Tetratricopeptide Repeat 2 Is a Quantitative Trait Locus That Controls Seed Size
Abstract
1. Introduction
2. Results
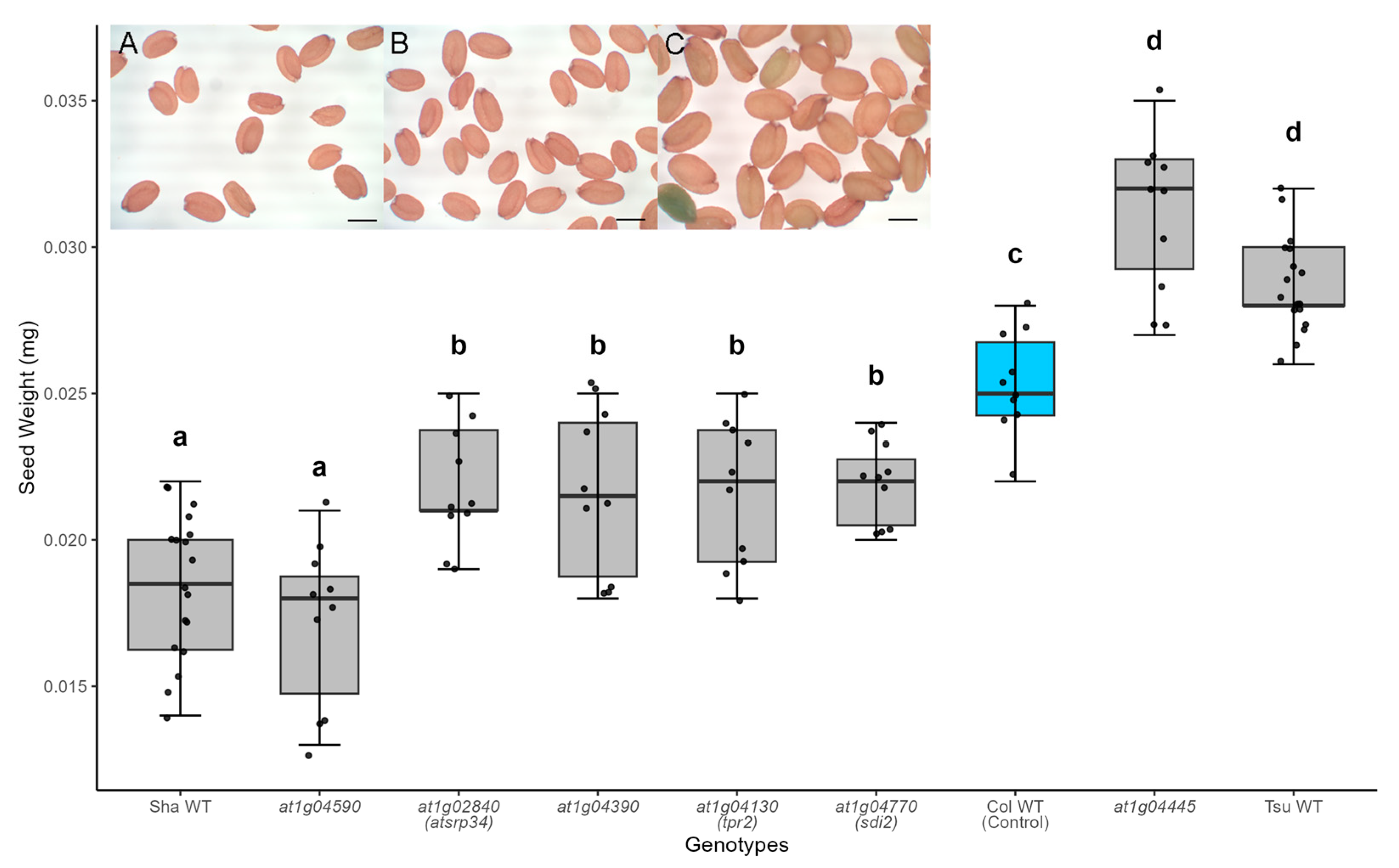
