FACS-Based Assessment of Human Hematopoietic Stem and Progenitor Cells
Abstract
1. Introduction
2. Experimental Design
3. Materials and Equipment
- Only use polypropylene (PP) tubes for the whole procedure (including FACS tubes). HSCs tend to stick to polystyrene (PS).
- Some staining protocols require commercially purchased staining buffers. These alternatives can be used instead of Horizon™ Brilliant Stain Buffer.
- Alternative products can be purchased from different providers at similar prices. The listed catalog numbers and providers are only a recommendation based on personal experience.
- MACS washing buffer: Dilute MACS® BSA Stock Solution at a ratio of 1:20 with autoMACS® Rinsing Solution; a temperature of 2–8 °C should be maintained.
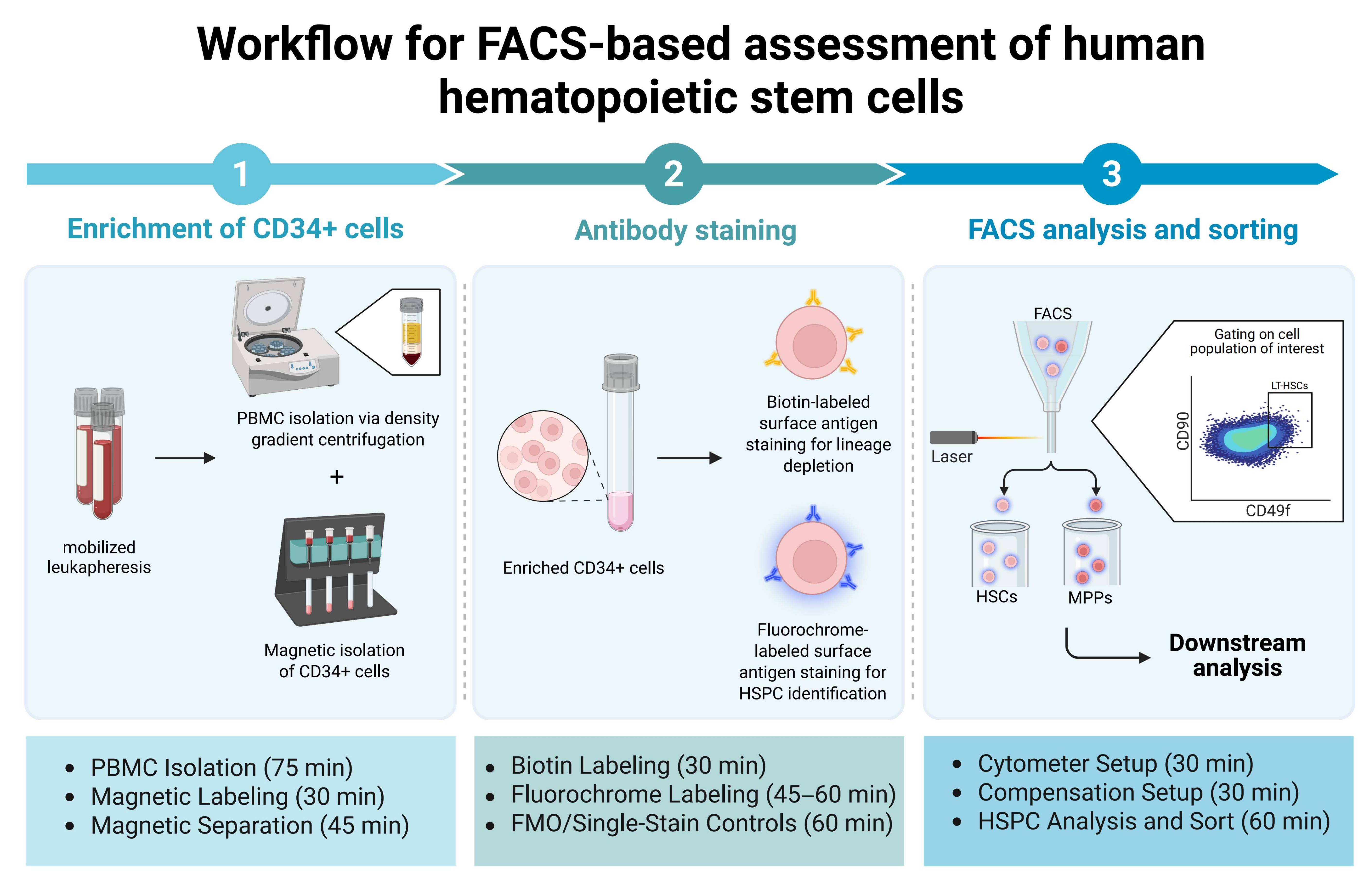
4. Protocol
4.1. Enrichment of CD34+ Cells from Leukapheresis Products After Mobilization
4.1.1. Isolation of PBMCs from Fresh Leukapheresis
- Dilute the leukapheresis products with PBS (1×) at ratios of 1:1 or 1:2.
- Aliquot 3 mL of Pancoll into a 15 mL falcon tube and bring to room temperature (RT).
- Top 3 mL aliquoted Pancoll with cell suspension.
- Centrifuge at 400× g for 30 min at RT without break.
- Isolate PMBCs from the interphase between the plasma and Pancoll layer. Transfer the interphase into a fresh 15 mL falcon tube.
- Wash isolated PMBCs 2 times with 5–7 mL PBS (1×) for 10 min at 200× g, 20 °C.
- Resuspend washed mononuclear cells in 5 mL MACS® washing buffer. Take 10 µL of the cell suspension to determine cell number. Centrifuge PMBCs again for 7 min at 300× g, 4 °C.
- Resuspend cells in 300 µL MACS® washing buffer for up to 108 cells (if this cell number is exceeded, increase the reagent volume for magnetic cell labeling).
- During density gradient centrifugation, it is crucial to avoid mixing the layers of the leukapheresis product and Pancoll solution. This careful separation allows the effective isolation of lymphocytes and monocytes from other types of blood cells. During centrifugation, erythrocytes are aggregated by polysucrose and rapidly sediment, whereas mononuclear cells remain at the plasma interphase. Erythrocytes pellet to the bottom of the centrifuge tube. Due to their higher density, granulocytes will not remain in the interphase and will move toward the bottom of the centrifuge tube.
