Genome-Wide Analysis and Expression Profiles of Auxin Response Factors in Ginger (Zingiber officinale Roscoe)
Abstract
1. Introduction
2. Results
2.1. Identification and Sequence Analysis of ZoARF Family Genes
2.2. Phylogenetic Analysis of the ZoARF Gene Family
2.3. Gene Structure and Motif Composition of the ZoARF Family
2.4. Chromosomal Localization of ZoARF Family Genes
2.5. Analysis of cis-Regulatory Elements in the Promoter Region of the ZoARF Gene Family
2.6. GO Annotation of ZoARF Protein Sequences
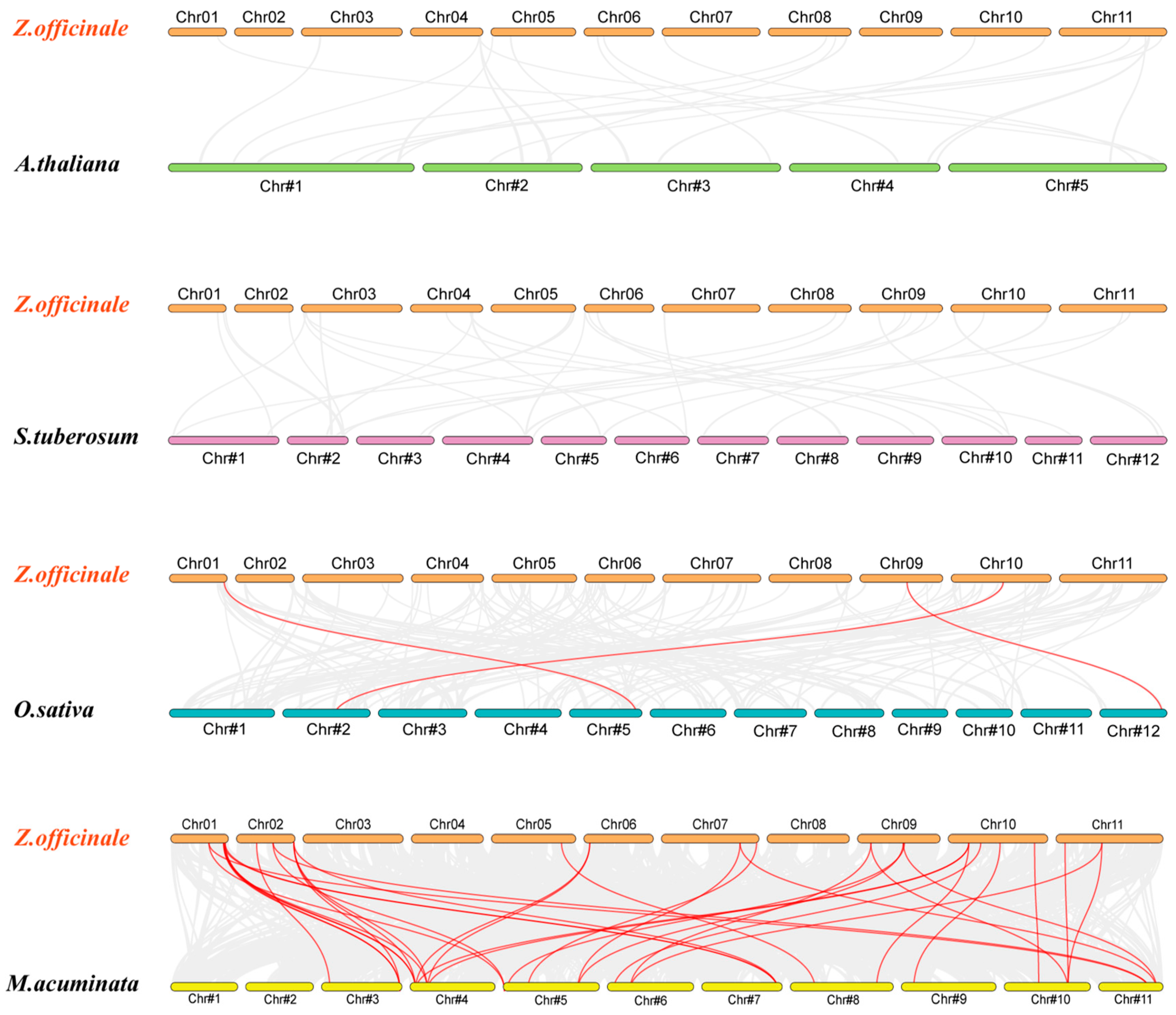
2.7. Gene Duplication and Synteny Analysis of the ZoARF Gene Family
2.8. Evolutionary Analysis of ZoARF Genes
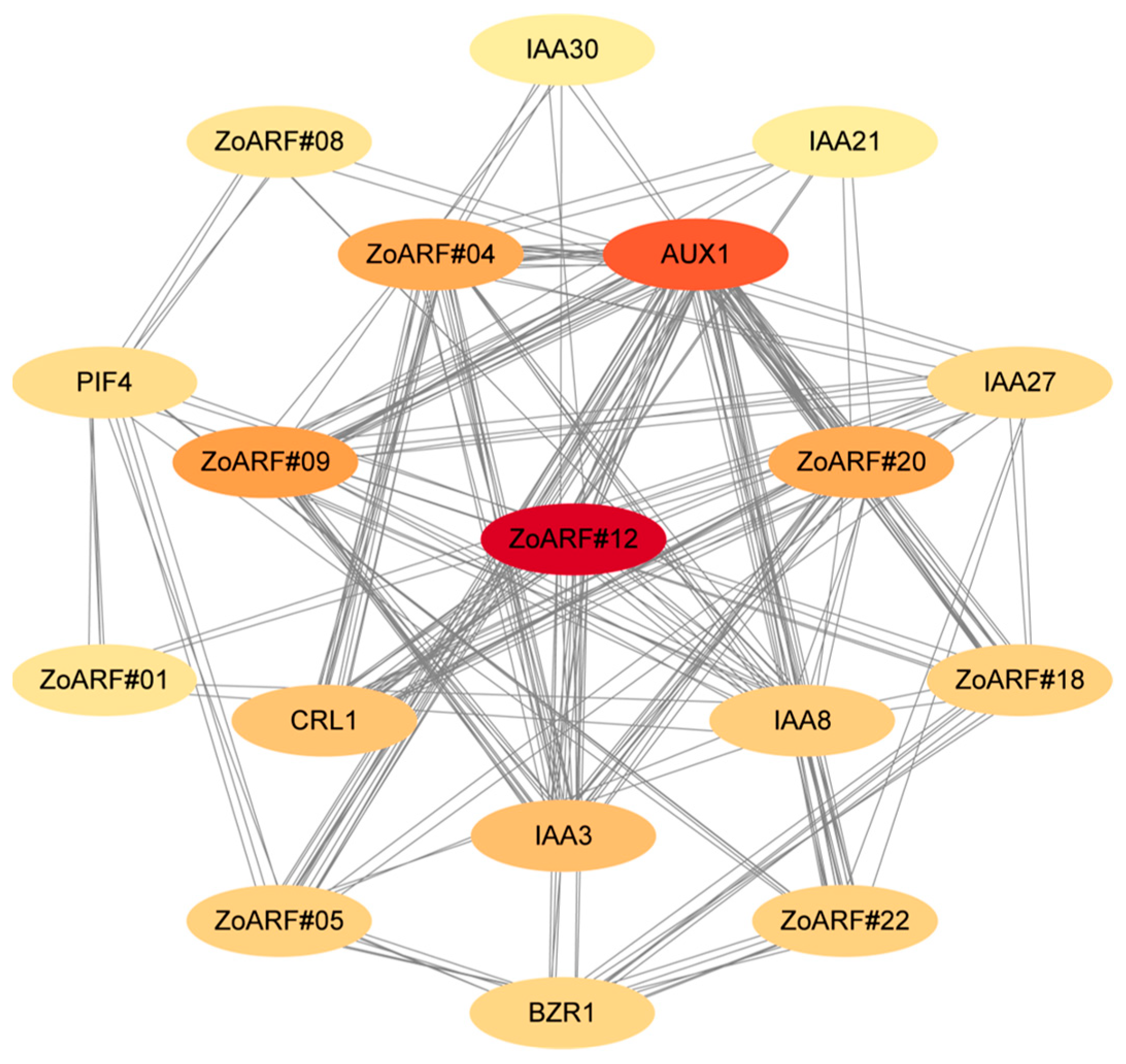
2.9. Protein-Protein Interaction Network Analysis
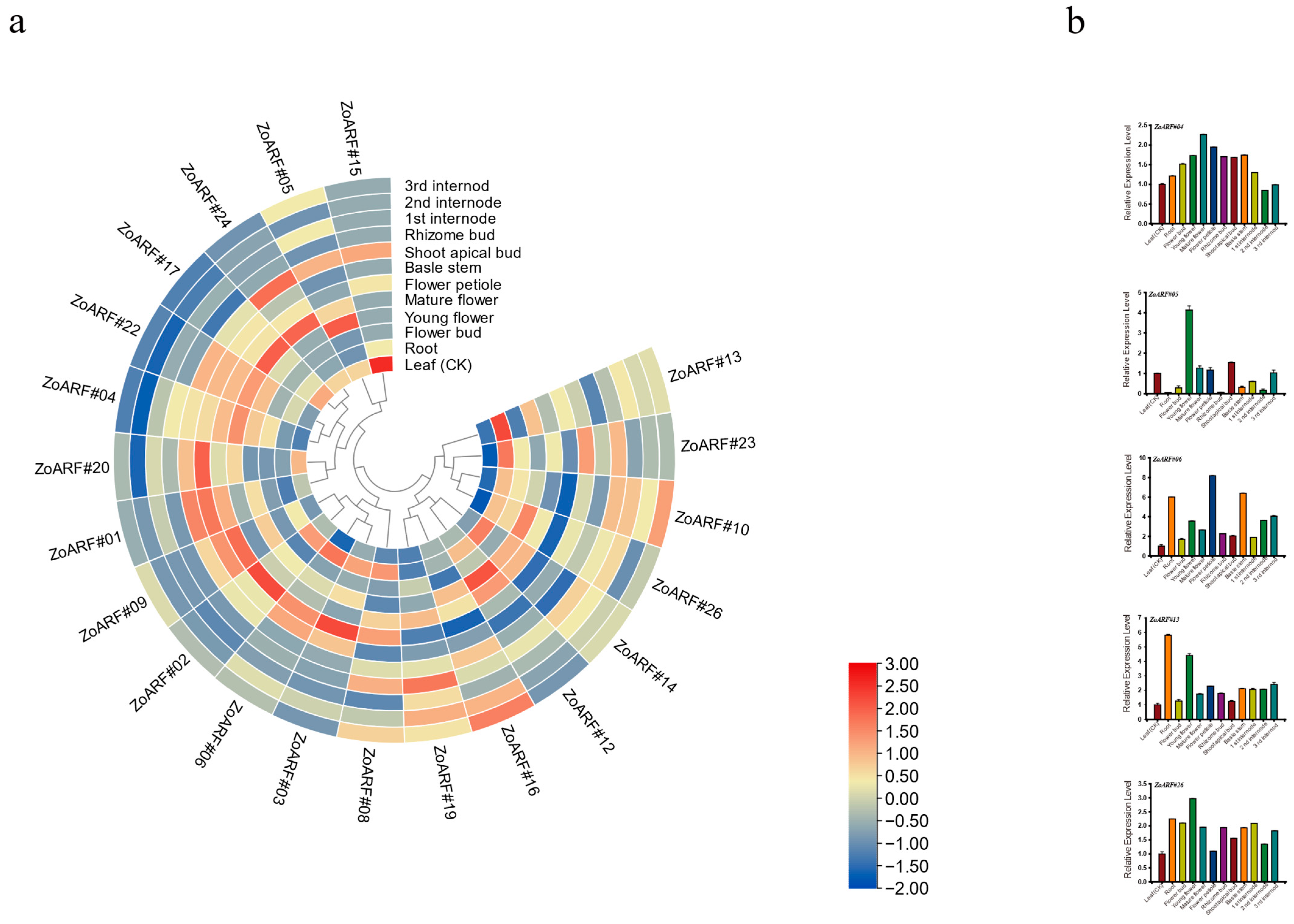
2.10. Expression Profiling of Ginger ZoARF Genes in Different Tissues
2.11. Expression Profiles of ZoARF Genes Under Abiotic Stress Conditions
3. Discussion
4. Materials and Methods
4.1. Identification Analysis of ZoARFs
4.2. Analysis of the Physicochemical Properties of the ZoARFs Gene Family
4.3. Phylogenetic Analysis, Gene Structure, and Motif Composition of the ZoARFs
4.4. Analysis of cis-Acting Elements
4.5. Chromosomal Distribution, Genomic Duplication, and Ka/Ks Ratios of ZoARF Genes
4.6. Protein Interaction Analysis
4.7. Plant Materials
4.8. Expression Analysis of ZoARF Genes by RNA-Seq and qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kou, X.; Zhao, X.; Wu, B.; Wang, C.; Wu, C.; Yang, S.; Zhou, J.; Xue, Z. Auxin Response Factors Are Ubiquitous in Plant Growth and Development, and Involved in Crosstalk between Plant Hormones: A Review. Appl. Sci. 2022, 12, 1360. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Estelle, M. Auxin signaling and regulated protein degradation. Trends Plant Sci. 2004, 9, 302–308. [Google Scholar] [CrossRef]
