Multi-Stage Transcriptome Analysis Identifies Key Molecular Pathways for Soybean Under Phosphorus-Limited Conditions
Abstract
1. Introduction
2. Results
2.1. Different Patterns of Phosphorus Distribution in Soybean Organs
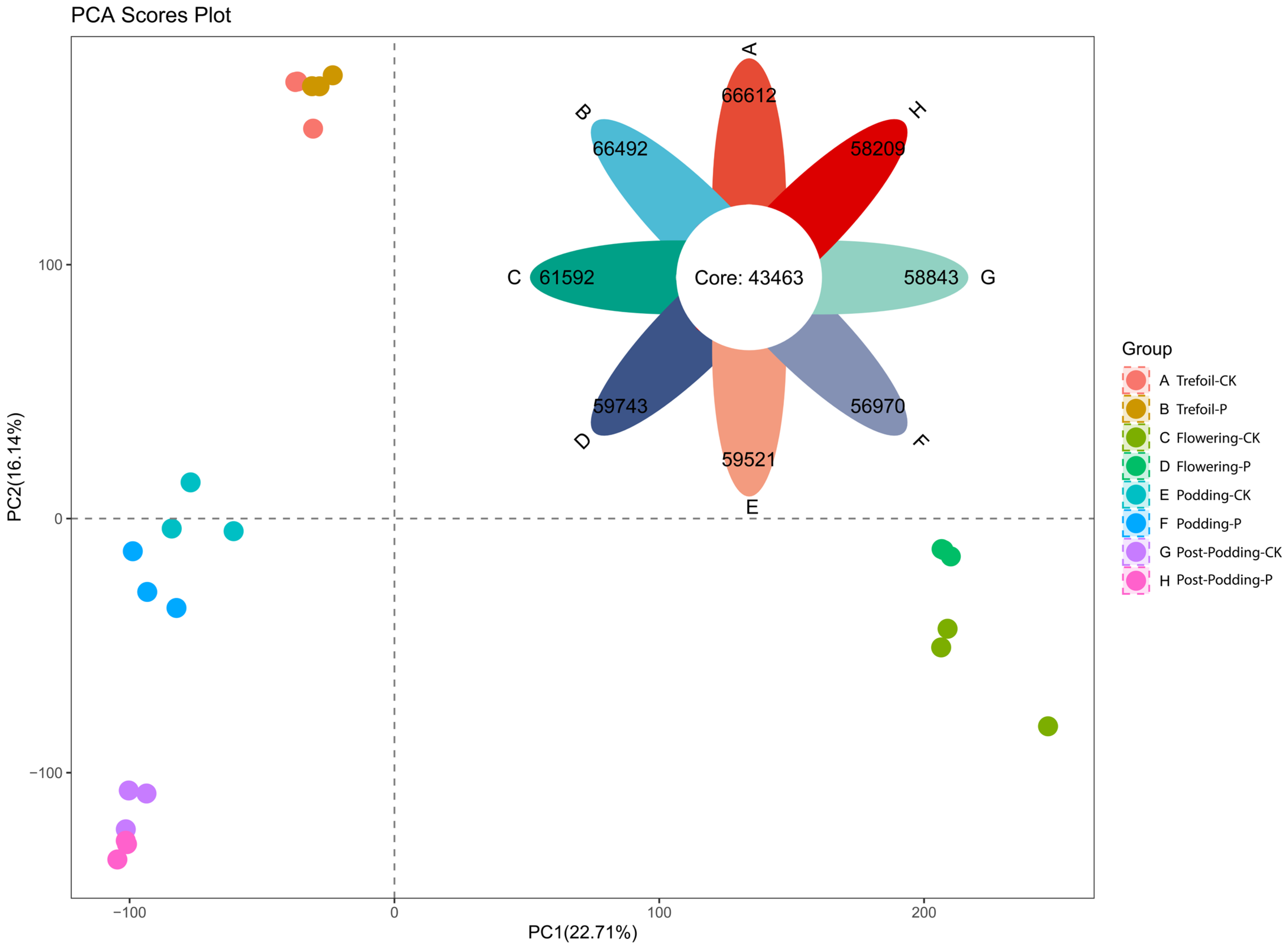
2.2. Transcriptome Sequencing and Quality Assessment
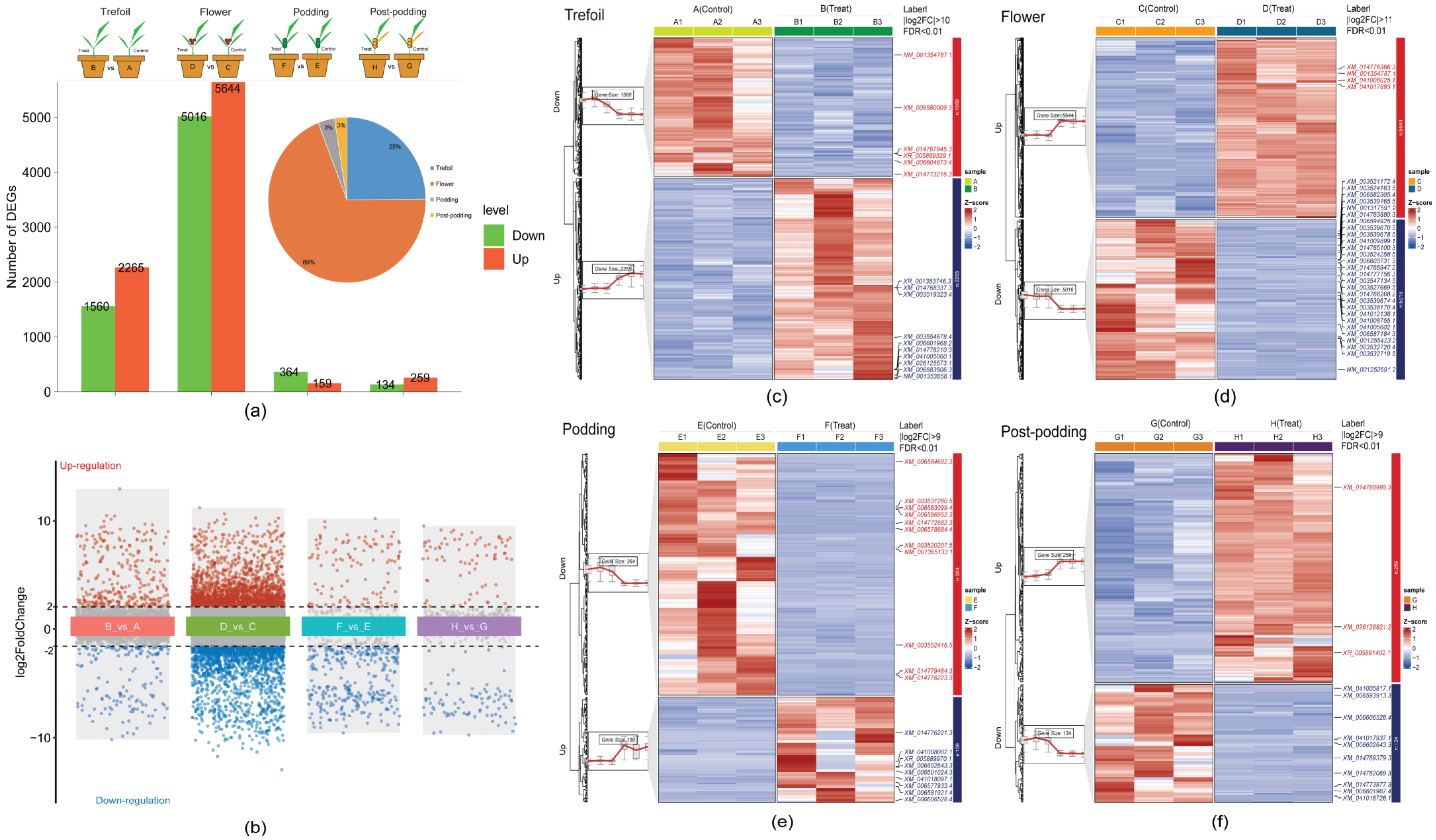
2.3. Differentially Expressed Genes Across Developmental Stages
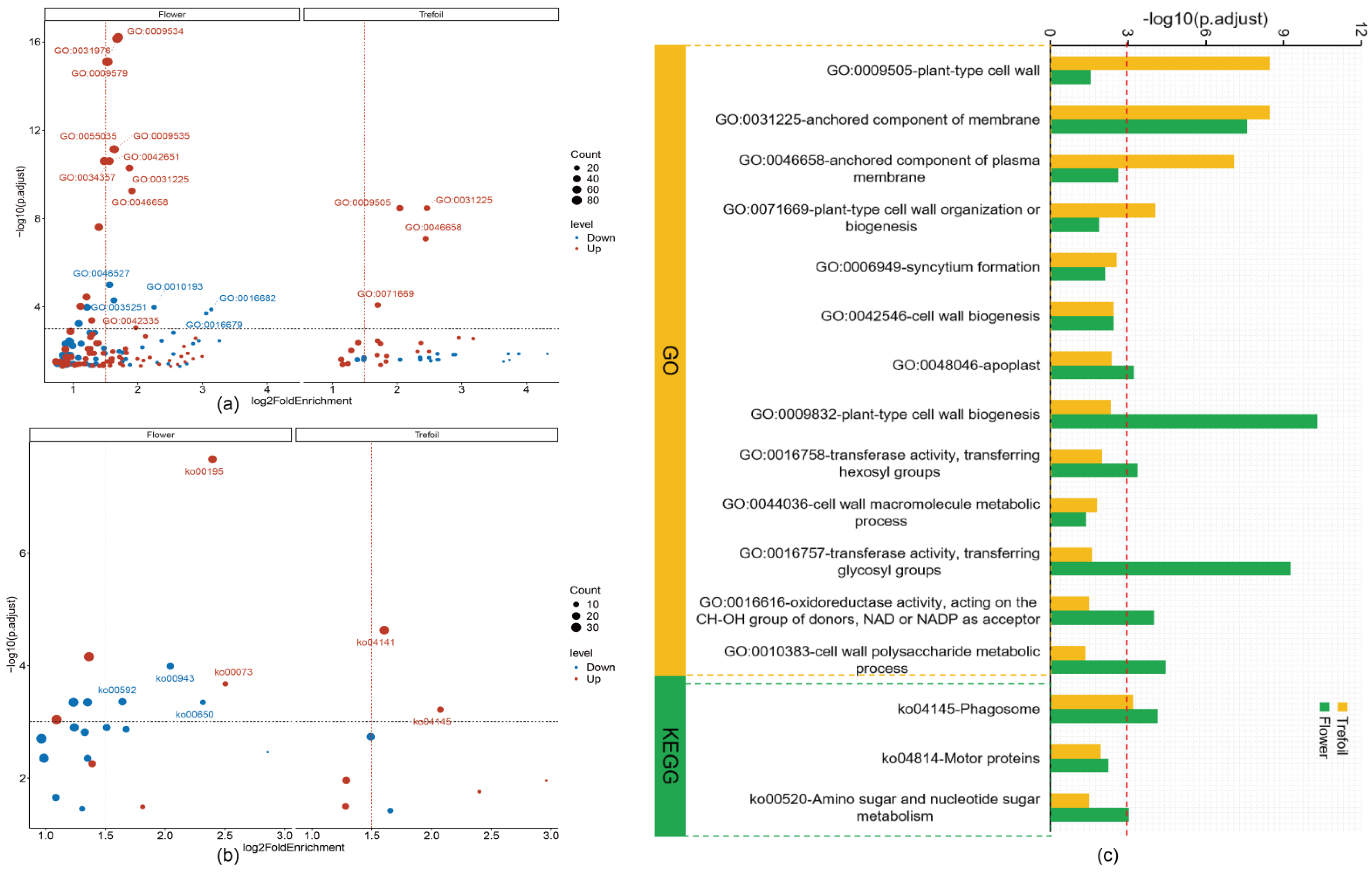
2.4. Functional Enrichment Analysis of DEGs
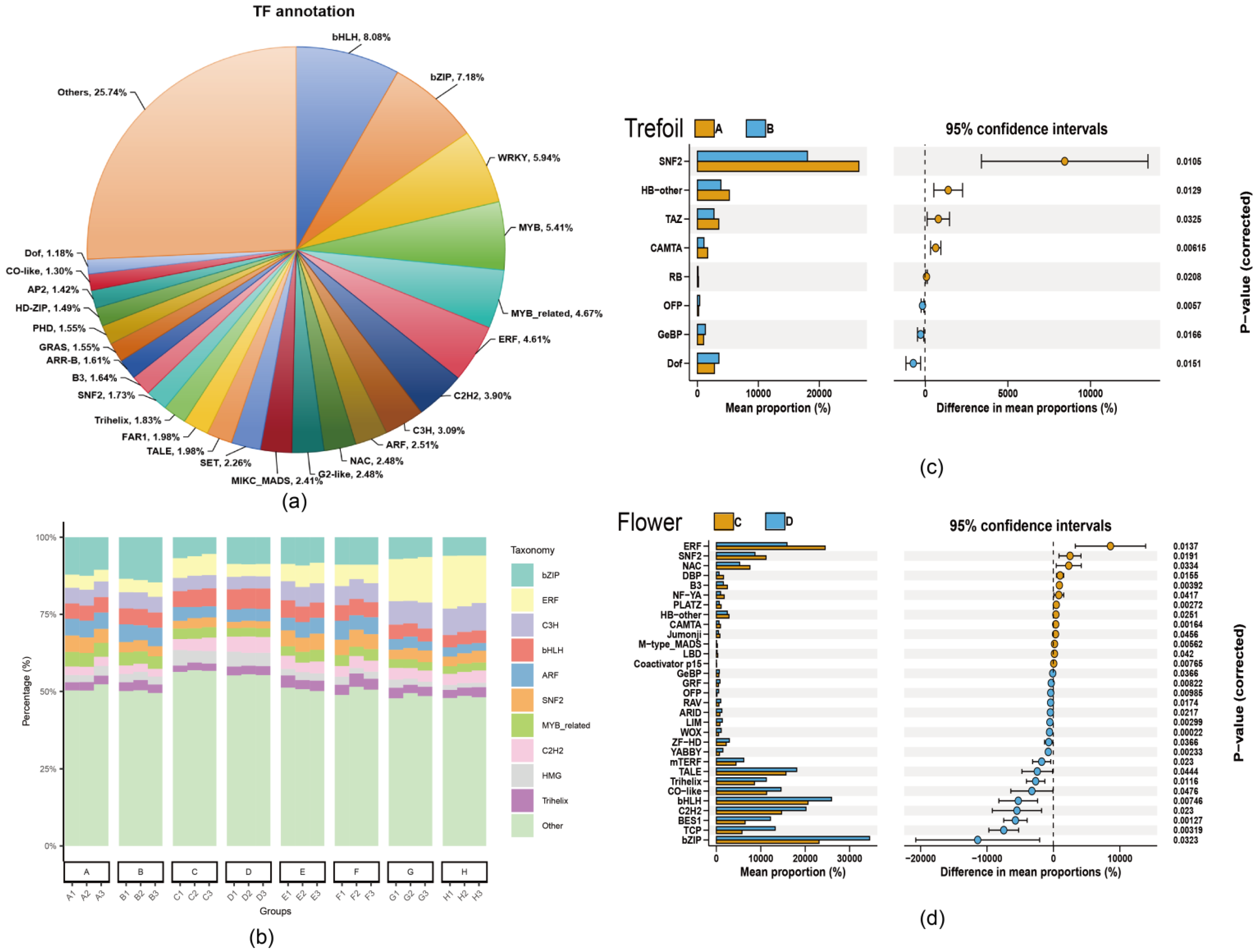
2.5. Transcription Factor Dynamics Under Low-Phosphorus Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Sample Collection and Preparation
4.3. RNA Extraction and Quality Control
4.4. Library Construction and RNA Sequencing
4.5. Functional Enrichment Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giri, J.; Bhosale, R.; Huang, G.; Pandey, B.K.; Parker, H.; Zappala, S.; Yang, J.; Dievart, A.; Bureau, C.; Ljung, K. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018, 9, 1408, Correction in Nat. Commun. 2018, 9, 1810. [Google Scholar] [CrossRef]
- Peng, W.; Wu, W.; Peng, J.; Li, J.; Lin, Y.; Wang, Y.; Tian, J.; Sun, L.; Liang, C.; Liao, H. Characterization of the soybean GmALMT family genes and the function of GmALMT5 in response to phosphate starvation. J. Integr. Plant Biol. 2018, 60, 216–231. [Google Scholar] [CrossRef]