2.1. Mutations Altering Seed Size in the SSQ1 Interval
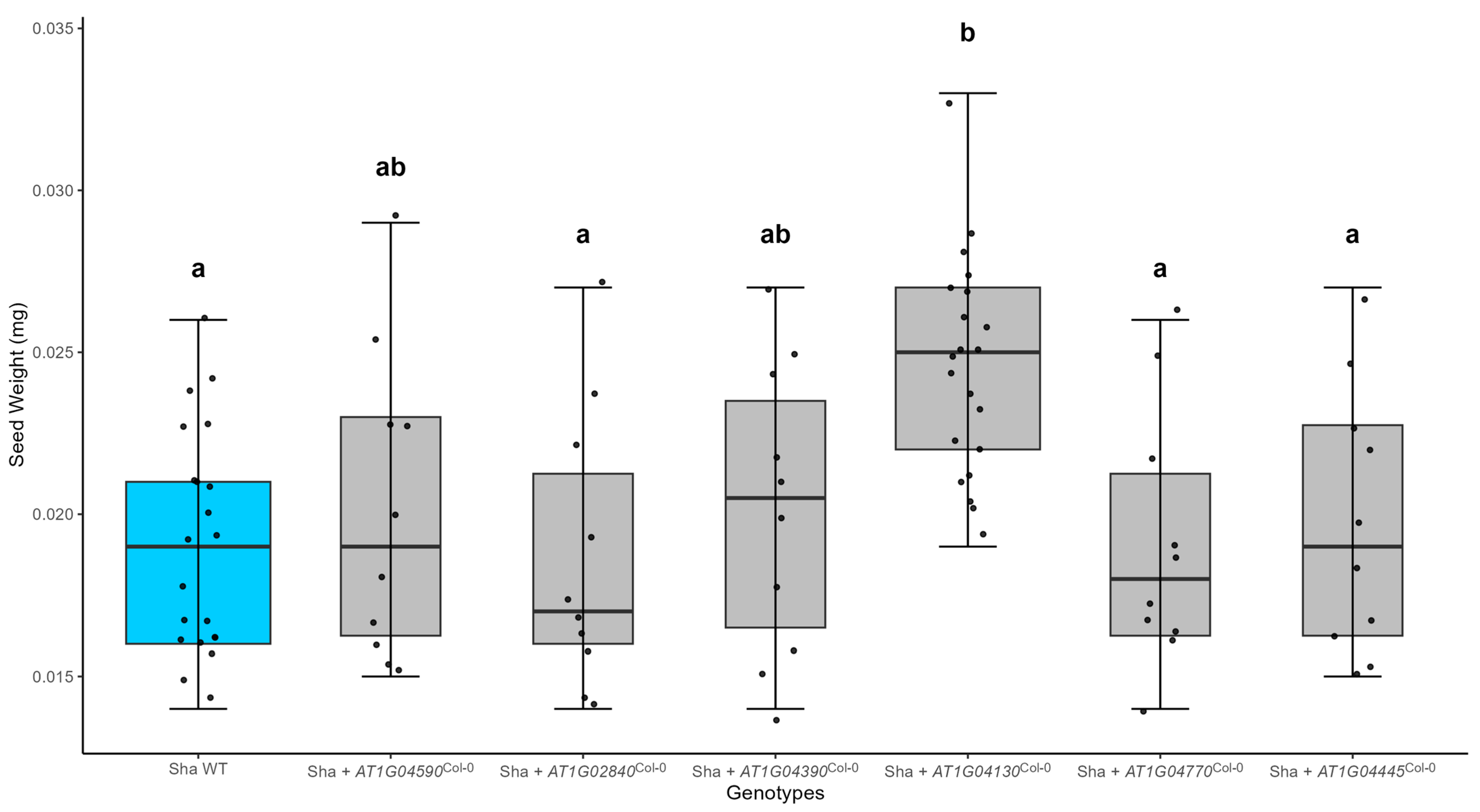
2.2. A Cosmid with the TPR2Col-0 