- Choudhary, P.; Aggarwal, P.R.; Salvi, P.; Muthamilarasan, M. Molecular insight into auxin signaling and associated network modulating stress responses in rice. Plant Physiol. Biochem. 2025, 219, 109452. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Ulmasov, T.; Hagen, G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 1998, 54, 619–627. [Google Scholar] [CrossRef]
- Liu, K.; Yuan, C.; Li, H.; Lin, W.; Yang, Y.; Shen, C.; Zheng, X. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genom. 2015, 16, 901. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, R.; Yu, Z.; Huang, X.; Wang, H.; Xu, W.; Yu, X. Genome-wide identification and functional analysis of the ARF gene family in tetraploid potato reveal its potential role in anthocyanin biosynthesis. BMC Plant Biol. 2025, 25, 342. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, J.; Ha, X.; Ma, H. Genome-wide identification and expression analysis of the Auxin-Response factor (ARF) gene family in Medicago sativa under abiotic stress. BMC Genom. 2023, 24, 498. [Google Scholar] [CrossRef]
- Zhang, H.X.; Cao, N.; Dong, C.J.; Shang, Q.M. Genome-wide Identification and Expression of ARF Gene Family during Adventitious Root Development in Hot Pepper (Capsicum annuum). Hortic. Plant J. 2017, 3, 151–164. [Google Scholar] [CrossRef]
- Guilfoyle, T.J. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell 2015, 27, 33–43. [Google Scholar] [CrossRef]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Li, S.B.; OuYang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-wide identification of the ARF (auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Hu, W.; Zuo, J.; Hou, X.; Yan, Y.; Wei, Y.; Liu, J.; Li, M.; Xu, B.; Jin, Z. The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 2015, 6, 742. [Google Scholar] [CrossRef]
- Yan, M.; Yan, Y.; Wang, P.; Wang, Y.; Piao, X.; Di, P.; Yang, D.-C. Genome-Wide Identification and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Panax ginseng Indicates Its Possible Roles in Root Development. Plants 2023, 12, 3943. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin Response Factors Mediate Arabidopsis Organ Asymmetry via Modulation of KANADI Activity. Plant Cell. 2005, 17, 2899–2910. [Google Scholar] [CrossRef]
