.0) | 58 (6.7) | 0.946 |
| MT-ND1 | 23 (5.7) | 49 (5.7) | 1 |
| MT-ND3 | 9 (2.2) | 18 (2.1) | 1 |
| Group | Heteroplasmy | Homoplasmy | ||
|---|---|---|---|---|
| Total Vars n (%) | MAF (Range) | Total Vars n (%) | MAF (Range) | |
| Controls | 85 (21.2) | 63.3% (22.2–95.0%) | 316 (78.8) | 99.8% (95–100%) |
| Cases | 576 (66.5) | 10.8% (0.02–94.8%) | 290 (33.5) | 99.7% (95.1–100%) |
| p-Value a | <2.2 × 10−16 * | <2.2 × 10−16 * | ||
| Mutational Burden | Controls (N = 75) | Cases (N = 92) | OR [CI] | p-Value a |
|---|---|---|---|---|
| n (%) | n (%) | |||
| High (>34 variants) | 35 (46.67) | 71 (77.17) | 3.83 [1.89–7.95] | 5.3 × 10−5 * |
| Low (≤34 variants) | 40 (53.33) | 21 (22.83) |
| Variant | Gene/Region | Functional Effect | Controls (N = 75) n (%) | Cases (N = 92) n (%) | OR [CI] | p-Value a | p-adj b |
|---|---|---|---|---|---|---|---|
| A263G | D-Loop | Regulatory: OHR | 38 (50.7) | 16 (17.4) | 0.2 [0.1–0.4] | 5.8 × 10−6 | 1.5 × 10−4 |
| A16183C | D-Loop | NC | 2 (2.6) | 21 (22.8) | 10.7 [2.5–97.3] | 9.3 × 10−5 | 2.3 × 10−4 |
| C14766T | MT-CYB | Missense: T/I | 62 (82.6) | 91 (98.9) | 18.8 [2.7–816.3] | 1.4 × 10−4 | 3.5 × 10−3 |
| C7028T | MT-CO1 | Synonymous: A | 63 (84.0) | 91 (98.9) | 17.1 [2.4–746.2] | 6 × 10−4 | 0.02 |
| A235G | D-Loop | Regulatory: OHR, mtTF1 BSX, CSB1 | 39 (52) | 24 (26.1) | 0.32 [0.2–0.6] | 7.3 × 10−4 | 0.02 |
| G4820A | MT-ND2 | Synonymous: E | 9 (12) | 28 (30.4) | 3.2 [1.3–8.3] | 4.8 × 10−3 | 0.12 |
| G11719A | MT-ND4 | Synonymous: G | 65 (86.6) | 90 (97.8) | 6.9 [1.4–66.5] | 6.5 × 10−3 | 0.16 |
| T16519C | D-Loop | NC | 25 (33.3) | 50 (54.3) | 2.4 [1.2–4.7] | 7.9 × 10−3 | 0.16 |
| T16189C | D-Loop | NC | 6 (8) | 21 (22.8) | 3.4 [1.2–10.8] | 0.01 | 0.27 |
| C6473T | MT-CO1 | Synonymous: I | 9 (12) | 26 (28.3) | 2.9 [1.2–7.5] | 0.01 | 0.32 |
| CT16188C | D-Loop | NC | 1 (1.3) | 11 (11.9) | 9.9 [1.4–437.4] | 0.01 | 0.32 |
| G11914A | MT-ND4 | Synonymous: T | 9 (12.0) | 25 (27.2) | 2.7 [1.1–7.1] | 0.01 | 0.40 |
| T4977C | MT-ND2 | Synonymous: L | 9 (12.0) | 24 (26.1) | 2.6 [1.1–6.8] | 0.03 | 0.77 |
| A10398G | MT-ND3 | Missense: T/A | 13 (17.3) | 30 (32.6) | 2.3 [1.1–4.8] | 0.03 | 0.81 |
| T16325C | D-Loop | NC | 13 (17.3) | 30 (32.6) | 2.3 [1.1–5.3] | 0.03 | 0.81 |
| C10400T | MT-ND3 | Synonymous: T | 11 (14.7) | 26 (28.3) | 2.3 [1.0–5.6] | 0.04 | 1 |
| T9950C | MT-CO3 | Synonymous: V | 11 (14.7) | 26 (28.3) | 2.3 [1.0–5.6] | 0.04 | 1 |
| A8701G | MT-ATP6 | Missense: T/A | 17 (22.7) | 35 (38.0) | 2.1 [1.0–4.4] | 0.04 | 1 |
| C15535T | MT-CYB | Synonymous: N | 12 (16.0) | 27 (29.3) | 2.2 [1.0–5.1] | 0.04 | 1 |
| T10873C | MT-ND4 | Synonymous: P | 9 (12.0) | 23 (25.0) | 2.4 [1.0–6.4] | 0.04 | 1 |
| A302AC | D-Loop | Regulatory: OHR, mtTF1 BSY, CSB2 | 3 (4.0) | 12 (13.0) | 3.6 [1.0–20.5] | 0.05 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Amado, C.J.; Bazan-Cordoba, A.; Gómez-Romero, L.; Ramírez-Bello, J.; Bautista-Piña, V.; Tenorio-Torres, A.; Ruvalcaba-Limón, E.; Villegas-Carlos, F.; Mendiola-Soto, D.K.; Hidalgo-Miranda, A.; et al. Mitogenomic Alterations in Breast Cancer: Identification of Potential Biomarkers of Risk and Prognosis. Int. J. Mol. Sci. 2025, 26, 8456. https://doi.org/10.3390/ijms26178456
Pérez-Amado CJ, Bazan-Cordoba A, Gómez-Romero L, Ramírez-Bello J, Bautista-Piña V, Tenorio-Torres A, Ruvalcaba-Limón E, Villegas-Carlos F, Mendiola-Soto DK, Hidalgo-Miranda A, et al. Mitogenomic Alterations in Breast Cancer: Identification of Potential Biomarkers of Risk and Prognosis. International Journal of Molecular Sciences. 2025; 26(17):8456. https://doi.org/10.3390/ijms26178456
Chicago/Turabian StylePérez-Amado, Carlos Jhovani, Amellalli Bazan-Cordoba, Laura Gómez-Romero, Julian Ramírez-Bello, Verónica Bautista-Piña, Alberto Tenorio-Torres, Eva Ruvalcaba-Limón, Felipe Villegas-Carlos, Diana Karen Mendiola-Soto, Alfredo Hidalgo-Miranda, and et al. 2025. "Mitogenomic Alterations in Breast Cancer: Identification of Potential Biomarkers of Risk and Prognosis" International Journal of Molecular Sciences 26, no. 17: 8456. https://doi.org/10.3390/ijms26178456
APA StylePérez-Amado, C. J., Bazan-Cordoba, A., Gómez-Romero, L., Ramírez-Bello, J., Bautista-Piña, V., Tenorio-Torres, A., Ruvalcaba-Limón, E., Villegas-Carlos, F., Mendiola-Soto, D. K., Hidalgo-Miranda, A., & Jiménez-Morales, S. (2025). Mitogenomic Alterations in Breast Cancer: Identification of Potential Biomarkers of Risk and Prognosis. International Journal of Molecular Sciences, 26(17), 8456. https://doi.org/10.3390/ijms26178456