Transgene Increases Seed Size
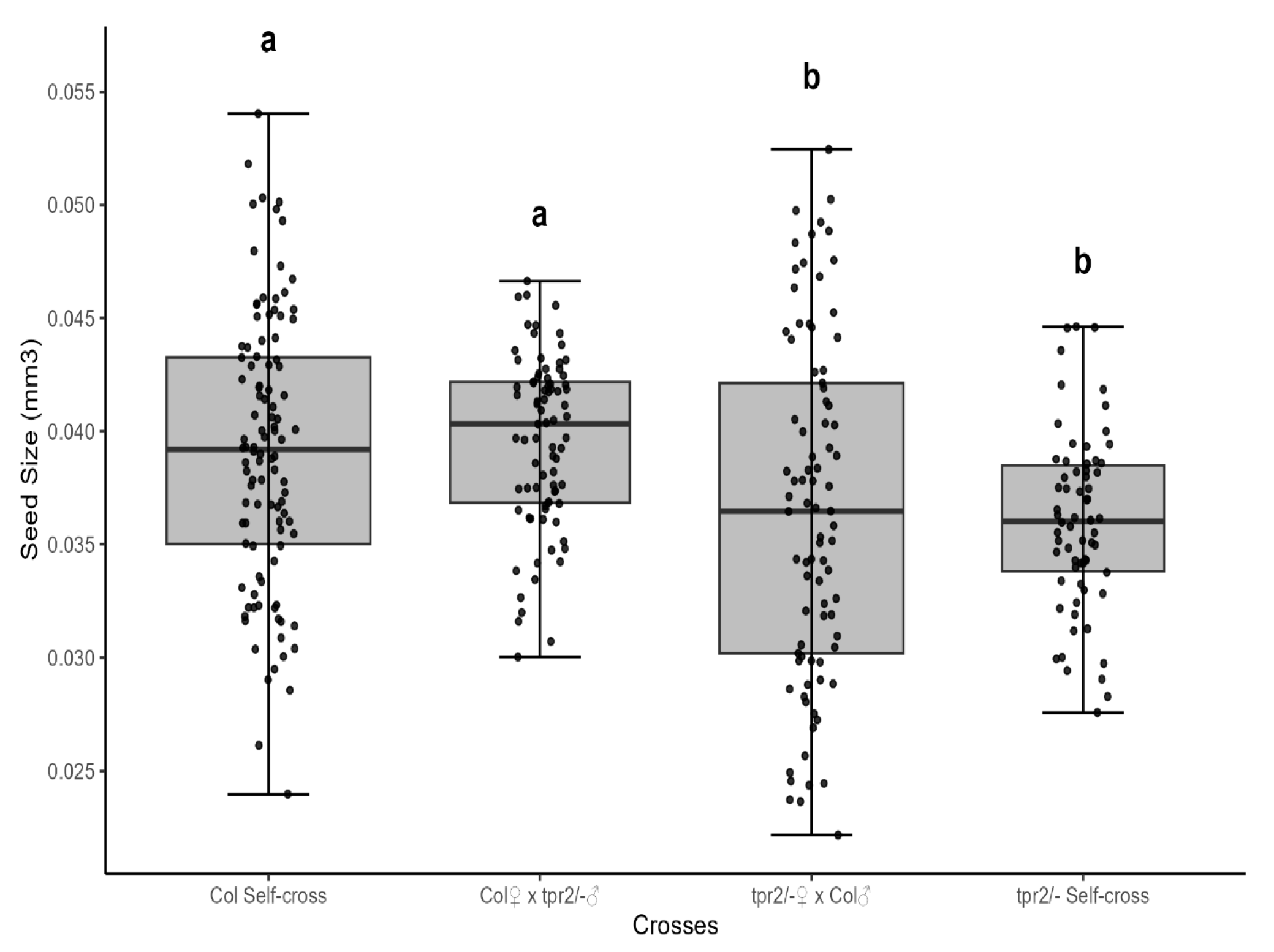
2.3. TPR2 Acts Maternally to Regulate Seed Size
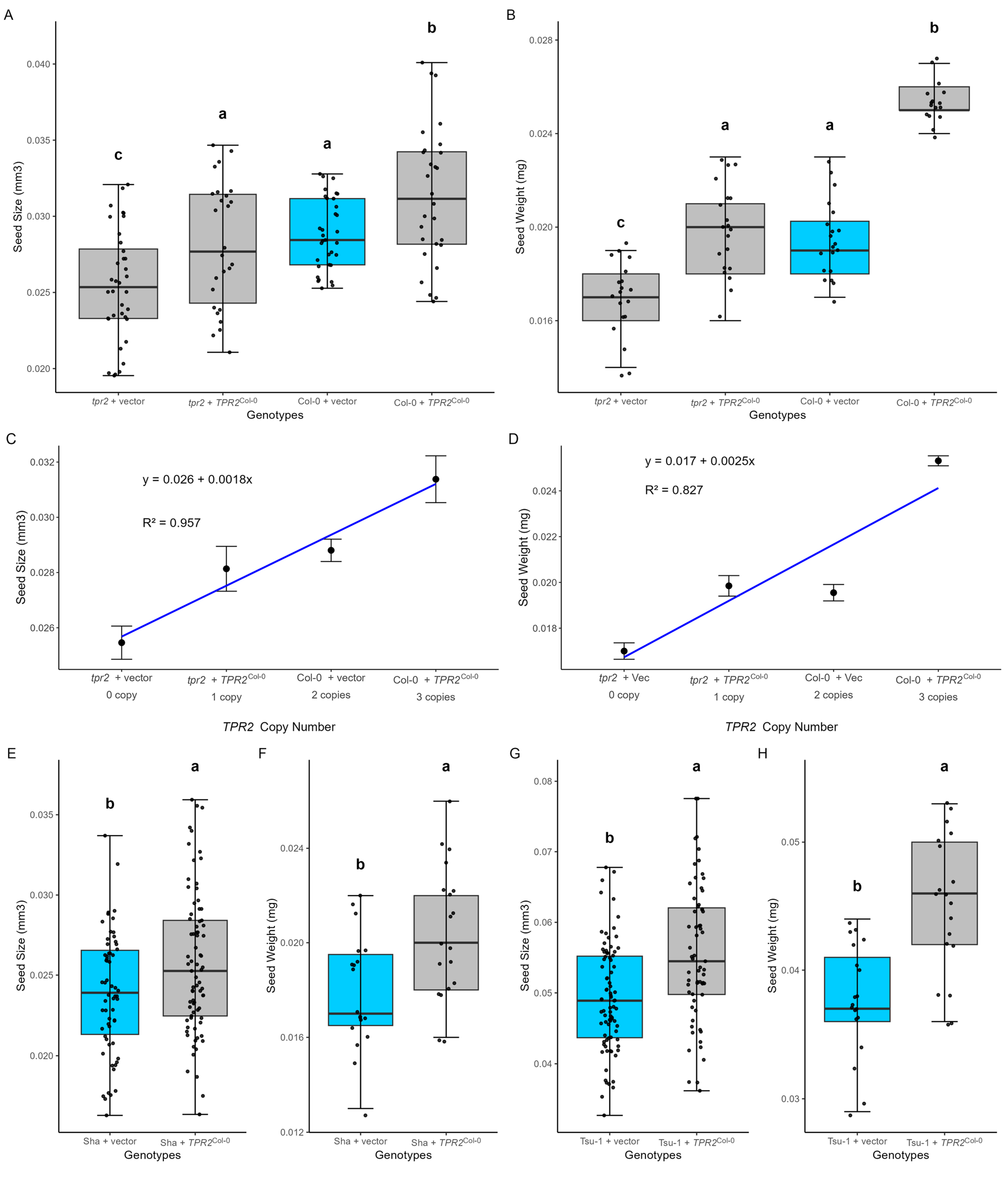