- Krogan, N.T.; Ckurshumova, W.; Marcos, D.; Caragea, A.E.; Berleth, T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012, 194, 391–401. [Google Scholar] [CrossRef]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Wilmoth, J.C.; Wang, S.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130. [Google Scholar] [CrossRef]
- Attia, K.A.; Abdelkhalik, A.F.; Ammar, M.H.; Wei, C.; Yang, J.; Lightfoot, D.A.; El-Sayed, W.M.; El-Shemy, H.A. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 2009, 11, i29–i34. [Google Scholar] [PubMed]
- Li, Y.; Han, S.; Qi, Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2023, 65, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, Y.; Zhu, X.; Yang, S.; Liao, X.; Li, H.; Li, Z.; Liao, Q.; Tang, J.; Zhao, G.; et al. Integrated analyses of metabolomics and transcriptomics reveal the potential regulatory roles of long non-coding RNAs in gingerol biosynthesis. BMC Genom. 2023, 24, 490. [Google Scholar] [CrossRef]
- Koo, H.; McDowell, E.T.; Ma, X.; Greer, K.A.; Kapteyn, J.; Xie, Z.; Descour, A.; Kim, H.; Yu, Y.; Kudrna, D.; et al. Ginger and turmeric expressed sequence tags identify signature genes for rhizome identity and development and the biosynthesis of curcuminoids, gingerols and terpenoids. BMC Plant Biol. 2013, 13, 27. [Google Scholar] [CrossRef]
- Edo, G.I.; Igbuku, U.A.; Makia, R.S.; Isoje, E.F.; Gaaz, T.S.; Yousif, E.; Jikah, A.N.; Zainulabdeen, K.; Akpoghelie, P.O.; Opiti, R.A.; et al. Phytochemical profile, therapeutic potentials, nutritional composition, and food applications of ginger: A comprehensive review. Discov. Food 2025, 5, 25. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Luo, D.; Ma, Y.; Zhang, J.; Li, M.; Yao, L.; Shi, X.; Liu, X.; Yang, K. Ginger for health care: An overview of systematic reviews. Complement. Ther. Med. 2019, 45, 114–123. [Google Scholar] [CrossRef]
