Identification of Inflammation Markers as Novel Potential Predictors of the HIV-DNA Reservoir Size
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Patients
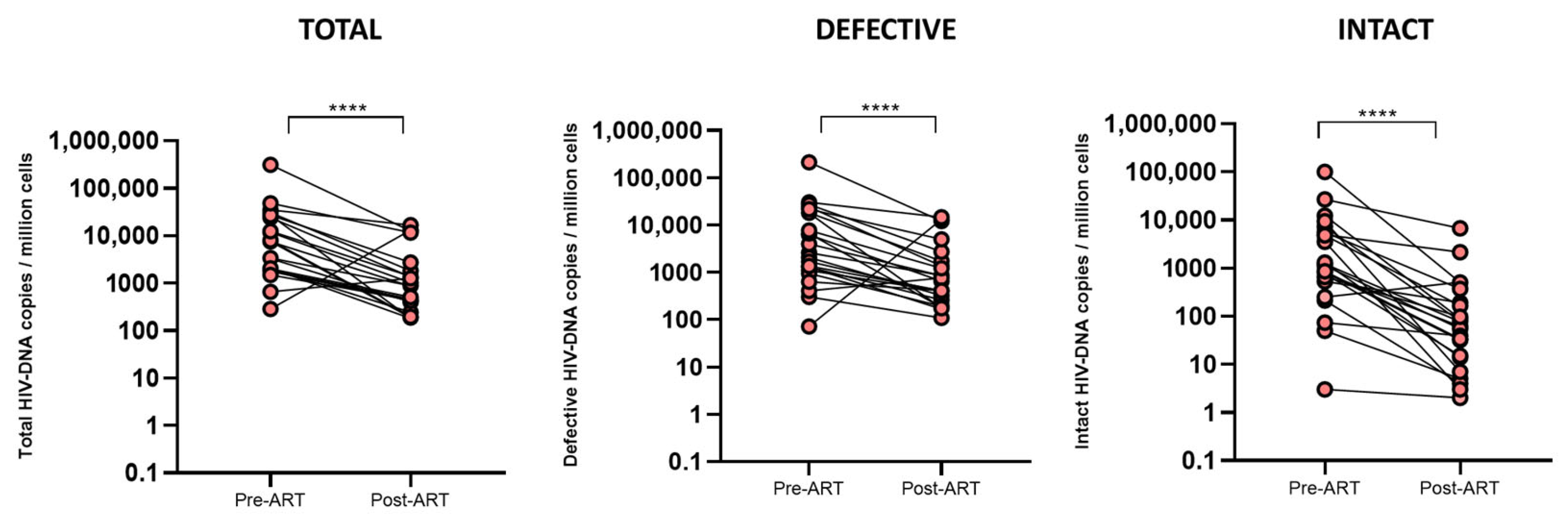
2.2. Levels of HIV-DNA Reservoir and Inflammatory Markers Drastically Reduce During ART Initiation
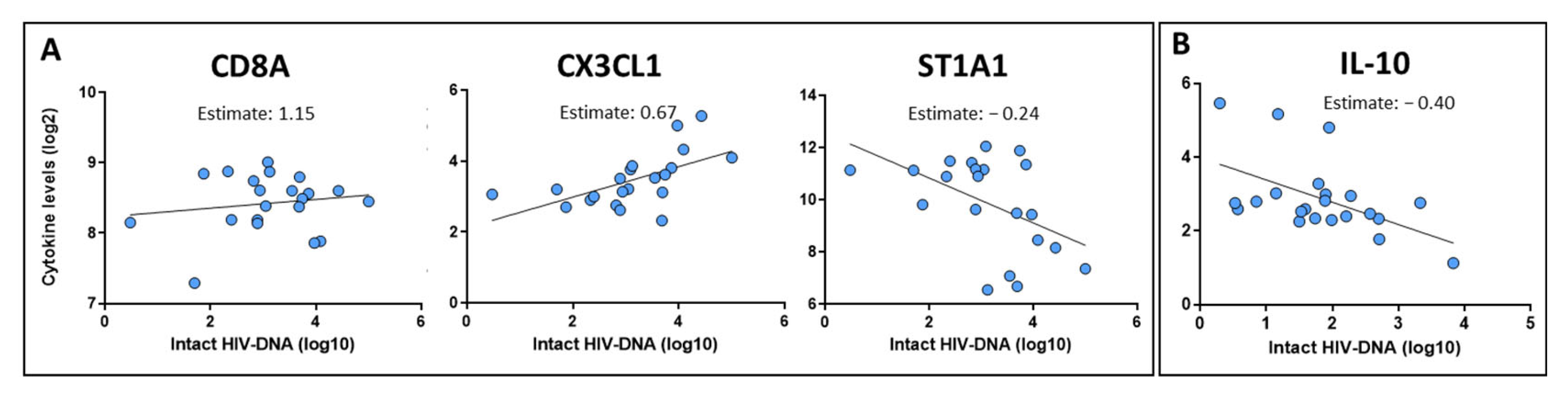
2.3. Identification of Inflammatory Markers Associated with the HIV Reservoir Size at Pre-ART and Post-ART Time Points
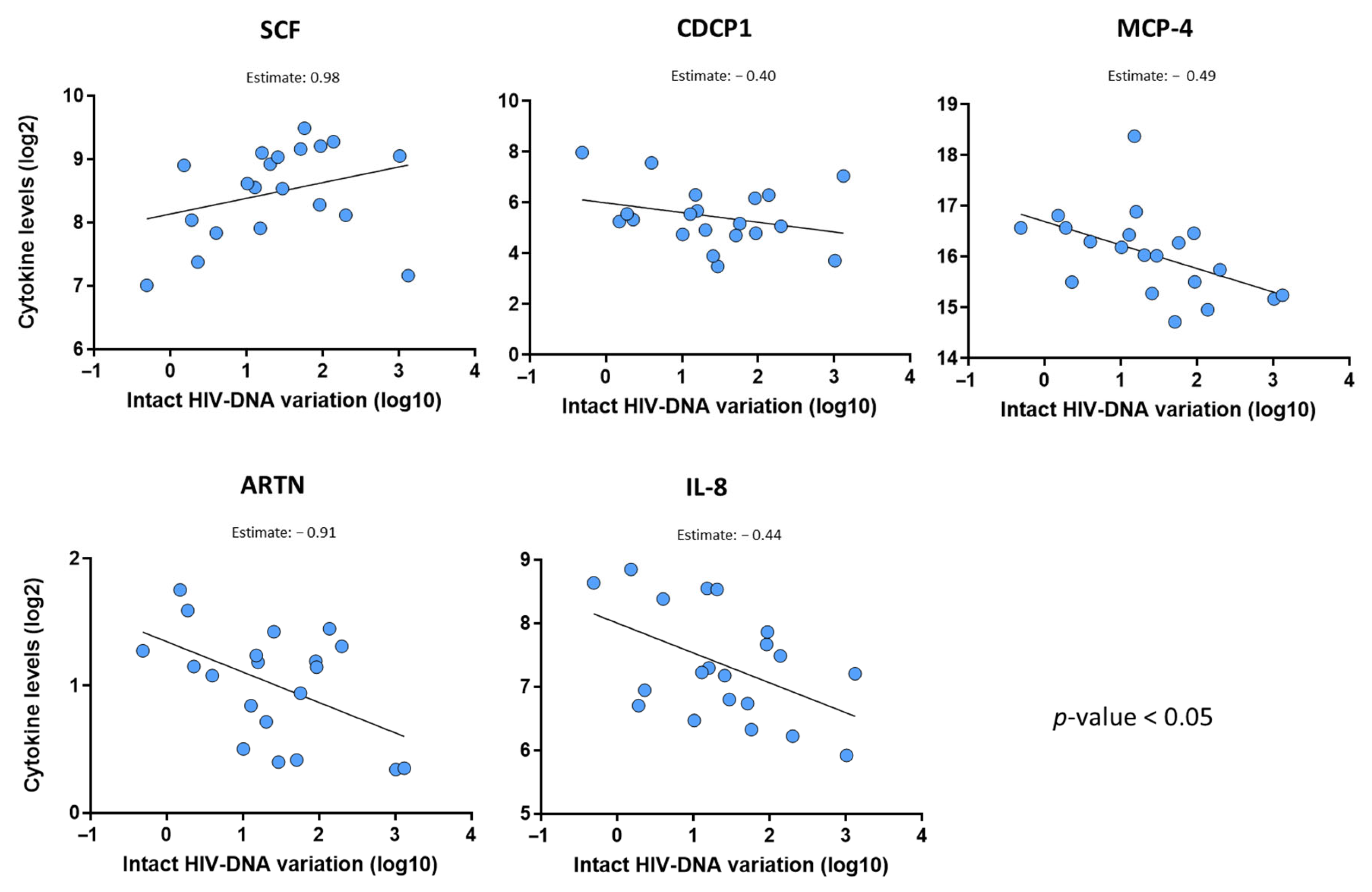
2.4. Baseline Level of Stem Cell Factor Is the Strongest Predictor of the Intact HIV-DNA Reservoir Decline
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Determination of the HIV-DNA Reservoir Size
4.3. Evaluation of Inflammation Marker Levels
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HIV | Human Immunodeficiency Virus |
| ART | Antiretroviral therapy |
| IPDA | Intact proviral DNA assay |
| PEA | Proximity extension assay |
| SCF | Stem cell factor |
| CXCL | C-X-C motif ligand |
Appendix A
Appendix A.1. CoRIS Members
Appendix A.2. Centers and Investigators Involved in the CoRIS Cohort Are Listed Below
References
- Deeks, S.G.; Archin, N.; Cannon, P.; Collins, S.; Jones, R.B.; De Jong, M.A.W.P.; Lambotte, O.; Lamplough, R.; Ndung’u, T.; Sugarman, J.; et al. Research Priorities for an HIV Cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 2021, 27, 2085–2098. [Google Scholar] [CrossRef]
- Pitman, M.C.; Lau, J.S.Y.; McMahon, J.H.; Lewin, S.R. Barriers and Strategies to Achieve A Cure for HIV. Lancet HIV 2018, 5, e317–e328. [Google Scholar] [CrossRef]
- Belmonti, S.; Di Giambenedetto, S.; Lombardi, F. Quantification of Total HIV DNA as a Marker to Measure Viral Reservoir: Methods and Potential Implications for Clinical Practice. Diagnostics 2021, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Sharaf, R.R.; Li, J.Z. The Alphabet Soup of HIV Reservoir Markers. Curr. HIV/AIDS Rep. 2017, 14, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.; Jenike, K.M.; et al. A Quantitative Approach for Measuring the Reservoir of Latent HIV-1 Proviruses. Nature 2019, 566, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Falcinelli, S.D.; Kilpatrick, K.W.; Read, J.; Murtagh, R.; Allard, B.; Ghofrani, S.; Kirchherr, J.; James, K.S.; Stuelke, E.; Baker, C.; et al. Longitudinal Dynamics of Intact HIV Proviral DNA and Outgrowth Virus Frequencies in a Cohort of Individuals Receiving Antiretroviral Therapy. J. Infect. Dis. 2021, 224, 92–100. [Google Scholar] [CrossRef]
- Chen, W.; Berkhout, B.; Pasternak, A.O. Phenotyping Viral Reservoirs to Reveal HIV-1 Hiding Places. Curr. HIV/AIDS Rep. 2025, 22, 15. [Google Scholar] [CrossRef]
- Corley, M.J.; Pang, A.P.S.; Rasmussen, T.A.; Tolstrup, M.; Søgaard, O.S.; Ndhlovu, L.C. Candidate Host Epigenetic Marks Predictive for HIV Reservoir Size, Responsiveness to Latency Reversal, and Viral Rebound. AIDS 2021, 35, 2269–2279. [Google Scholar] [CrossRef]
- Sun, W.; Gao, C.; Hartana, C.A.; Osborn, M.R.; Einkauf, K.B.; Lian, X.; Bone, B.; Bonheur, N.; Chun, T.-W.; Rosenberg, E.S.; et al. Phenotypic Signatures of Immune Selection in HIV-1 Reservoir Cells. Nature 2023, 614, 309–317. [Google Scholar] [CrossRef]
- McGowan, I.; Elliott, J.; Fuerst, M.; Taing, P.; Boscardin, J.; Poles, M.; Anton, P. Increased HIV-1 Mucosal Replication is Associated with Generalized Mucosal Cytokine Activation. JAIDS J. Acquir. Immune Defic. Syndr. 2004, 37, 1228–1236. [Google Scholar] [CrossRef]
- De Clercq, J.; De Scheerder, M.-A.; Mortier, V.; Verhofstede, C.; Vandecasteele, S.J.; Allard, S.D.; Necsoi, C.; De Wit, S.; Gerlo, S.; Vandekerckhove, L. Longitudinal Patterns of Inflammatory Mediators After Acute HIV Infection Correlate to Intact and Total Reservoir. Front. Immunol. 2024, 14, 1337316. [Google Scholar] [CrossRef]
- Massanella, M.; Fromentin, R.; Chomont, N. Residual Inflammation and Viral Reservoirs: Alliance against an HIV Cure. Curr. Opin. HIV AIDS 2016, 11, 234–241. [Google Scholar] [CrossRef]