- Centrifugation below RT can lead to impaired separation of macromolecules in the leukapheresis sample.
- After density gradient separation, the isolated PBMC solution will contain platelets. Washing at low speed, 20 °C for 10 min, helps remove platelets from cell suspension and increase purity.
- Cell number should be obtained manually via microscopy. We recommend counting cells in a Neubauer chamber. For this protocol, 10 µL of cell suspension is mixed with 10 µL of trypan blue (diluted at 1:1 with 1× PBS). C-Chips are loaded with 10 µL of pre-mixed cell/trypan blue solution.
4.1.2. Processing of Frozen PBMCs
- Thaw the previously isolated and frozen PBMCs from leukapheresis (mob LPs) by immersing the vials in a 37 °C water bath.
- Transfer completely thawed cells into a 15 mL falcon tube filled with 10 mL of thawing media at RT.
- Centrifuge cells at 300× g for 7 min at RT.
- Resuspend cells in 10 mL StemStan™ SFEM II media and wash again at 300× g for 7 min at RT.
- Determine cell number and viability and resuspend in appropriate volume of ml MACS® washing buffer.
4.1.3. Magnetic Labeling and Separation of Isolated PBMCs
- To obtain pre-separation single-cell solution, pass cells through 30 µm nylon mesh.
- Add 100 µL of FcR Blocking Reagent.
- Add 100 µL of CD34 MicroBeads UltraPure.
- Mix the solution and incubate for 30 min at 4 °C.
- Wash cells with 5–10 mL buffer, centrifuge for 10 min at 300× g, 4 °C.
- Resuspend cells in 500 µL buffer for up to 108 cells.
- Avoid air bubbles, since they can block the column.
- Pre-separation filtering removes cell clumps and debris. The single-cell solution increases labeling performance and prevents clogging of columns during magnetic separation.
- FcR blocking reagent is added to avoid unwanted binding of antibodies to human Fc receptors, which would lead to unspecific signals in flow cytometry.
- If cell number exceeds 108 cells, increase the reagent volume for magnetic cell labeling according to the manufacturer’s protocol.
- LS columns are placed in MACS® Quadro Separator.
- Prepare the column by rinsing it with 3 mL MACS® washing buffer.
- Filter the samples again through 30 µm pre-separator filter to remove aggregated cells and prevent clogging of columns.
- Apply 5 mL cell suspension onto the column. Collect the flow-through containing unlabeled cells.
- Wash magnetically bound cells in the column by applying 3 × 3 mL of MACS® washing buffer.
- Remove the column from the separator and place it into 5 mL PP tube.
- Add 3–5 mL of MACS® washing buffer. Immediately flush out the magnetically labeled cells by firmly pushing the plunger into the column.
- Take 100 µL aliquot as unstained control for cytometer and FACS analysis.
- Determine cell number and centrifuge cells at 300× g for 7 min.
- The required rinsing volume varies according to the specific column and MACS Separator employed. Use an appropriate amount and adapt volumes and consumables according to your experimental layout.
- Purity can be increased by repeating the separation procedure with eluted fraction.
- Unstained cells function as setup control for adjusted photomultiplier tube (PMT) voltages.
4.2. Antibody Staining
4.2.1. Biotin-Labeled Antibody Staining of CD34+ HSPCs
- Resuspend cells in 50 µL MACS® buffer. Purified CD34+ cells are stained with biotin-labeled antibodies against lineage markers CD2, CD3, CD14, CD16, CD19, CD56 and CD235a to exclude lineage-positive cells (Table 2).
- Incubate cells for 15 min in a refrigerator.
- Wash cells with 1× PBS via centrifugation at 300× g for 7 min at 4 °C to remove unbound antibody.
- Discard the supernatant and resuspend cells in 50 µL of Horizon™ Brilliant Stain Buffer for fluorochrome-labeled antibody staining.
4.2.2. Fluorochrome-Labeled Antibody Staining of CD34+ HSPCs
- Stain 50 µL of purified CD34+ cells with fluorochrome-labeled antibody mixture (Table 3).
- Mix cells by gently pipetting the solution up and down at least 5 times.
- Incubate the cell solution for 30 min on ice without light exposure.
- Wash cells with 3–5 mL of 1× PBS via centrifugation at 300× g for 7 min at 4 °C to remove unbound antibodies.
- Remove the supernatant and resuspend cells in 500 µL of MACS® buffer.
- The samples are ready for analysis in flow cytometry.
- The exposure of light leads to degradation of the fluorochromes and decreased signals for the following experimental analysis. Incubating cells in the dark is therefore recommended.
- Antibody volumes are calculated for 5 × 106 cells. For increased cell numbers, increase the indicated volumes accordingly.
- It is advisable to use fluorochromes with strong fluorescence intensities for detecting antigens with low expression levels while reserving fluorochromes with lower fluorescence intensities for markers with high expression levels.
- Titration of the new antibodies is needed prior to the experiment. This helps optimize the concentrations and combinations of conjugated antibodies.
- Some staining protocols require commercially purchased staining buffers. These alternatives can be used instead of the Horizon™ Brilliant Stain Buffer.
4.2.3. Fluorescence-Minus-One (FMO) Controls for Gate Setting
- Label the FACS tubes for each FMO control, as illustrated in Table 4.
- Aliquot 1 × 105 CD34+ cells into each labeled FACS tube.
- Add 1 μL of each antibody conjugate to the associated labeled FACS tube.
- Mix the FMO controls by vortexing and incubate tubes for 30 min on ice without light exposure.
- Add 3–5 mL of PBS (1×) and centrifuge tubes at 300× g for 7 min.
- Discard the supernatant and resuspend FMO controls in 200 µL MACS® buffer.
- The FMO controls are ready for analysis in the cytometer.
- This step may not be required once the panel has been established and validated.
- FMO controls can assist in setting gates, but they should not be used exclusively for defining gates in FACS experiments. Relying strictly on FMO controls for gating can result in the exclusion of antigens that are expressed at low levels.
4.2.4. Single Antibody Stain Control for Compensation
- Experiments involving multiple parameters can lead to spectral spillovers of a fluorochrome into another detector. To prevent false positive signals caused by spectral spillover, single-antibody-stained controls should be used for compensation setup.