- Mao, Q.-Q.; Xu, X.-Y.; Cao, S.-Y.; Gan, R.-Y.; Corke, H.; Beta, T.; Li, H.-B. Bioactive Compounds and Bioactivities of Ginger (Zingiber officinale Roscoe). Foods 2019, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, M.; Gupta, A.; Vaidya, D.; Gupta, M. Postharvest Management and Value Addition of Ginger (Zingiber Officinale Roscoe): A Review. Int. J. Environ. Agric. Biotechnol. 2017, 2, 397–412. [Google Scholar] [CrossRef]
- Li, H.L.; Wu, L.; Dong, Z.; Jiang, Y.; Jiang, S.; Xing, H.; Li, Q.; Liu, G.; Tian, S.; Wu, Z.; et al. Haplotype-resolved genome of diploid ginger (Zingiber officinale) and its unique gingerol biosynthetic pathway. Hortic. Res. 2021, 8, 189, Erratum in Hortic. Res. 2021, 8, 242. https://doi.org/10.1038/s41438-021-00700-1. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Nosaki, S.; Mitsuda, N.; Sakamoto, S.; Kusubayashi, K.; Yamagami, A.; Xu, Y.; Bui, T.B.C.; Terada, T.; Miura, K.; Nakano, T.; et al. Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout. Nat. Plants 2022, 8, 1440–1452. [Google Scholar] [CrossRef]
- Zhao, H.; Bao, Y. PIF4: Integrator of light and temperature cues in plant growth. Plant Sci. 2021, 313, 111086. [Google Scholar] [CrossRef]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Luo, X.C.; Sun, M.H.; Xu, R.R.; Shu, H.R.; Wang, J.W.; Zhang, S.Z. Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef]
- Zhang, M.-J.; Xue, Y.-Y.; Xu, S.; Jin, X.-R.; Man, X.-C. Identification of ARF genes in Cucurbita pepo L and analysis of expression patterns, and functional analysis of CpARF22 under drought, salt stress. BMC Genom. 2024, 25, 112. [Google Scholar] [CrossRef]
- Zouine, M.; Fu, Y.; Chateigner-Boutin, A.-L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.-P.; Bouzayen, M. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Shi, Y.; Miao, N.; Bian, Y.; Yin, Z. Diversification, phylogeny and evolution of auxin response factor (ARF) family: Insights gained from analyzing maize ARF genes. Mol. Biol. Rep. 2012, 39, 2401–2415. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, N.; Gao, Z.; Hua, Y.; Cao, Y.; Yin, D.; Jia, Q.; Wang, D. Systematic analysis of the ARF gene family in Fagopyrum dibotrys and its potential roles in stress tolerance. Genetica 2024, 152, 159–178. [Google Scholar] [CrossRef]