- Vandergeeten, C.; Fromentin, R.; Chomont, N. The Role of Cytokines in the Establishment, Persistence and Eradication of the HIV Reservoir. Cytokine Growth Factor. Rev. 2012, 23, 143–149. [Google Scholar] [CrossRef]
- Serrano-Villar, S.; Gala, A.; Bacchetti, P.; Hoh, R.; Di Germanio, C.; Cohn, L.B.; Henrich, T.J.; Hunt, P.W.; Laird, G.M.; Pillai, S.K.; et al. Galectin 9 Levels as a Potential Predictor of Intact HIV Reservoir Decay. J. Infect. Dis. 2025, 231, 156–164. [Google Scholar] [CrossRef]
- De La Torre Tarazona, E.; Moraga, E.; Vaquer, R.; Sánchez-Palomino, S.; De Lazzari, E.; Luna, L.; Vicens-Artés, S.; García Fraile, L.J.; Peraire, J.; Garcia-Gasalla, M.; et al. Impact of the Initial Administration of an Antiretroviral Drug with Latency Reversal Properties on the HIV Reservoir Size. Sci. Rep. 2025, 15, 25306. [Google Scholar] [CrossRef]
- Chomont, N.; Okoye, A.A.; Favre, D.; Trautmann, L. Wake Me up Before You Go: A Strategy to Reduce the Latent HIV Reservoir. AIDS 2018, 32, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Buchholtz, N.V.E.J.; Hermans, L.E.; Umunnakwe, C.N.; Nühn, M.M.; Voss, R.; Need, E.; Kootstra, N.A.; Maurer, I.; De Jong, D.C.M.; Symons, J.; et al. Defective Proviruses Significantly Impact Viral Transcription and Immune Activation in Men and Women with HIV-1 Subtype C in Rural South Africa. Front. Immunol. 2024, 15, 1484358. [Google Scholar] [CrossRef] [PubMed]
- Hileman, C.O.; Funderburg, N.T. Inflammation, Immune Activation, and Antiretroviral Therapy in HIV. Curr. HIV/AIDS Rep. 2017, 14, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Osuji, F.N.; Onyenekwe, C.C.; Ahaneku, J.E.; Ukibe, N.R. The Effects of Highly Active Antiretroviral Therapy on the Serum Levels of Pro-Inflammatory and Anti-Inflammatory Cytokines in HIV Infected Subjects. J. Biomed. Sci. 2018, 25, 88. [Google Scholar] [CrossRef]