2.4. TPR2 Increases Seed Size and Mass in All Accessions
2.5. Additional TPR2Col-0 Alleles Add ~10% to Seed Size
2.6. TPR2 Transcript Abundance Similar Among Arabidopsis Accessions
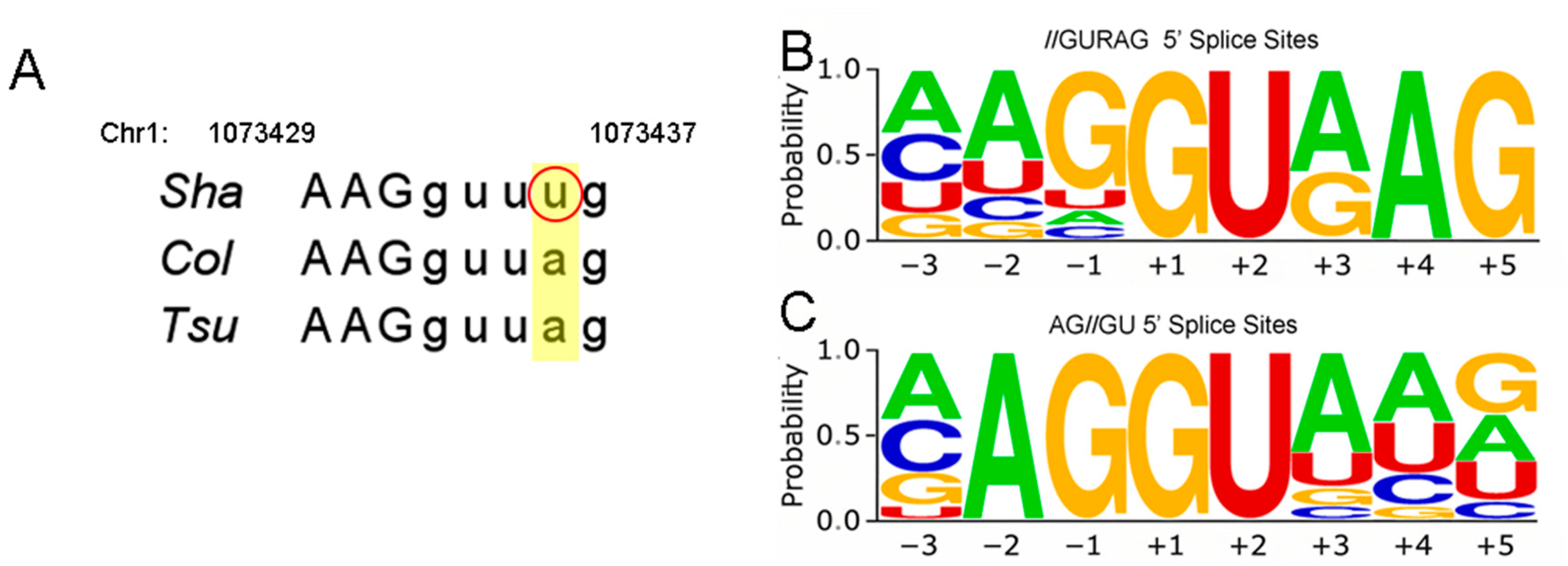
2.7. RNA Variants Among TPR2 Alleles