- Mo, X.; Zhang, M.; Liang, C.; Cai, L.; Tian, J. Integration of metabolome and transcriptome analyses highlights soybean roots responding to phosphorus deficiency by modulating phosphorylated metabolite processes. Plant Physiol. Biochem. 2019, 139, 697–706. [Google Scholar] [CrossRef]
- Yamaji, N.; Takemoto, Y.; Miyaji, T.; Mitani-Ueno, N.; Yoshida, K.T.; Ma, J.F. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 2017, 541, 92–95, Erratum in Nature 2017, 543, 136. [Google Scholar] [CrossRef]
- Chu, S.; Zhang, X.; Yu, K.; Lv, L.; Sun, C.; Liu, X.; Zhang, J.; Jiao, Y.; Zhang, D. Genome-wide analysis reveals dynamic epigenomic differences in soybean response to low-phosphorus stress. Int. J. Mol. Sci. 2020, 21, 6817. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, D. Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 2008, 50, 849–859. [Google Scholar] [CrossRef]
- Fixen, P.E.; Johnston, A.M. World fertilizer nutrient reserves: A view to the future. J. Sci. Food Agric. 2012, 92, 1001–1005. [Google Scholar] [CrossRef]
- Han, Y.; White, P.J.; Cheng, L. Mechanisms for improving phosphorus utilization efficiency in plants. Ann. Bot. 2022, 129, 247–258. [Google Scholar] [CrossRef]
- Cong, W.-F.; Suriyagoda, L.D.; Lambers, H. Tightening the phosphorus cycle through phosphorus-efficient crop genotypes. Trends Plant Sci. 2020, 25, 967–975. [Google Scholar] [CrossRef]