- Yin, X.; Wang, Z.; Wu, T.; Ma, M.; Zhang, Z.; Chu, Z.; Hu, Q.; Ding, H.; Han, X.; Xu, J.; et al. The Combination of CXCL9, CXCL10 and CXCL11 Levels During Primary HIV Infection Predicts HIV Disease Progression. J. Transl. Med. 2019, 17, 417. [Google Scholar] [CrossRef]
- Cao, W.; Mehraj, V.; Kaufmann, D.E.; Li, T.; Routy, J. Elevation and Persistence of CD8 T-cells in HIV Infection: The Achilles Heel in the ART Era. J. Int. AIDS Soc. 2016, 19, 20697. [Google Scholar] [CrossRef] [PubMed]
- Soares-Schanoski, A.; Sauerwald, N.; Goforth, C.W.; Periasamy, S.; Weir, D.L.; Lizewski, S.; Lizewski, R.; Ge, Y.; Kuzmina, N.A.; Nair, V.D.; et al. Asymptomatic SARS-CoV-2 Infection Is Associated With Higher Levels of Serum IL-17C, Matrix Metalloproteinase 10 and Fibroblast Growth Factors Than Mild Symptomatic COVID-19. Front. Immunol. 2022, 13, 821730. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.-Y.; Huang, H.-H.; Zhen, C.; Chen, S.-Y.; Song, B.; Cao, W.-J.; Shen, L.-L.; Zhou, M.-J.; Zhang, X.-C.; Xu, R.; et al. Distinct Inflammation-Related Proteins Associated with T Cell Immune Recovery During Chronic Hiv-1 Infection. Emerg. Microbes Infect. 2023, 12, 2150566. [Google Scholar] [CrossRef] [PubMed]
- Mastroianni, C.M.; Liuzzi, G.M. Matrix Metalloproteinase Dysregulation in HIV infection: Implications for Therapeutic Strategies. Trends Mol. Med. 2007, 13, 449–459. [Google Scholar] [CrossRef]
- Porichis, F.; Hart, M.G.; Zupkosky, J.; Barblu, L.; Kwon, D.S.; McMullen, A.; Brennan, T.; Ahmed, R.; Freeman, G.J.; Kavanagh, D.G.; et al. Differential Impact of PD-1 and/or Interleukin-10 Blockade on HIV-1-Specific CD4 T Cell and Antigen-Presenting Cell Functions. J. Virol. 2014, 88, 2508–2518. [Google Scholar] [CrossRef]
- Fombellida-Lopez, C.; Aguilar Ortmans, D.; Moutschen, M.; Pasternak, A.O.; Darcis, G. No Associations Between HIV Reservoir and Inflammation in Long-Term Virally Suppressed Dolutegravir-Based ART-Treated Individuals. Front. Immunol. 2025, 16, 1628086. [Google Scholar] [CrossRef]