- Label the FACS tubes for each antibody conjugate that will be used in the experiment, as illustrated in Table 5.
- Mix the compensation beads by vigorously inverting at least 10 times or vortexing.
- Aliquot one drop of compensation beads into each labeled FACS tube.
- Add 1 μL of each antibody conjugate to the respective FACS tube.
- Mix the single staining solution by vortexing and incubate tubes for 20 min on ice without light exposure.
- Add 3–5 mL of PBS (1×) and centrifuge tubes at 300× g for 7 min.
- Discard the supernatant and resuspend single staining solutions in 200 µL MACS® buffer.
- The compensations are ready for analysis in the cytometer.
- Single-cell staining controls for compensation can also be performed using cells that positively express the relevant antigens to determine spectral overlap. If the cells of interest do not express the antigen, compensation beads are a suitable alternative.
- Single antibody staining for viability dye compensation should be performed using cells. A positive population of dead cells can be generated by inducing cell death through heat shock.
4.3. FACS Analysis and Sorting
4.3.1. Setting up FACS Machine
- The workflow outlined in this protocol details the process of sorting LT-HSCs and MPPs from human mob LPs using the FACSAria™ III Cell Sorter (BD) and is based on the manufacturer’s manual. Other FACS instruments with similar configurations may also be appropriate.
- Fluidics startup is performed according to cytometer instructions.
- Sorting human HSPCs can be performed using a 70 µm nozzle.
- Turn on the stream and optimize the breakoff by adjusting the amplitude. The gap size adjusts automatically according to your set amplitude. For performing sorts using the 70 µm nozzle, the gap size should be valued around 6–7.
- Run a CST performance check before every sort by using BD®CS&T Beads (BD). Laser delay depends on the daily performance of the lasers and can vary from day to day.
- Turn on the sweat spot to enable stable stream.
- Set the drop delay manually or automatically using BD® FACS Accudrop technology. The drop delay is the time interval required for the drop drive energy to be applied to the stream, breaking it into highly uniform droplets containing the particles of interest, and it is crucial for efficient sorting.
- Adjust photomultiplier tubes (PMTs) to an autofluorescent area by using the unstained control. The FACSAria™ III (BD) uses PMTs to convert scattered light and all fluorescence channels into an electrical signal called pulse. To optimize the PMT settings, unstained cells can be used to detect the levels of autofluorescence.
- The fluorescent signals are measured on a five-decade log scale; therefore, stained cells can be up to five decades brighter than unstained cells. Single-antibody-stained controls indicate the detection range of the fluorescent signal and are used to adjust PMTs.
- Adjust the side streams, so that cells gently slide into the sorting buffer. Directly hitting the liquid surface increases the risk of cell death.
- The 70 µm nozzle is appropriate for cells < 12 µm in size. If working with larger or fragile cells, a larger nozzle size and lower pressure are recommended to exclude the risk of exposing cells to excessive shear stress.
- Check the deflection plates before turning on the stream; residual salts can affect the sorting performance.
- Modern cell sorters can measure signals across up to a seven-decade log scale. Adjust the fluorescent signals according to your specific cytometry setup.
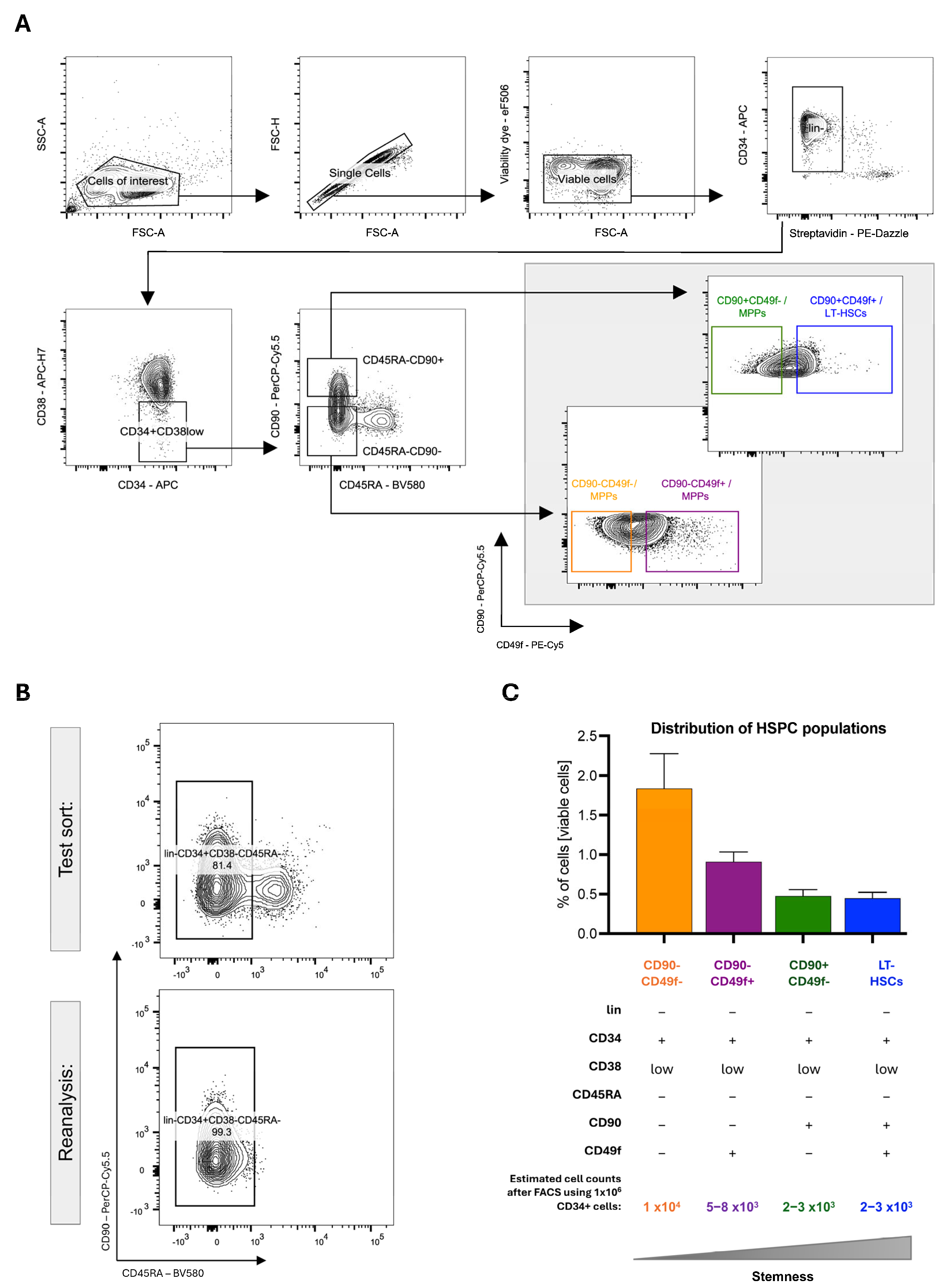
4.3.2. Gating Strategy for Discriminating Human MPPs and HSCs
- 1.