- Zhang, D.; Song, H.; Cheng, H.; Hao, D.; Wang, H.; Kan, G.; Jin, H.; Yu, D. The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet. 2014, 10, e1004061. [Google Scholar] [CrossRef]
- Cai, Z.; Cheng, Y.; Xian, P.; Ma, Q.; Wen, K.; Xia, Q.; Zhang, G.; Nian, H. Acid phosphatase gene GmHAD1 linked to low phosphorus tolerance in soybean, through fine mapping. Theor. Appl. Genet. 2018, 131, 1715–1728. [Google Scholar] [CrossRef]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Young, I.M.; Hallett, P.D.; White, P.J.; George, T.S. Root hairs improve root penetration, root–soil contact, and phosphorus acquisition in soils of different strength. J. Exp. Bot. 2013, 64, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Shane, M.W.; Cramer, M.D.; Pearse, S.J.; Veneklaas, E.J. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann. Bot. 2006, 98, 693–713. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- De Bang, T.C.; Husted, S.; Laursen, K.H.; Persson, D.P.; Schjoerring, J.K. The molecular–physiological functions of mineral macronutrients and their consequences for deficiency symptoms in plants. New Phytol. 2021, 229, 2446–2469. [Google Scholar] [CrossRef]
- Zha, B.; Zhang, C.; Yuan, R.; Zhao, K.; Sun, J.; Liu, X.; Wang, X.; Zhang, F.; Zhang, B.; Lamlom, S.F. Integrative QTL mapping and candidate gene analysis for main stem node number in soybean. BMC Plant Biol. 2025, 25, 422. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, K.; Ren, H.; Lamlom, S.F.; Liu, X.; Wang, X.; Zhang, F.; Yuan, R.; Wang, J. Comparative study of isoflavone synthesis genes in two wild soybean varieties using transcriptomic analysis. Agriculture 2023, 13, 1164. [Google Scholar] [CrossRef]
- Zhang, C.; Zha, B.; Yuan, R.; Zhao, K.; Sun, J.; Liu, X.; Wang, X.; Zhang, F.; Zhang, B.; Lamlom, S.F. Identification of Quantitative Trait Loci for Node Number, Pod Number, and Seed Number in Soybean. Int. J. Mol. Sci. 2025, 26, 2300. [Google Scholar] [CrossRef]
- Wilson, J.D.; Zheljazkov, V.D.; Rathgeber, B.; Caldwell, C.D.; Burton, D.L. Reduction in phosphorus availability in poultry and diary manure by mineral amendments. Soil Sci. Plant Nutr. 2008, 54, 600–605. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef]
- Li, X.; Gai, J.; Chang, W.; Zhang, C. Identification of phosphorus starvation tolerant soybean (Glycine max) germplasms. Front. Agric. China 2010, 4, 272–279. [Google Scholar] [CrossRef]
- Tesfaye, A.; Githiri, M.; Derera, J.; Debele, T. Genetic variability in soybean (Glycine max L.) for low soil phosphorus tolerance. Ethiop. J. Agric. Sci. 2017, 27, 1–15. [Google Scholar]
- He, J.; Jin, Y.; Du, Y.-L.; Wang, T.; Turner, N.C.; Yang, R.-P.; Siddique, K.H.; Li, F.-M. Genotypic variation in yield, yield components, root morphology and architecture, in soybean in relation to water and phosphorus supply. Front. Plant Sci. 2017, 8, 1499. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Temple, G.; Temple, S.J.; Beschow, H.; Vance, C.P. Nitrogen fixation by white lupin under phosphorus deficiency. Ann. Bot. 2006, 98, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.Q.; Saleem, M.F.; Khan, H.Z.; Anjum, S.A. Performance of soybean (Glycine max L.) under different phosphorus levels and inoculation. Pak. J. Agric. Sci. 2009, 46, 237–241. [Google Scholar]
- Zeng, H.; Zhang, X.; Zhang, X.; Pi, E.; Xiao, L.; Zhu, Y. Early transcriptomic response to phosphate deprivation in soybean leaves as revealed by RNA-sequencing. Int. J. Mol. Sci. 2018, 19, 2145. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Lin, Y.; Lin, P.; Lu, Y.; Zheng, J.; Zheng, Y.; Huang, X.; Zhao, X.; Cui, L. Post-Translational Modifications of RNA-Modifying Proteins in Cellular Dynamics and Disease Progression. Adv. Sci. 2024, 11, 2406318. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Wen, R.; Yang, N.; Zhang, T.-N.; Liu, C.-F. Roles of protein post-translational modifications in glucose and lipid metabolism: Mechanisms and perspectives. Mol. Med. 2023, 29, 93. [Google Scholar] [CrossRef]
- Luo, D.; Wang, L.; Nan, H.; Cao, Y.; Wang, H.; Kumar, T.V.; Wang, C. Phosphorus adsorption by functionalized biochar: A review. Environ. Chem. Lett. 2023, 21, 497–524. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.