- Broudy, V. Stem cell factor and hematopoiesis. Blood J. Am. Soc. Hematol 1997, 90, 1345–1364. [Google Scholar]
- Mann, Z.; Sengar, M.; Verma, Y.K.; Rajalingam, R.; Raghav, P.K. Hematopoietic Stem Cell Factors: Their Functional Role in Self-Renewal and Clinical Aspects. Front. Cell Dev. Biol. 2022, 10, 664261. [Google Scholar] [CrossRef]
- Boulassel, M.-R.; Chomont, N.; Pai, N.P.; Gilmore, N.; Sékaly, R.-P.; Routy, J.-P. CD4 T Cell Nadir Independently Predicts the Magnitude of the HIV Reservoir After Prolonged Suppressive Antiretroviral Therapy. J. Clin. Virol. 2012, 53, 29–32. [Google Scholar] [CrossRef]
- Rasmussen, T.A.; Ahuja, S.K.; Kuwanda, L.; Vjecha, M.J.; Hudson, F.; Lal, L.; Rhodes, A.; Chang, J.; Palmer, S.; Auberson-Munderi, P.; et al. Antiretroviral Initiation at ≥800 CD4+ Cells/mm3 Associated With Lower Human Immunodeficiency Virus Reservoir Size. Clin. Infect. Dis. 2022, 75, 1781–1791. [Google Scholar] [CrossRef]
- Sobrino-Vegas, P.; Gutiérrez, F.; Berenguer, J.; Labarga, P.; García, F.; Alejos-Ferreras, B.; Muñoz, M.A.; Moreno, S.; Del Amo, J. La cohorte de la red española de investigación en sida y su biobanco: Organización, principales resultados y pérdidas al seguimiento. Enfermedades Infecc. Microbiol. Clín. 2011, 29, 645–653. [Google Scholar] [CrossRef]

| Baseline Clinical and Socio-Demographic Parameters | |
|---|---|
| Number of individuals | 23 |
| Viral load (HIV RNA copies/mL plasma) | 166,003 (19,657–853,933) |
| Pre-ART CD4 counts (cells/mm3) | 339 (91–575) |
| Post-ART CD4 counts (cells/mm3) | 600 (422–777) |
| Follow-up upon ART initiation (weeks) | 84 (72–110) |
| Age (years) | 33 (25–40) |
| AIDS diagnosis Yes (n, %) No (n, %) | 6 (26%) 17 (74%) |
| HCV status Yes No Unknown | 1 (4%) 4 (17%) 18 (89%) |
| Sex Male (n, %) Female (n, %) | 21 (91%) 2 (9%) |
| Mode of transmission MSM (n, %) Others (HSX, unknown) (n, %) | 18 (78%) 5 (22%) |