- After setting up the cytometer, a gating strategy must be established. This involves using single antibody controls to compensate for spectral spillover and FMO controls to set gate boundaries. The hierarchical gating strategy depends on the experimental layout. Figure 2A shows the hierarchical gating strategy of the analysis template displaying consecutive 2D FACS contour-dot plots. The individual FACS plots of the template are shortly explained here.
- 2.
- Cells of interest: Forward scatter (FSC-A) and sideward scatter (SSC-A) are used to exclude debris on a linear scale. The gate can be established by measuring the unstained control.
- 3.
- Singlets: Doublets causing unspecific signals can be excluded by analyzing the samples in FSC-Area (FSC-A) scaling versus FSC-Height (FSC-H) scaling.
- 4.
- Viable cells: Dead cells are excluded by a viability gate.
- 5.
- Lin- (CD2-, CD3-, CD14-, CD16-, CD19-, CD56- and CD235a-): Exclusion of the remaining lineage-positive progenitors is achieved via streptavidin–secondary antibody recognition of biotinylated antibodies.
- Lin-CD34+CD38low
- Lin-CD34+CD38lowCD45RA-CD90-
- Lin-CD34+CD38lowCD45RA-CD90+
- 6.
- HSPC populations including LT-HSCs can be identified using specific marker combinations adapted from the publication of Kaufmann et al. (Figure 2A) [44]:
- CD90-CD49f-/MPPs (Lin- CD34+CD38-CD45RA-CD90-CD49f-),
- CD90-CD49f+/MPPs (Lin- CD34+CD38-CD45RA-CD90-CD49f+),
- CD90+CD49f-/MPPs (Lin- CD34+CD38-CD45RA-CD90+CD49f-),
- CD90+CD49f+/LT-HSCs (Lin- CD34+CD38-CD45RA-CD90+CD49f+).
4.3.3. FACS Sorting of Human MPPs and LT-HSCs
- Once the hierarchical gating is established, cells can be sorted.
- For sorting two or more populations, the sort precision mode should be set to “Purity” or “4-Way Purity”.
- Reanalysis of the sorted cell populations is essential to ensure reliable cell separation. Sufficient cell numbers for reanalysis can be obtained by using a more abundant cell population higher up in the hierarchy; therefore, Lin-CD34+CD38low/-CD45RA- are used to validate the sort efficiency and cytometer settings. The sorting purity should be >95% (Figure 2B).
- After sorting MPPs and LT-HSCs, viable cells should be counted microscopically and can be processed for downstream analysis.
- Data analysis can be performed using Diva 8.1 or FlowJo software (BD).
- The yield mask, purity mask and phase mask settings address different types of conflicts during sorting. These settings work together to define the sort precision modes, which are detailed in the manufacturer’s manual.
- The recommended flow rate should not exceed 8000–10,000 events per second, as higher flow rates can increase the risk of nozzle O-ring occlusion and decrease sorting efficiency.
- During reanalysis, the sorted cells may have lost signal intensities and are localized near the boundaries of the set gates due to fluorochrome bleaching. Therefore, correctly sorted cells can fall outside the initial gates, necessitating slight gate adjustments to account for bleaching effects.
5. Expected Results and Limitations
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CB | Cord blood |
| CD | Cluster of differentiation |
| CITE | Cellular indexing of transcriptome and epitope |
| EPCR | Endothelial protein C receptor |
| FACS | Fluorescence-activated cell sorting |
| FMO | Fluorescence-minus-one |
| G-CSF | Granulocyte colony-stimulating factor |
| GvHD | Graft-versus-host disease |
| HSC | Hematopoietic stem cell |
| HSCT | Hematopoietic stem cell transplantation |
| HSPC | Hematopoietic stem and progenitor cell |
| LT-HSC | Long-term repopulating hematopoietic stem cell |
| mob LPs | Mobilized CD34+ cells from leukapheresis products |
| mPB | Mobilized peripheral blood |
| MPP | Multipotent progenitor |
| NOD/SCID | Non-obese diabetic/severe combined immunodeficient mice |
| PBMCs | Peripheral blood mononuclear cells |
| PMT | Photomultiplier tube |
| PP | Polypropylene |
| PS | Polystyrene |
References
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J.E. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef]
- Laurenti, E.; Göttgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553, 418–426. [Google Scholar] [CrossRef]
- Lee-Six, H.; Øbro, N.F.; Shepherd, M.S.; Grossmann, S.; Dawson, K.; Belmonte, M.; Osborne, R.J.; Huntly, B.J.P.; Martincorena, I.; Anderson, E.; et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018, 561, 473–478. [Google Scholar] [CrossRef]
- Spangrude, G.J.; Heimfeld, S.; Weissman, I.L. Purification and characterization of mouse hematopoietic stem cells. Science 1988, 241, 58–62, Erratum in Science 1989, 244. [Google Scholar] [CrossRef]
- Till, J.E.; McCulloch, E.A. Hemopoietic stem cell differentiation. Biochim. Biophys. Acta 1980, 605, 431–459. [Google Scholar] [CrossRef]