-P.; Yang, Y.; Yu, M.; Wang, C.; Yan, J. Interactive effects of nitrogen and phosphorus additions on plant growth vary with ecosystem type. Plant Soil 2019, 440, 523–537. [Google Scholar] [CrossRef]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1; 8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A constitutive expressed phosphate transporter, OsPht1; 1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Zhang, X.; Fan, H.; Gu, M.; Qu, H.; Xu, G. Phosphate transporter OsPht1; 8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 2015, 230, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Siddique, A.B.; Shabala, S.; Zhou, M.; Zhao, C. Phosphorus plays key roles in regulating plants’ physiological responses to abiotic stresses. Plants 2023, 12, 2861. [Google Scholar] [CrossRef]
- Sánchez-Calderón, L.; Chacon-López, A.; Pérez-Torres, C.-A.; Herrera-Estrella, L. Phosphorus: Plant strategies to cope with its scarcity. In Cell Biology of Metals and Nutrients; Springer: Berlin/Heidelberg, Germany, 2010; Volume 17, pp. 173–198. [Google Scholar] [CrossRef]
- Dunja, B. Phosphorous Uptake and Utilization Efficiency in Cluster Root and Non-Cluster Root Forming Species of the Core Cape Subregion, South Africa. Master’s Thesis, University of Cape Town, Faculty of Science, Department of Biological Sciences, Cape Town, South Africa, 2015. Available online: http://hdl.handle.net/11427/15470 (accessed on 18 July 2025).