| Treatment regimen INI-based (n, %) NNRTI-based (n, %) PI-based (n, %) | 5 (22%) 14 (61%) 4 (17%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De La Torre Tarazona, E.; Moraga, E.; Fons-Contreras, M.; Vaquer, R.; Sánchez-Palomino, S.; Vallejo-Palma, G.; Calderón-Vicente, S.; Vicens-Artés, S.; Aldamiz-Echevarria, T.; Ciudad Sañudo, M.; et al. Identification of Inflammation Markers as Novel Potential Predictors of the HIV-DNA Reservoir Size. Int. J. Mol. Sci. 2025, 26, 8430. https://doi.org/10.3390/ijms26178430
De La Torre Tarazona E, Moraga E, Fons-Contreras M, Vaquer R, Sánchez-Palomino S, Vallejo-Palma G, Calderón-Vicente S, Vicens-Artés S, Aldamiz-Echevarria T, Ciudad Sañudo M, et al. Identification of Inflammation Markers as Novel Potential Predictors of the HIV-DNA Reservoir Size. International Journal of Molecular Sciences. 2025; 26(17):8430. https://doi.org/10.3390/ijms26178430
Chicago/Turabian StyleDe La Torre Tarazona, Erick, Elisa Moraga, María Fons-Contreras, Raúl Vaquer, Sonsoles Sánchez-Palomino, Germán Vallejo-Palma, Sergio Calderón-Vicente, Sònia Vicens-Artés, Teresa Aldamiz-Echevarria, Marianela Ciudad Sañudo, and et al. 2025. "Identification of Inflammation Markers as Novel Potential Predictors of the HIV-DNA Reservoir Size" International Journal of Molecular Sciences 26, no. 17: 8430. https://doi.org/10.3390/ijms26178430
APA StyleDe La Torre Tarazona, E., Moraga, E., Fons-Contreras, M., Vaquer, R., Sánchez-Palomino, S., Vallejo-Palma, G., Calderón-Vicente, S., Vicens-Artés, S., Aldamiz-Echevarria, T., Ciudad Sañudo, M., Moreno, C., Armenteros-Yeguas, I., Tiraboschi, J., Reus Bañuls, S., Alcamí, J., Serrano-Villar, S., Moreno, S., & on behalf of the CoRIS cohort. (2025). Identification of Inflammation Markers as Novel Potential Predictors of the HIV-DNA Reservoir Size. International Journal of Molecular Sciences, 26(17), 8430. https://doi.org/10.3390/ijms26178430