- Tombuloglu, H. Genome-wide analysis of the auxin response factors (ARF) gene family in barley (Hordeum vulgare L.). J. Plant Biochem. Biotechnol. 2019, 28, 14–24. [Google Scholar] [CrossRef]
- Chen, D.; Wang, W.; Wu, Y.; Xie, H.; Zhao, L.; Zeng, Q.; Zhan, Y. Expression and Distribution of the Auxin Response Factors in Sorghum bicolor During Development and Temperature Stress. Int. J. Mol. Sci. 2019, 20, 4816. [Google Scholar] [CrossRef]
- Zhai, Y.; Shen, X.; Sun, Y.; Liu, Q.; Ma, N.; Zhang, X.; Jia, Q.; Liang, Z.; Wang, D. Genome-wide investigation of ARF transcription factor gene family and its responses to abiotic stress in Coix (Coix lacryma-jobi L.). Protoplasma 2023, 260, 1389–1405. [Google Scholar] [CrossRef]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Ricke, D.; Lan, T.-H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2012, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Jin, L.; Yarra, R.; Zhou, L.; Cao, H. The auxin response factor (ARF) gene family in Oil palm (Elaeis guineensis Jacq.): Genome-wide identification and their expression profiling under abiotic stresses. Protoplasma 2022, 259, 47–60. [Google Scholar] [CrossRef]
- Lin, J.X.; Ali, A.; Chu, N.; Fu, H.-Y.; Huang, M.-T.; Mbuya, S.N.; Gao, S.-J.; Zhang, H.-L. Identification of ARF transcription factor gene family and its defense responses to bacterial infection and salicylic acid treatment in sugarcane. Front. Microbiol. 2023, 14, 1257355. [Google Scholar] [CrossRef]
- Mo, Z.H.; Zhang, Y.; Hu, L.J.; Zhai, M.; Xuan, J.P. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in pecan indicates its possible roles during graft union formation. Sci. Hortic. 2023, 322, 112401. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.; Wang, J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.-E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell. 2011, 23, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell. 2009, 21, 3119–3132. [Google Scholar] [CrossRef]