- Till, J.E.; McCulloch, E.A. A Direct Measurement of the Radiation Sensitivity of Normal Mouse Bone Marrow Cells. Radiat. Res. 1961, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Siminovitch, L.; Mcculloch, E.A.; Till, J.E. The distribution of colony-forming cells among spleen colonies. J. Cell Comp. Physiol. 1963, 62, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Lemischka, I.R. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes. Dev. 1990, 4, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Hawley, R.; Mintz, B. Long-and Short-Lived Murine Hematopoietic Stem Cell Clones Individually Identified with Retroviral Integration Markers. Blood 1990, 75, 2267–2270. [Google Scholar] [CrossRef]
- Keller, G.; Paige, C.; Gilboa, E.; Wagner, E.F. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature 1985, 318, 149–154. [Google Scholar] [CrossRef]
- Abramson, S.; Miller, R.G.; Phillips, R.A. The identification in adult bone marrow of pluripotent and restricted stem cells of the myeloid and lymphoid systems. J. Exp. Med. 1977, 145, 1567. [Google Scholar] [CrossRef]
- Thomas, E.D.; Lochte, H.L.; Lu, W.C.; Ferrebee, J.W. Intravenous infusion of bone marrow in patients receiving radiation and chemotherapy. N. Engl. J. Med. 1957, 257, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Shizuru, J.A.; Negrin, R.S.; Weissman, I.L. Hematopoietic stem and progenitor cells: Clinical and preclinical regeneration of the hematolymphoid system. Annu. Rev. Med. 2005, 56, 509–538. [Google Scholar] [CrossRef] [PubMed]
- Steward, C.G.; Jarisch, A. Haemopoietic stem cell transplantation for genetic disorders. Arch. Dis. Child. 2005, 90, 1259–1263. [Google Scholar] [CrossRef]
- Gatti, R.A.; Meuwissen, H.J.; Allen, H.D.; Hong, R.; Good, R.A. Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet 1968, 2, 1366–1369. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.M.; Weissman, I.L.; Tsukamoto, A.S.; Buckle, A.M.; Peault, B. Isolation of a candidate human hematopoietic stem-cell population. Proc. Natl. Acad. Sci. USA 1992, 89, 2804–2808. [Google Scholar] [CrossRef]
- Majeti, R.; Park, C.Y.; Weissman, I.L. Identification of a Hierarchy of Multipotent Hematopoietic Progenitors in Human Cord Blood. Cell Stem Cell 2007, 1, 635–645. [Google Scholar] [CrossRef]
- Jassinskaja, M.; Gonka, M.; Kent, D.G. Resolving the hematopoietic stem cell state by linking functional and molecular assays. Blood 2023, 142, 543–552. [Google Scholar] [CrossRef]
- Link, H.; Arseniev, L.; Bähre, O.; Kadar, J.G.; Diedrich, H.; Poliwoda, H. Transplantation of allogeneic CD34+ blood cells. Blood 1996, 87, 4903–4909. [Google Scholar] [CrossRef]
- Michallet, M.; Thomas, X.; Vernant, J.; Kuentz, M.; Socié, G.; Espérou-Bourdeau, H.; Milpied, N.; Blaise, D.; Rio, B.; Reiffers, J.; et al. Long-term outcome after allogeneic hematopoietic stem cell transplantation for advanced stage acute myeloblastic leukemia: A retrospective study of 379 patients reported to the Société Française de Greffe de Moelle (SFGM). Bone Marrow Transpl. 2000, 26, 1157–1163. [Google Scholar] [CrossRef]
- Krause, D.; Fackler, M.; Civin, C.; May, W. CD34: Structure, Biology, and Clinical Utility. Blood 1996, 87, 1–13. [Google Scholar] [CrossRef]
- McCune, J.M.; Péault, B.; Streeter, P.R.; Rabin, L. Preclinical Evaluation of Human Hematolymphoid Function in the SCID-hu Mouse. Immunol. Rev. 1991, 124, 45–62. [Google Scholar] [CrossRef]
- Terstappen, L.; Huang, S.; Safford, M.; Lansdorp, P.; Loken, M. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38- progenitor cells. Blood 1991, 77, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Craig, W.; Kay, R.; Cutler, R.L.; Lansdorp, P.M. Expression of Thy-1 on human hematopoietic progenitor cells. J. Exp. Med. 1993, 177, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Mayani, H.; Dragowska, W.; Lansdorp, P.M. Characterization of functionally distinct subpopulations of CD34+ cord blood cells in serum-free long-term cultures supplemented with hematopoietic cytokines. Blood 1993, 82, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Murray, L.J.; Tsukamoto, A.; Hoffman, R. CD34+Thy-1+Lin- stem cells from mobilized peripheral blood. Leuk. Lymphoma 1996, 22, 37–42. [Google Scholar] [CrossRef]
- Hao, Q.L.; Shah, A.J.; Thiemann, F.T.; Smogorzewska, E.M.; Crooks, G.M. A functional comparison of CD34+CD38- cells in cord blood and bone marrow. Blood 1995, 86, 3745–3753. [Google Scholar] [CrossRef]
- Bhatia, M.; Wang, J.C.Y.; Kapp, U.; Bonnet, D.; Dick, J.E. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc. Natl. Acad. Sci. USA 1997, 94, 5320–5325. [Google Scholar] [CrossRef]
- Benveniste, P. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 2010, 6, 48–58. [Google Scholar] [CrossRef]
- Notta, F.; Doulatov, S.; Laurenti, E.; Poeppl, A.; Jurisica, I.; Dick, J.E. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 2011, 333, 218–221. [Google Scholar] [CrossRef]
- Bonner, W.A.; Hulett, H.R.; Sweet, R.G.; Herzenberg, L.A. Fluorescence Activated Cell Sorting. Rev. Sci. Instrum. 1972, 43, 404–409. [Google Scholar] [CrossRef]
- Hulett, H.R.; Bonner, W.A.; Barrett, J.; Herzenberg, L.A. Cell sorting: Automated separation of mammalian cells as a function of intracellular fluorescence. J. Immunol. 2014, 193, 2045–2047. [Google Scholar] [CrossRef]