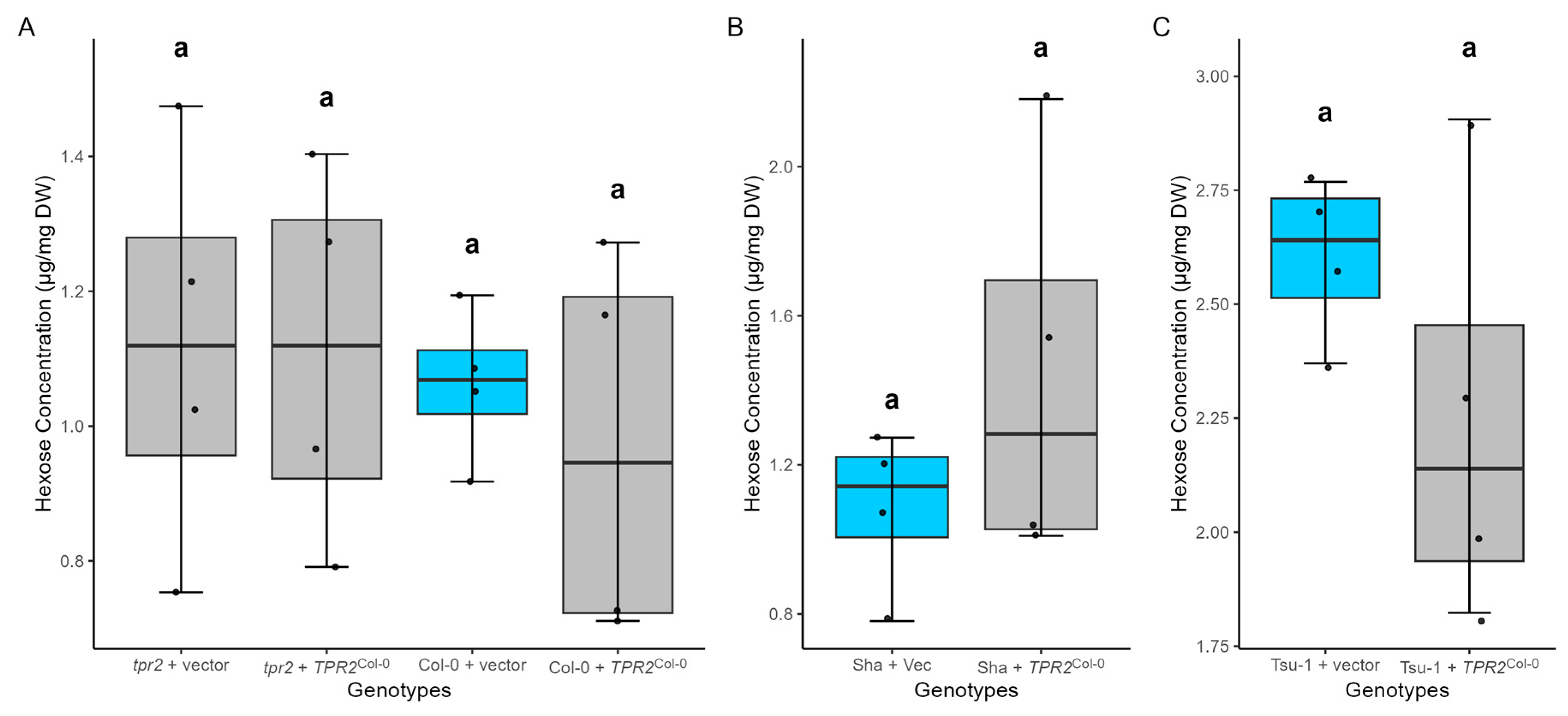
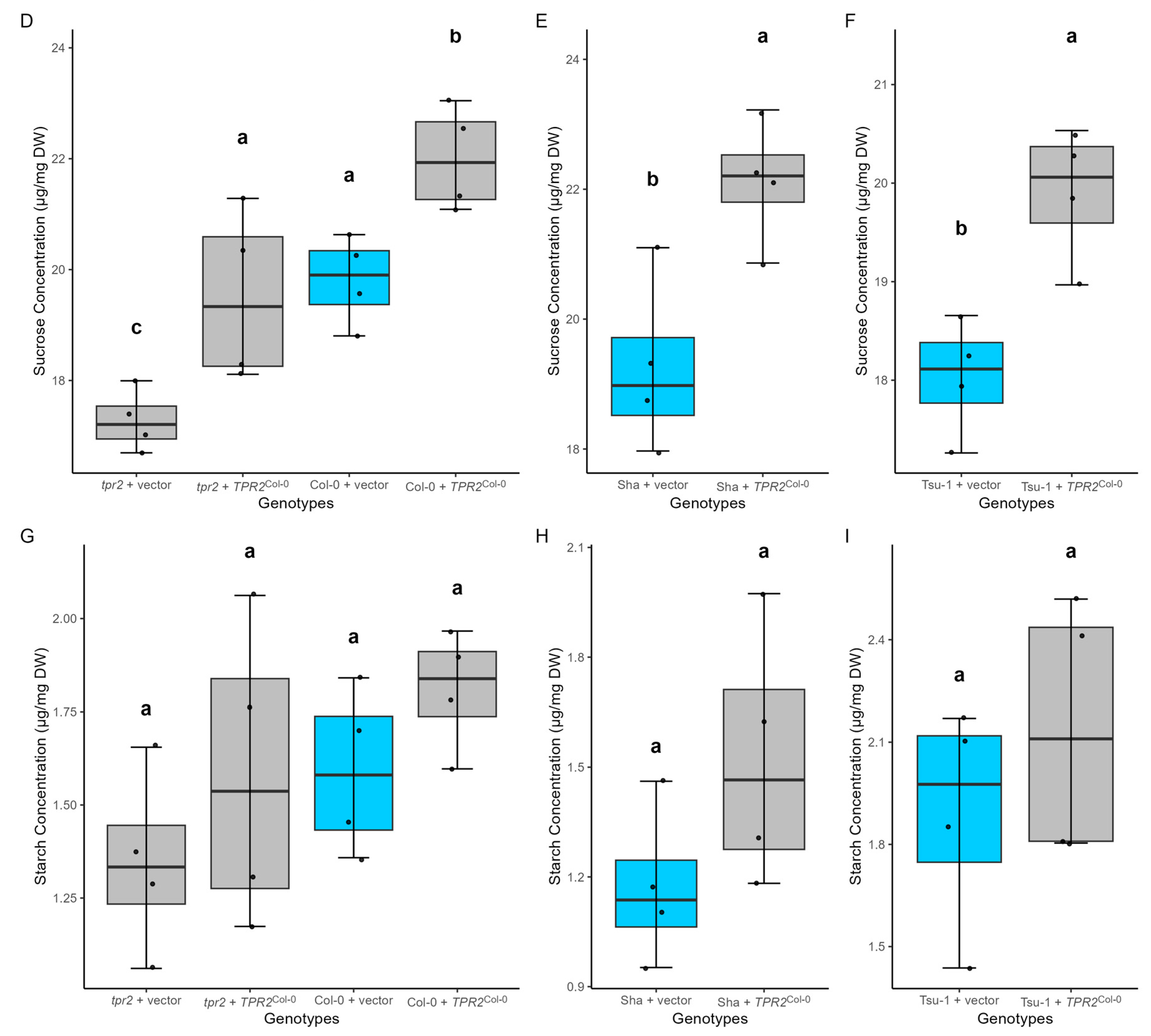
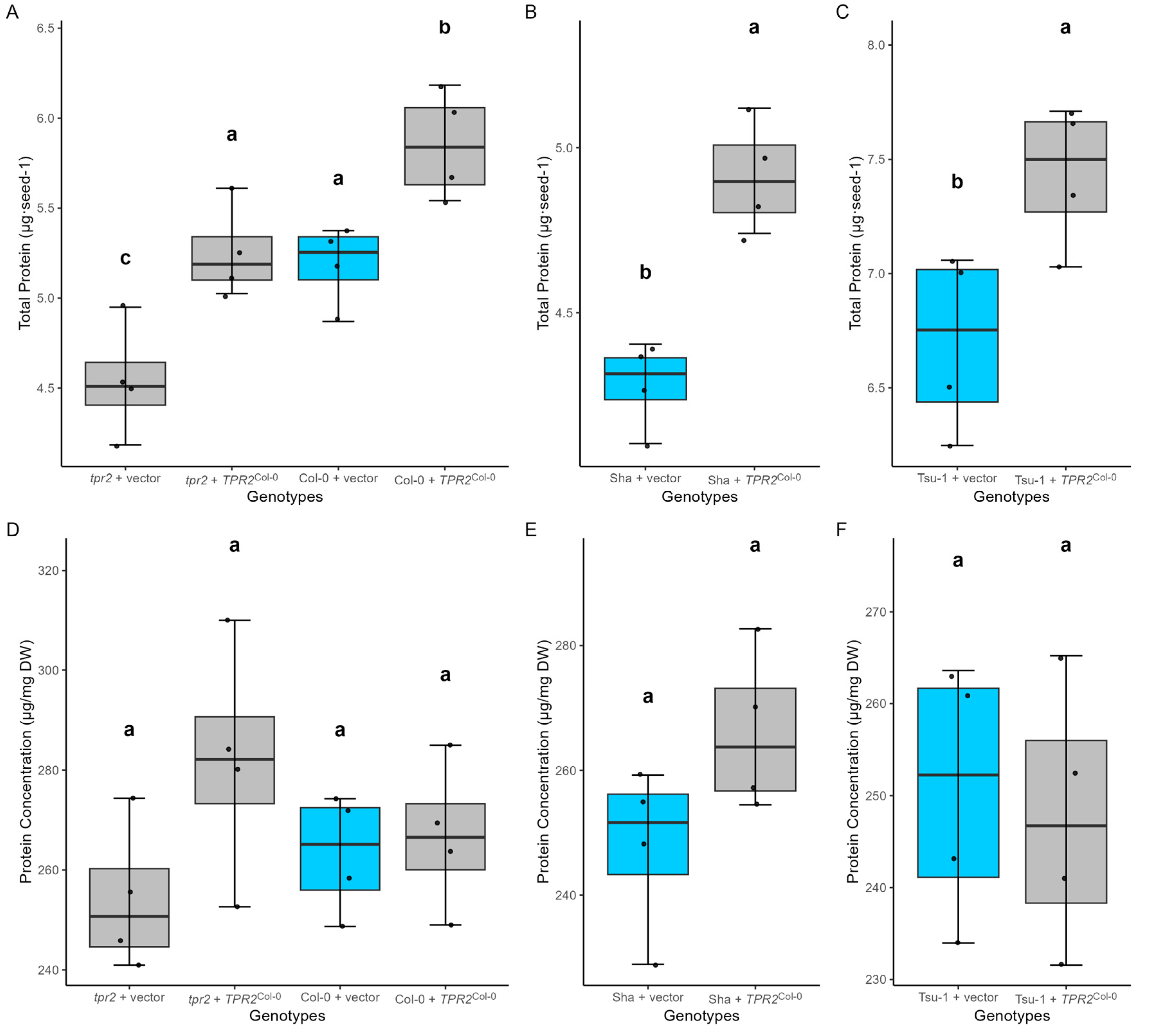
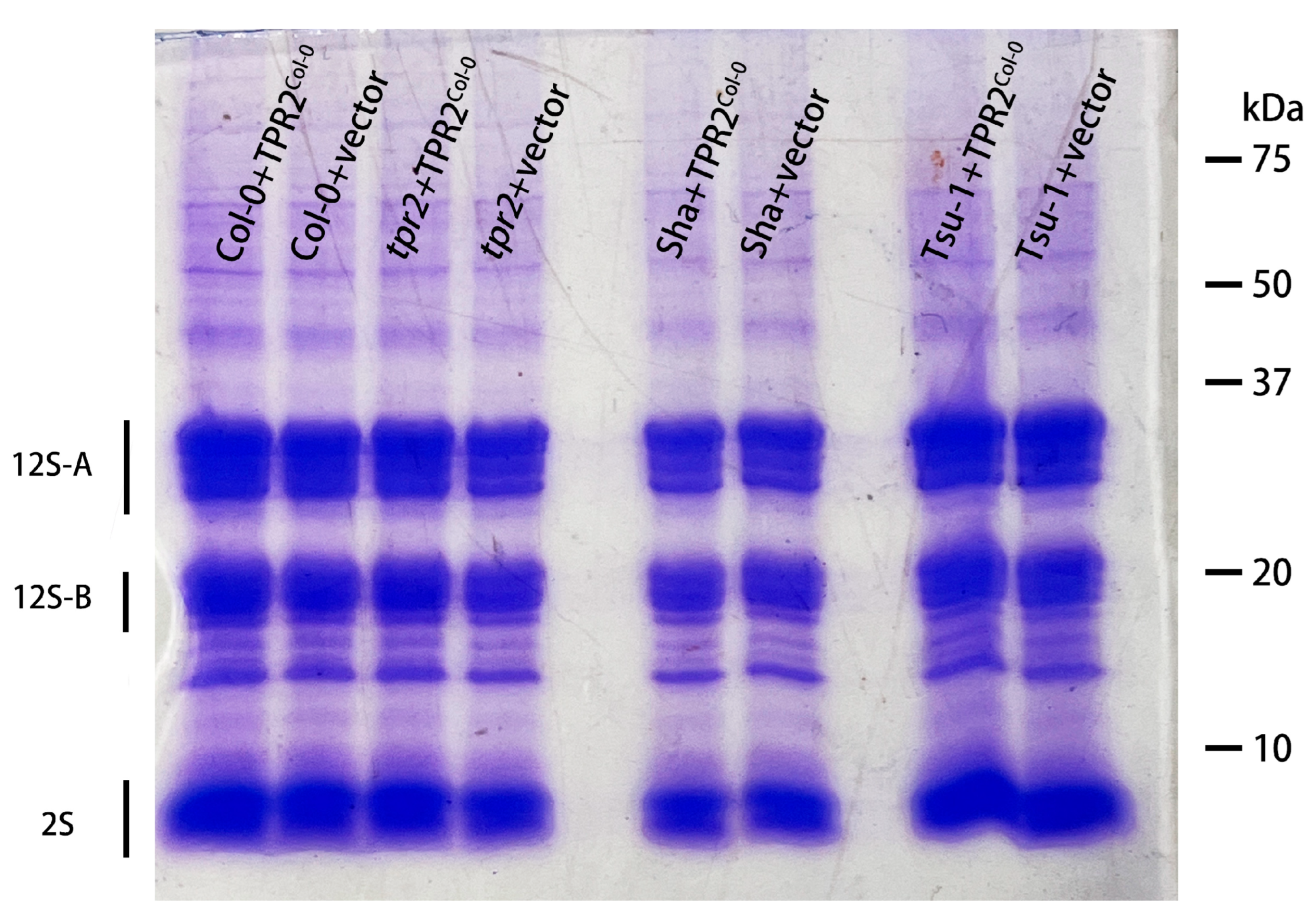