- Diao, R.; Zhao, M.; Liu, Y.; Zhang, Z.; Zhong, B. The advantages of crosstalk during the evolution of the BZR1-ARF6-PIF4 (BAP) module. J. Integr. Plant Biol. 2023, 65, 2631–2644. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.; Jiao, Y.; Qin, Y.; Liu, X.; He, K.; Chen, C.; Ma, L.; Wang, J.; Xiong, L.; et al. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol. Biol. 2007, 63, 591–608. [Google Scholar] [CrossRef]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.Z.; Hong, X.H.; Gong, Z.Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef]
- Sims, K.; Abedi-Samakush, F.; Szulc, N.; Honti, M.G.M.; Mattsson, J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021, 22, 4089. [Google Scholar] [CrossRef]
- Ma, H.; Cai, L.; Lin, J.; Zhou, K.; Li, Q.Q. Divergence in the Regulation of the Salt Tolerant Response Between Arabidopsis thaliana and Its Halophytic Relative Eutrema salsugineum by mRNA Alternative Polyadenylation. Front. Plant Sci. 2022, 13, 866054. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
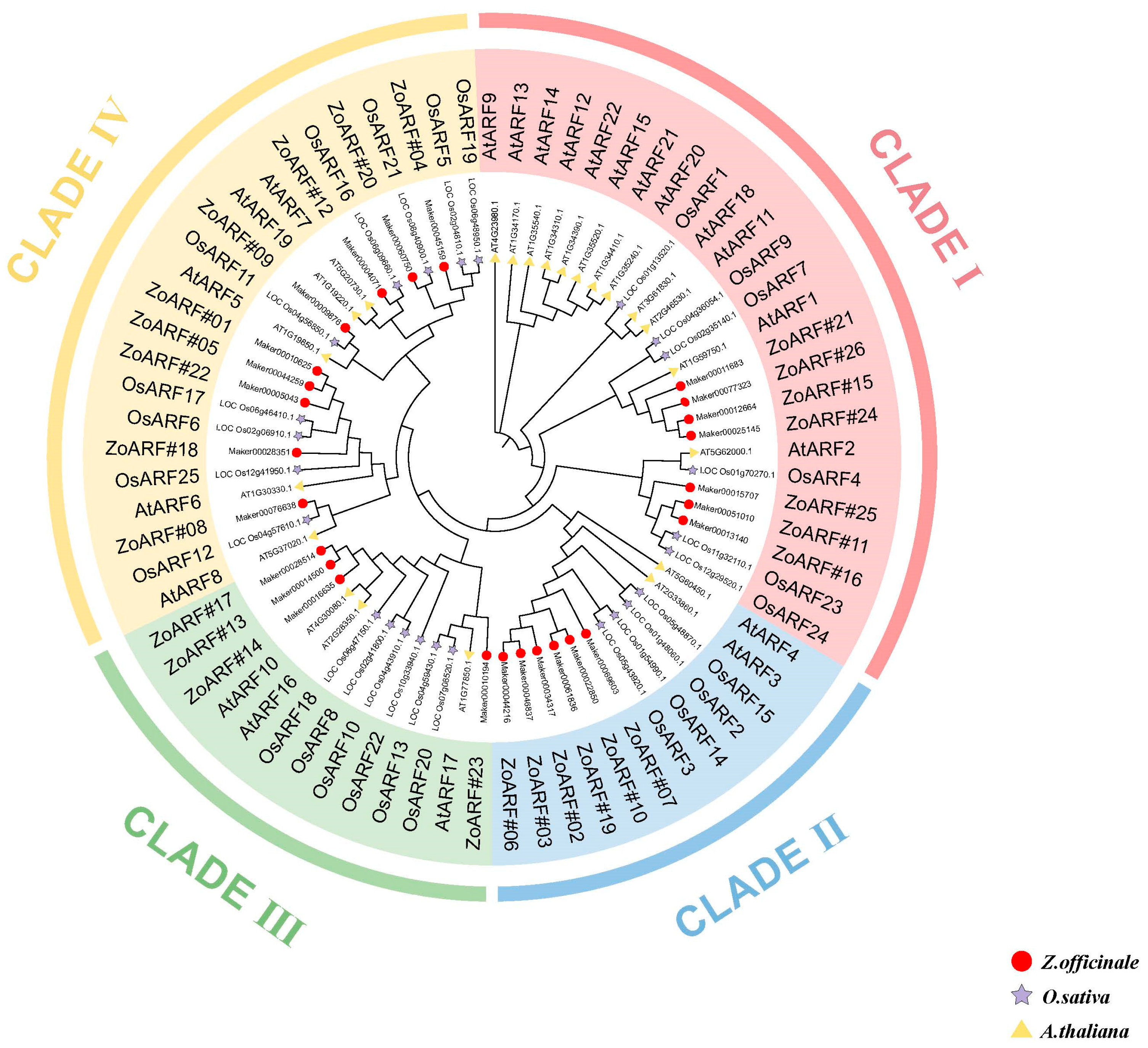
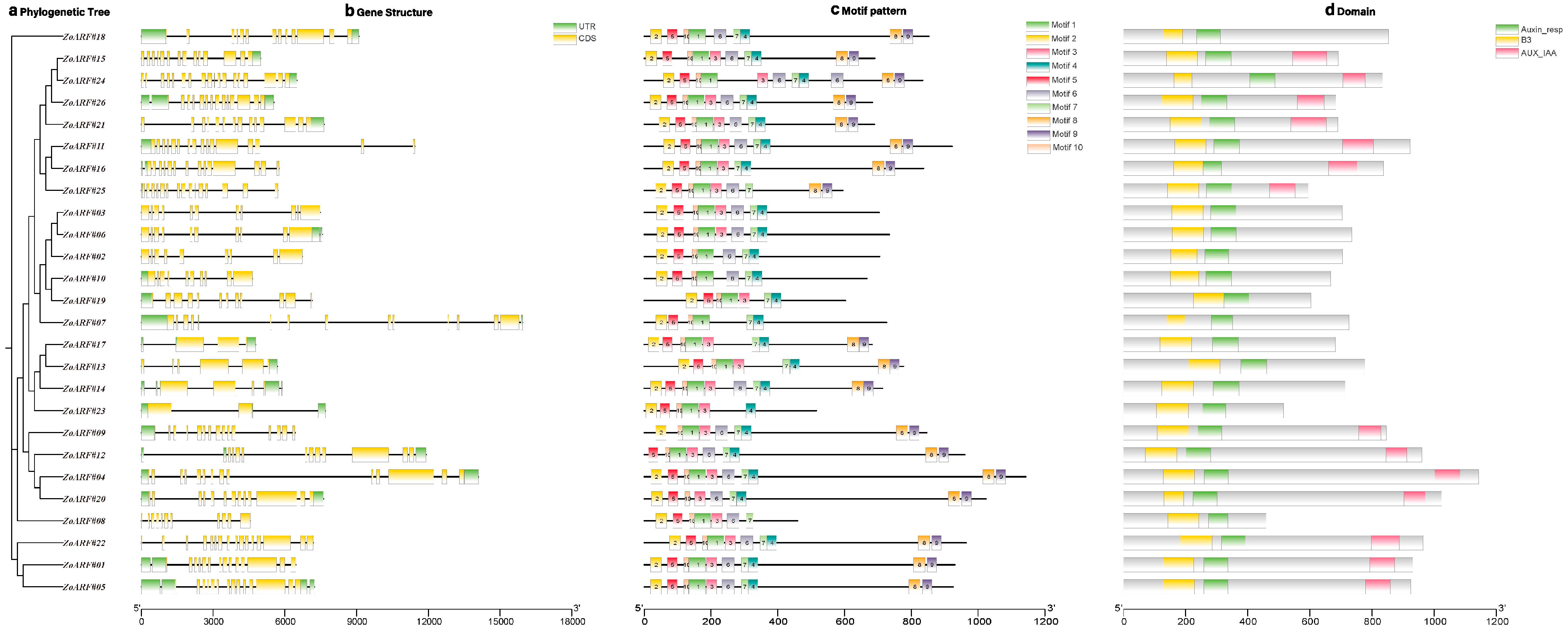
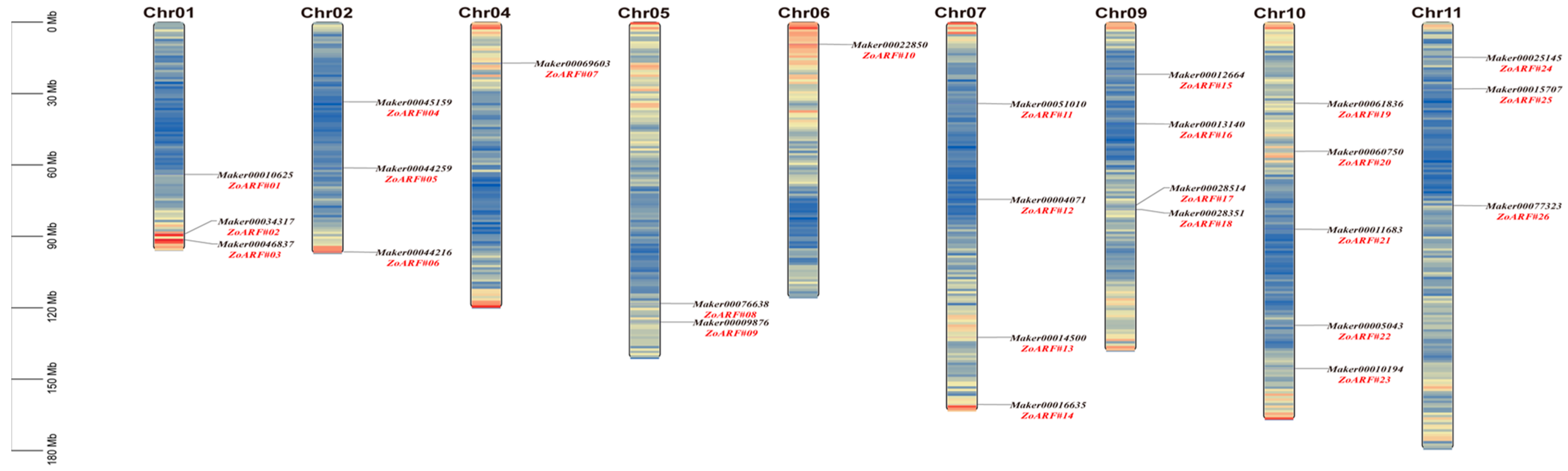
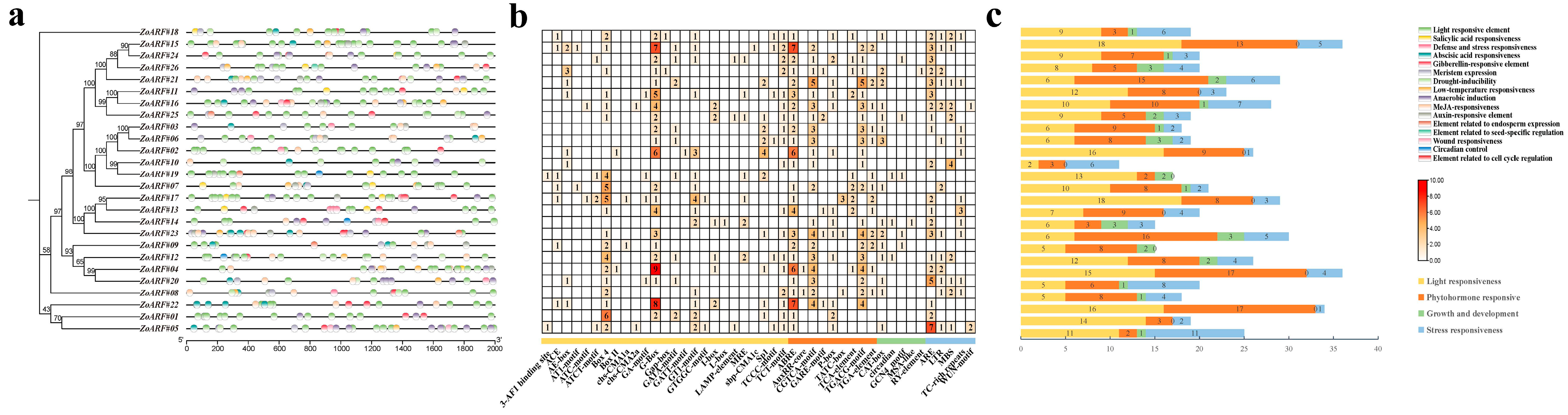


| Gene ID | Rename | Group | Chromsome | mRNA Start | mRNA End | Strand | No. of Exon | No. of Intron | CDS Length | Amino Acid Length | Isoelectric Point | Molecular Weight | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maker00034317 | ZoARF#02 | II | Chr01 | 88858607 | 88865333 | − | 9 | 8 | 2115 | 704 | 6.09 | 76,431.35 | nucleus |
| Maker00010625 | ZoARF#01 | IV | Chr01 | 63966592 | 63973047 | − | 14 | 13 | 2793 | 930 | 5.96 | 103,376.9 | nucleus |
| Maker00046837 | ZoARF#03 | II | Chr01 | 91467289 | 91474763 | + | 11 | 10 | 2112 | 703 | 6.49 | 77,384.55 | nucleus |