- Julius, M.H.; Masuda, T.; Herzenberg, L.A. Demonstration That Antigen-Binding Cells Are Precursors of Antibody-Producing Cells After Purification with a Fluorescence-Activated Cell Sorter. In Proceedings of the National Academy of Sciences, Washington, DC, USA, 24–26 April 1972; Volume 69, pp. 1934–1938. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P. The evolution of spectral flow cytometry. Cytom. Part A 2022, 101, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Font, L.; Mayer, J.U.; Old, S.; Hermans, I.F.; Irish, J.; Price, K.M. High-Dimensional Data Analysis Algorithms Yield Comparable Results for Mass Cytometry and Spectral Flow Cytometry Data. Cytom. Part A 2020, 97, 824–831. [Google Scholar] [CrossRef]
- Ferrer-Font, L.; Pellefigues, C.; Mayer, J.U.; Small, S.J.; Jaimes, M.C.; Price, K.M. Panel Design and Optimization for High-Dimensional Immunophenotyping Assays Using Spectral Flow Cytometry. Curr. Protoc. Cytom. 2020, 92, e70. [Google Scholar] [CrossRef]
- Fox, A.; Dutt, T.S.; Karger, B.; Obregón-Henao, A.; Anderson, G.B.; Henao-Tamayo, M. Acquisition of High-Quality Spectral Flow Cytometry Data. Curr. Protoc. Cytom. 2020, 93, e74. [Google Scholar] [CrossRef]
- Schmutz, S.; Valente, M.; Cumano, A.; Novault, S. Spectral Cytometry Has Unique Properties Allowing Multicolor Analysis of Cell Suspensions Isolated from Solid Tissues. PLoS ONE 2016, 11, e0159961. [Google Scholar] [CrossRef]
- Solomon, M.; DeLay, M.; Reynaud, D. Phenotypic Analysis of the Mouse Hematopoietic Hierarchy Using Spectral Cytometry: From Stem Cell Subsets to Early Progenitor Compartments. Cytom. Part A 2020, 97, 1057–1065. [Google Scholar] [CrossRef]
- Nagler, A.; Labopin, M.; Shimoni, A.; Niederwieser, D.; Mufti, G.J.; Zander, A.R.; Arnold, R.; Greinix, H.; Cornelissen, J.J.; Jackson, G.H.; et al. Mobilized Peripheral Blood Stem Cells Compared with Bone Marrow as the Stem Cell Source for Unrelated Donor Allogeneic Transplantation with Reduced-Intensity Conditioning in Patients with Acute Myeloid Leukemia in Complete Remission: An Analysis from the Acute Leukemia Working Party of the European Group for Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2012, 18, 1422–1429. [Google Scholar] [CrossRef]
- Anasetti, C.; Logan, B.R.; Lee, S.J.; Waller, E.K.; Weisdorf, D.J.; Wingard, J.R.; Cutler, C.S.; Westervelt, P.; Woolfrey, A.; Couban, S.; et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. N. Engl. J. Med. 2012, 367, 1487–1496. [Google Scholar] [CrossRef]
- Lee, S.J.; Logan, B.; Westervelt, P.; Cutler, C.; Woolfrey, A.; Khan, S.P.; Waller, E.K.; Maziarz, R.T.; Wu, J.; Shaw, B.E.; et al. Comparison of Patient-Reported Outcomes in 5-Year Survivors Who Received Bone Marrow vs Peripheral Blood Unrelated Donor Transplantation: Long-term Follow-up of a Randomized Clinical Trial. JAMA Oncol. 2016, 2, 1583–1589. [Google Scholar] [CrossRef]
- Kaufmann, K.B.; Zeng, A.G.X.; Coyaud, E.; Garcia-Prat, L.; Papalexi, E.; Murison, A.; Laurent, E.M.N.; Chan-Seng-Yue, M.; Gan, O.I.; Pan, K.; et al. A latent subset of human hematopoietic stem cells resists regenerative stress to preserve stemness. Nat. Immunol. 2021, 22, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Laurenti, E.; Oser, G.; van der Wath, R.C.; Blanco-Bose, W.; Jaworski, M.; Offner, S.; Dunant, C.F.; Eshkind, L.; Bockamp, E.; et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008, 135, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Zandi, S.; Takayama, N.; Dobson, S.; Gan, O.I.; Wilson, G.; Kaufmann, K.B.; McLeod, J.; Laurenti, E.; Dunant, C.F.; et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016, 351, aab2116. [Google Scholar] [CrossRef]
- Huntsman, H.D.; Bat, T.; Cheng, H.; Cash, A.; Cheruku, P.S.; Fu, J.-F.; Keyvanfar, K.; Childs, R.W.; Dunbar, C.E.; Larochelle, A. Human hematopoietic stem cells from mobilized peripheral blood can be purified based on CD49f integrin expression. Blood 2015, 126, 1631–1633. [Google Scholar] [CrossRef]
- Laurenti, E. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 2015, 16, 302–313. [Google Scholar] [CrossRef]
- Wang, T.Y.; Chang, S.J.; Chang, M.D.T.; Wang, H.W. Unique biological properties and application potentials of CD34+ CD38- stem cells from various sources. Taiwan. J. Obs. Gynecol. 2009, 48, 356–369. [Google Scholar] [CrossRef]
- Wang, J.C.Y.; Doedens, M.; Dick, J.E. Primitive Human Hematopoietic Cells Are Enriched in Cord Blood Compared with Adult Bone Marrow or Mobilized Peripheral Blood as Measured by the Quantitative In Vivo SCID-Repopulating Cell Assay. Blood 1997, 89, 3919–3924. [Google Scholar] [CrossRef]
- van der Loo, J.C.M.; Hanenberg, H.; Cooper, R.J.; Luo, F.-Y.; Lazaridis, E.N.; Williams, D.A. Nonobese Diabetic/Severe Combined Immunodeficiency (NOD/SCID) Mouse as a Model System to Study the Engraftment and Mobilization of Human Peripheral Blood Stem Cells. Blood 1998, 92, 2556–2570. [Google Scholar] [CrossRef]