- Tang, H.; Wang, Y.; Niu, L.; Wei, J.; Chen, Y. Enhanced adaptation to low-p stress by altering rhizosphere exudation and P-uptake rate other than root morphological traits in two maize genotypes. Preprints 2019. [Google Scholar] [CrossRef]
- George, T.S.; White, P.J. Advances in understanding crop use of phosphorus. In Achieving Sustainable Crop Nutrition; Taylor & Francis Group: Abingdon, UK, 2020; pp. 83–114. [Google Scholar]
- Murata, Y.; Takahashi, M. An alternative electron transfer pathway mediated by chloroplast envelope. Plant Cell Physiol. 1999, 40, 1007–1013. [Google Scholar] [CrossRef]
- Liu, J.; Xu, L.; Wang, Y.; Han, L. Ethephon treatment reduced Mondo grass (Ophiopogon japonicus) gas exchange rate and gene expression of Rbcs. Eur. J. Hortic. Sci. 2019, 84, 106–112. [Google Scholar] [CrossRef]
- Wasternack, C. Action of jasmonates in plant stress responses and development—Applied aspects. Biotechnol. Adv. 2014, 32, 31–39. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Chiou, T.-J.; Lin, S.-I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Huang, T.-K.; Kuo, H.-F.; Chiou, T.-J. Role of vacuoles in phosphorus storage and remobilization. J. Exp. Bot. 2017, 68, 3045–3055. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, S.; Chen, C.; Yang, J.; Cheng, L.; Ma, F.; Widemann, E.; Sun, Q. DNA topoisomerase 1 prevents R-loop accumulation to modulate auxin-regulated root development in rice. Mol. Plant 2017, 10, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, Y.; Chu, H.; Wang, L.; Fu, Y.; Liu, P.; Upadhyaya, N.; Chen, C.; Mou, T.; Feng, Y. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 2015, 11, e1005464. [Google Scholar] [CrossRef]
- Gu, M.; Zhang, J.; Li, H.; Meng, D.; Li, R.; Dai, X.; Wang, S.; Liu, W.; Qu, H.; Xu, G. Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3-type MYB transcription factor in rice. J. Exp. Bot. 2017, 68, 3603–3615. [Google Scholar] [CrossRef]
- Slabaugh, E.; Held, M.; Brandizzi, F. Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 2011, 67, 395–405. [Google Scholar] [CrossRef]
- Yi, K.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 2010, 42, 264–267. [Google Scholar] [CrossRef]
- Pires, N.D.; Yi, K.; Breuninger, H.; Catarino, B.; Menand, B.; Dolan, L. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 9571–9576. [Google Scholar] [CrossRef]
- Breuninger, H.; Thamm, A.; Streubel, S.; Sakayama, H.; Nishiyama, T.; Dolan, L. Diversification of a transcription factor family led to the evolution of antagonistically acting genetic regulators of root hair growth. Curr. Biol. 2016, 26, 1622–1628. [Google Scholar] [CrossRef]
- Xu, P.; Cai, X.-T.; Wang, Y.; Xing, L.; Chen, Q.; Xiang, C.-B. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J. Exp. Bot. 2014, 65, 4285–4295. [Google Scholar] [CrossRef]
- Bhosale, R.; Giri, J.; Pandey, B.K.; Giehl, R.F.; Hartmann, A.; Traini, R.; Truskina, J.; Leftley, N.; Hanlon, M.; Swarup, K. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 2018, 9, 1409, Correction in Nat. Commun. 2018, 9, 1818. [Google Scholar] [CrossRef]
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef]
- Le Hir, R.; Bellini, C. The plant-specific Dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Lamlom, S.F.; Wang, X.; Zhang, C.; Zhang, F.; Zhao, K.; Yuan, R.; Zhang, B.; Ren, H. Multi-Stage Transcriptome Analysis Identifies Key Molecular Pathways for Soybean Under Phosphorus-Limited Conditions. Int. J. Mol. Sci. 2025, 26, 8385. https://doi.org/10.3390/ijms26178385
Liu X, Lamlom SF, Wang X, Zhang C, Zhang F, Zhao K, Yuan R, Zhang B, Ren H. Multi-Stage Transcriptome Analysis Identifies Key Molecular Pathways for Soybean Under Phosphorus-Limited Conditions. International Journal of Molecular Sciences. 2025; 26(17):8385. https://doi.org/10.3390/ijms26178385
Chicago/Turabian StyleLiu, Xiulin, Sobhi F. Lamlom, Xueyang Wang, Chunlei Zhang, Fengyi Zhang, Kezhen Zhao, Rongqiang Yuan, Bixian Zhang, and Honglei Ren. 2025. "Multi-Stage Transcriptome Analysis Identifies Key Molecular Pathways for Soybean Under Phosphorus-Limited Conditions" International Journal of Molecular Sciences 26, no. 17: 8385. https://doi.org/10.3390/ijms26178385
APA StyleLiu, X., Lamlom, S. F., Wang, X., Zhang, C., Zhang, F., Zhao, K., Yuan, R., Zhang, B., & Ren, H. (2025). Multi-Stage Transcriptome Analysis Identifies Key Molecular Pathways for Soybean Under Phosphorus-Limited Conditions. International Journal of Molecular Sciences, 26(17), 8385. https://doi.org/10.3390/ijms26178385