| Maker00044216 | ZoARF#06 | II | Chr02 | 96517654 | 96525217 | − | 10 | 9 | 2205 | 734 | 6.36 | 80,760.08 | nucleus |
| Maker00045159 | ZoARF#04 | IV | Chr02 | 33489395 | 33503493 | + | 14 | 13 | 3429 | 1142 | 6.29 | 127,342.96 | nucleus |
| Maker00044259 | ZoARF#05 | IV | Chr02 | 61251400 | 61258631 | − | 14 | 13 | 2775 | 924 | 6.21 | 102,663.2 | nucleus |
| Maker00069603 | ZoARF#07 | II | Chr04 | 17191057 | 17206998 | + | 14 | 13 | 1680 | 559 | 5.49 | 81,066.21 | nucleus |
| Maker00009876 | ZoARF#09 | IV | Chr05 | 125989767 | 125996191 | + | 15 | 14 | 2538 | 845 | 5.54 | 94,659.43 | nucleus |
| Maker00076638 | ZoARF#08 | IV | Chr05 | 118203804 | 118208349 | − | 11 | 10 | 1377 | 458 | 8.55 | 50,857.31 | cytoskeleton |
| Maker00022850 | ZoARF#10 | II | Chr06 | 9308738 | 9313381 | − | 10 | 9 | 2001 | 666 | 6.83 | 73,121.55 | nucleus |
| Maker00016635 | ZoARF#14 | III | Chr07 | 160616034 | 160621927 | − | 4 | 3 | 2142 | 713 | 7.53 | 78,081.88 | nucleus |
| Maker00051010 | ZoARF#11 | I | Chr07 | 34201967 | 34213404 | + | 16 | 15 | 2766 | 921 | 6.21 | 102,241.46 | nucleus |
| Maker00014500 | ZoARF#13 | III | Chr07 | 132442912 | 132448595 | + | 6 | 5 | 2331 | 776 | 7.03 | 86,413.33 | nucleus |
| Maker00004071 | ZoARF#12 | IV | Chr07 | 74447622 | 74459523 | − | 11 | 10 | 2883 | 960 | 6.07 | 107,389.35 | nucleus |
| Maker00028351 | ZoARF#18 | IV | Chr09 | 78769700 | 78778791 | − | 12 | 11 | 2559 | 852 | 5.81 | 94,688.59 | nucleus |
| Maker00013140 | ZoARF#16 | I | Chr09 | 42682558 | 42688310 | − | 14 | 13 | 2511 | 836 | 6.04 | 93,016.74 | nucleus |
| Maker00012664 | ZoARF#15 | I | Chr09 | 21899860 | 21904846 | + | 14 | 13 | 2073 | 690 | 6.36 | 77,079.79 | Endoplasmic reticulum |
| Maker00028514 | ZoARF#17 | III | Chr09 | 76701982 | 76706759 | + | 3 | 2 | 2046 | 681 | 6.97 | 75,501.6 | nucleus |
| Maker00060750 | ZoARF#20 | IV | Chr10 | 54271985 | 54279602 | − | 13 | 12 | 3072 | 1023 | 6.04 | 114,496.96 | nucleus |