- McCune, J.M.; Namikawa, R.; Kaneshima, H.; Shultz, L.D.; Lieberman, M.; Weissman, I.L. The SCID-hu mouse: Murine model for the analysis of human hematolymphoid differentiation and function. Science 1988, 241, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Lyons, B.L.; Burzenski, L.M.; Gott, B.; Chen, X.; Chaleff, S.; Kotb, M.; Gillies, S.D.; King, M.; Mangada, J.; et al. Human Lymphoid and Myeloid Cell Development in NOD/LtSz-scid IL2Rγnull Mice Engrafted with Mobilized Human Hemopoietic Stem Cells. J. Immunol. 2005, 174, 6477–6489. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Yasukawa, M.; Lyons, B.; Yoshida, S.; Miyamoto, T.; Yoshimoto, G.; Watanabe, T.; Akashi, K.; Shultz, L.D.; Harada, M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood 2005, 106, 1565. [Google Scholar] [CrossRef] [PubMed]
- Halkias, J.; Yen, B.; Taylor, K.T.; Reinhartz, O.; Winoto, A.; Robey, E.A.; Melichar, H.J. Conserved and divergent aspects of human T-cell development and migration in humanized mice. Immunol. Cell Biol. 2015, 93, 716. [Google Scholar] [CrossRef]
- Cosgun, K.N.; Rahmig, S.; Mende, N.; Reinke, S.; Hauber, I.; Schäfer, C.; Petzold, A.; Weisbach, H.; Heidkamp, G.; Purbojo, A.; et al. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell 2014, 15, 227–238. [Google Scholar] [CrossRef]
- Rahmig, S.; Kronstein-Wiedemann, R.; Fohgrub, J.; Kronstein, N.; Nevmerzhitskaya, A.; Bornhäuser, M.; Gassmann, M.; Platz, A.; Ordemann, R.; Tonn, T.; et al. Improved Human Erythropoiesis and Platelet Formation in Humanized NSGW41 Mice. Stem Cell Rep. 2016, 7, 591. [Google Scholar] [CrossRef]
- Kanate, A.S.; Majhail, N.S.; Savani, B.N.; Bredeson, C.; Champlin, R.E.; Crawford, S.; Giralt, S.A.; LeMaistre, C.F.; Marks, D.I.; Omel, J.L.; et al. Indications for Hematopoietic Cell Transplantation and Immune Effector Cell Therapy: Guidelines from the American Society for Transplantation and Cellular Therapy. Biol. Blood Marrow Transpl. 2020, 26, 1247–1256. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Sudo, K.; Yamazaki, S.; Wilkinson, A.C.; Nakauchi, H.; Nakamura, Y. Polyvinyl alcohol hydrolysis rate and molecular weight influence human and murine HSC activity ex vivo. Stem Cell Res. 2021, 56, 102531. [Google Scholar] [CrossRef]
- Cohen, S.; Roy, J.; Lachance, S.; Delisle, J.-S.; Marinier, A.; Busque, L.; Roy, D.-C.; Barabé, F.; Ahmad, I.; Bambace, N.; et al. Hematopoietic stem cell transplantation using single UM171-expanded cord blood: A single-arm, phase 1–2 safety and feasibility study. Lancet Haematol. 2020, 7, e134–e145. [Google Scholar] [CrossRef]
- Dumont-Lagacé, M.; Feghaly, A.; Meunier, M.-C.; Finney, M.; Hof, W.V.; Frenet, E.M.; Sauvageau, G.; Cohen, S. UM171 Expansion of Cord Blood Improves Donor Availability and HLA Matching for All Patients, Including Minorities. Transpl. Cell Ther. 2022, 28, 410.e1–410.e5, Erratum in Transpl. Cell Ther. 2023, 30, https://doi.org/10.1016/j.jtct.2023.08.024. [Google Scholar] [CrossRef]
- Fares, I.; Chagraoui, J.; Gareau, Y.; Gingras, S.; Ruel, R.; Mayotte, N.; Csaszar, E.; Knapp, D.J.; Miller, P.; Ngom, M.; et al. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 2014, 345, 1509–1512. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Song, B.; Carlino, M.J.; Li, G.; Ferchen, K.; Chen, M.; Thompson, E.N.; Kain, B.N.; Schnell, D.; Thakkar, K.; et al. An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors. Nat. Immunol. 2024, 25, 703–715. [Google Scholar] [CrossRef]
- Povinelli, B.J.; Rodriguez-Meira, A.; Mead, A.J. Single cell analysis of normal and leukemic hematopoiesis. Mol. Asp. Med. 2018, 59, 85. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Huang, W.; Guo, G. Studying hematopoiesis using single-cell technologies. J. Hematol. Oncol. 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 2395. [Google Scholar] [CrossRef]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir, E.-A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 2011, 332, 687. [Google Scholar] [CrossRef]
- Komic, H.; Schmachtel, T.; Simoes, C.; Külp, M.; Yu, W.; Jolly, A.; Nilsson, M.S.; Gonzalez, C.; Prosper, F.; Bonig, H.; et al. Continuous map of early hematopoietic stem cell differentiation across human lifetime. Nat. Commun. 2025, 16, 2287. [Google Scholar] [CrossRef]
- Anjos-Afonso, F.; Buettner, F.; Mian, S.A.; Rhys, H.; Perez-Lloret, J.; Garcia-Albornoz, M.; Rastogi, N.; Ariza-McNaughton, L.; Bonnet, D. Single cell analyses identify a highly regenerative and homogenous human CD34+ hematopoietic stem cell population. Nat. Commun. 2022, 13, 2048. [Google Scholar] [CrossRef]
| Consumables | Catalog Number | Company |
|---|---|---|
| Anti-Human CD14 [61D3] | 13-0149-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD16 [CB16] | 13-0168-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD19 [HIB19] | 13-0199-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD2 [RPA-2.10] | 13-0029-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD235a [HIR2] | 13-9987-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD3 [OKT3] | 13-0037-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD34 [8G12] | 345804 | BD Bioscience, Frankling Lakes, NJ, USA |
| Anti-Human CD38 [HB7] | 656646 | BD Bioscience, Frankling Lakes, NeJ, USA |
| Anti-Human CD45RA [HI100] | 304132 | BioLegend, San Diego, CA, USA |
| Anti-Human CD49f [GoH3] | 551129 | BD Bioscience, Frankling Lakes, NJ, USA |
| Anti-Human CD56 [NCAM] | 13-0567-82 | Thermo Fisher Scientific, Waltham, MA, USA |
| Anti-Human CD90/Thy1 [5E10] | 561557 | BD Bioscience, Frankling Lakes, New Jersey, USA |
| autoMACS® Rinsing Solution | 130-091-222 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| BD® Accudrop Beads | 345249 | BD Bioscience, Frankling Lakes, NJ, USA |
| BD® CS&T Research Beads | 655051 | BD Bioscience, Frankling Lakes, NJ, USA |
| C-Chip, Neubauer Improved Counting chamber | PK36.1 | Roth, Karlsruhe, Germany |
| CD34 MicroBead Kit UltraPure human | 130-100-453 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| Cellstar® sterile centrifuge tubes (15 mL) | 188271 | Greiner Bio-One, Frickenhausen, Germany |
| Centrifuge Rotana 460R | 5650SET1 | Hettich, Tuttlingen, Germany |
| eBioscience™ UltraComp eBeads | 01-2222-42 | Thermo Fisher Scientific, Waltham, MA, USA |
| FACSAria™ III Cell Sorter | BD Bioscience, Frankling Lakes, NJ, USA | |
| FACSDiva™ Software (version 8.1) | BD Bioscience, Frankling Lakes, NJ, USA | |
| Fixable Viability dye | 65-0866-14 | Thermo Fisher Scientific, Waltham, MA, USA |
| FlowJo™ Software (version 10.10.0) | BD Bioscience, Frankling Lakes, NJ, USA | |
| Gibco™ 1× PBS | 633801 | Thermo Fisher Scientific, Waltham, MA, USA |
| Horizon™ Brilliant Stain Buffer | 563794 | BD Bioscience, Frankling Lakes, NJ, USA |
| LS Columns | 130-042-401 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| LSRFortessa™ Cell Analyzer | BD Bioscience, Frankling Lakes, NJ, USA | |
| MACS®BSA Stock Solution | 130-091-376 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| Pancoll human, Density: 1.077 g/mL | P04-60500 | PAN-Biotech, Aidenbach, Germany |
| Polypropylene FACS tubes (5 mL) | 10171942 | Thermo Fisher Scientific, Waltham, MA, USA |
| Polypropylene round bottom tube (14 mL) | 10574991 | Thermo Fisher Scientific, Waltham, MA, USA |
| Polystyrene FACS tubes (5 mL) | 551.579 | Sarstedt, Nuembrecht, Germany |
| Polystyrene FACS tubes with cell-strainer cap (5 mL) | 10585801 | Thermo Fisher Scientific, Waltham, MA, USA |
| Pre-Separation Filters (70 µm) | 130-095-823 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| QuadroMACS™ Separator | 130-090-976 | Miltenyi Biotec, Bergisch Gladbach, Germany |
| StemStan™ SFEM II media | 9655 | STEMCELL Technologies, Vancouver, Canada |
| Streptavidin | 405248 | BioLegend, San Diego, CA, USA |
| Antibody [Clone] | Volume for 5 × 106 Cells (Predilution) |
|---|---|
| CD2 [RPA-2.10] | 0.5 µL |
| CD3 [OKT3] | 2.4 µL (1:10) |
| CD14 [61D3] | 0.5 µL |
| CD16 [CB16] | 0.5 µL (1:10) |
| CD19 [HIB19] | 1 µL |
| CD56 [NCAM] | 2.5 µL (1:50) |
| CD235a [HIR2] | 2.5 µL (1:100) |
| Antibody [Clone] | Fluorochrome | Volume for 5 × 106 Cells |
|---|---|---|
| Viability dye | ef506 | 2 µL |
| CD45RA [HI100] | BV580 | 5 µL |
| CD90/Thy1 [5E10] | PerCP-Cy5.5 | 5 µL |
| Streptavidin | PE-Dazzle 594 | 1.5 µL (of a 1:10 dilution) |
| CD49f [GoH3] | PE-Cy5 | 2.5 µL |
| CD34 [8G12] | APC | 2.5 µL |
| CD38 [HB7] | APC-H7 | 5 µL |
| Tube Label | Viability Dye eF506 | CD45RA BV580 | CD90 PerCP-Cy5.5 | Strep. PE-Dazzle | CD49f PE-Cy5 | CD34 APC | CD38 APC-H7 |
|---|---|---|---|---|---|---|---|
| FMO-eF506 | − | + | + | + | + | + | + |
| FMO-BV580 | + | − | + | + | + | + | + |
| FMO-PerCP-Cy5.5 | + | + | − | + | + | + | + |
| FMO-PE-Dazzle 594 | + | + | + | − | + | + | + |
| FMO-PE-Cy5 | + | + | + | + | − | + | + |
| FMO-APC | + | + | + | + | + | − | + |
| FMO-APC-H7 | + | + | + | + | + | + | − |
| Tube Label | Viability Dye eF506 | CD45RA BV580 | CD90 PerCP-Cy5.5 | Strep. PE-Dazzle | CD49f PE-Cy5 | CD34 APC | CD38 APC-H7 |
|---|---|---|---|---|---|---|---|
| eF506 | + | − | − | − | − | − | − |
| BV580 | − | + | − | − | − | − | − |
| PerCP-Cy5.5 | − | − | + | − | − | − | − |
| PE-Dazzle 594 | − | − | − | + | − | − | − |
| PE-Cy5 | − | − | − | − | + | − | − |
| APC | − | − | − | − | − | + | − |
| APC-H7 | − | − | − | − | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmachtel, T.; Bonig, H.; Rieger, M.A. FACS-Based Assessment of Human Hematopoietic Stem and Progenitor Cells. Int. J. Mol. Sci. 2025, 26, 8381. https://doi.org/10.3390/ijms26178381
Schmachtel T, Bonig H, Rieger MA. FACS-Based Assessment of Human Hematopoietic Stem and Progenitor Cells. International Journal of Molecular Sciences. 2025; 26(17):8381. https://doi.org/10.3390/ijms26178381
Chicago/Turabian StyleSchmachtel, Tessa, Halvard Bonig, and Michael A. Rieger. 2025. "FACS-Based Assessment of Human Hematopoietic Stem and Progenitor Cells" International Journal of Molecular Sciences 26, no. 17: 8381. https://doi.org/10.3390/ijms26178381
APA StyleSchmachtel, T., Bonig, H., & Rieger, M. A. (2025). FACS-Based Assessment of Human Hematopoietic Stem and Progenitor Cells. International Journal of Molecular Sciences, 26(17), 8381. https://doi.org/10.3390/ijms26178381