| Maker00010194 | ZoARF#23 | III | Chr10 | 145551145 | 145558840 | − | 2 | 1 | 1545 | 514 | 6.53 | 56,971.54 | cytoskeleton |
| Maker00011683 | ZoARF#21 | I | Chr10 | 87004599 | 87012232 | − | 14 | 13 | 2070 | 689 | 6.62 | 77,586.95 | mitochondrion |
| Maker00005043 | ZoARF#22 | IV | Chr10 | 127366956 | 127374139 | − | 16 | 15 | 2892 | 963 | 6.18 | 107,125.42 | nucleus |
| Maker00061836 | ZoARF#19 | II | Chr10 | 34100555 | 34107690 | + | 12 | 11 | 1809 | 602 | 7.83 | 67,207 | nucleus |
| Maker00015707 | ZoARF#25 | I | Chr11 | 28036173 | 28041877 | + | 15 | 14 | 1785 | 594 | 5.87 | 66,223.6 | nucleus |
| Maker00025145 | ZoARF#24 | I | Chr11 | 14800662 | 14807162 | + | 18 | 17 | 2502 | 833 | 6.02 | 93,122.76 | nucleus |
| Maker00077323 | ZoARF#26 | I | Chr11 | 77040284 | 77045821 | − | 14 | 13 | 2052 | 683 | 6.04 | 75,684.72 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, Y.; Xu, S.; Shi, J.; He, Y.; Li, H.-L.; Yu, T.; Zhang, S.; Xing, H.-T. Genome-Wide Analysis and Expression Profiles of Auxin Response Factors in Ginger (Zingiber officinale Roscoe). Int. J. Mol. Sci. 2025, 26, 8412. https://doi.org/10.3390/ijms26178412
Tong Y, Xu S, Shi J, He Y, Li H-L, Yu T, Zhang S, Xing H-T. Genome-Wide Analysis and Expression Profiles of Auxin Response Factors in Ginger (Zingiber officinale Roscoe). International Journal of Molecular Sciences. 2025; 26(17):8412. https://doi.org/10.3390/ijms26178412
Chicago/Turabian StyleTong, Yuanyuan, Sujuan Xu, Jiayu Shi, Yi He, Hong-Lei Li, Tian Yu, Sinian Zhang, and Hai-Tao Xing. 2025. "Genome-Wide Analysis and Expression Profiles of Auxin Response Factors in Ginger (Zingiber officinale Roscoe)" International Journal of Molecular Sciences 26, no. 17: 8412. https://doi.org/10.3390/ijms26178412
APA StyleTong, Y., Xu, S., Shi, J., He, Y., Li, H.-L., Yu, T., Zhang, S., & Xing, H.-T. (2025). Genome-Wide Analysis and Expression Profiles of Auxin Response Factors in Ginger (Zingiber officinale Roscoe). International Journal of Molecular Sciences, 26(17), 8412. https://doi.org/10.3390/ijms26178412