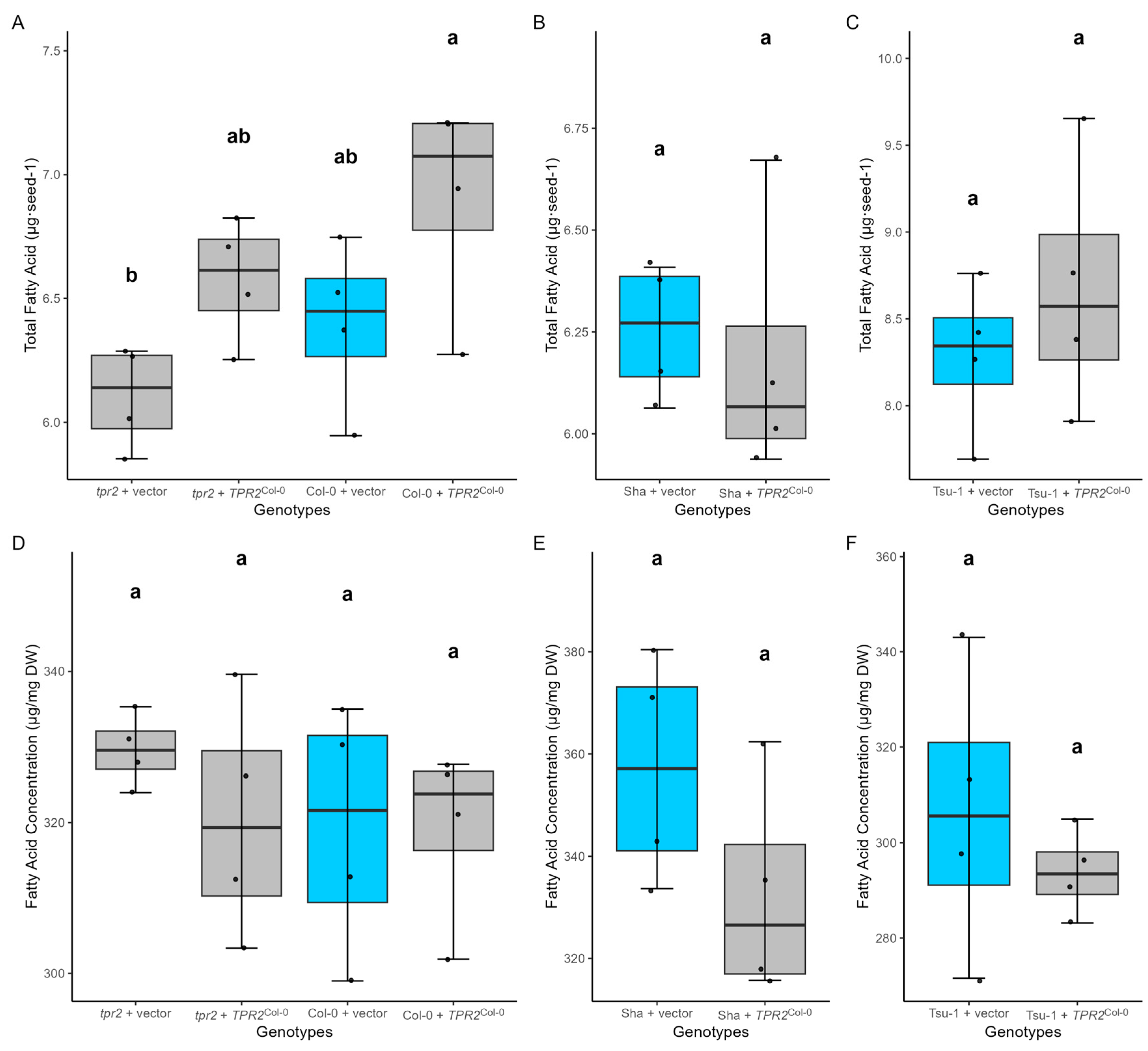
2.8. TPR2Col-0 Activity Affects Sucrose and Protein Levels in Seeds
3. Discussion
3.1. A Strategy to Find QTG Altering Seed Mass
3.2. TPR2 Splice Variants
3.3. TPR2 Effects on Sucrose and Protein Accumulation in Seeds Is Consistent with a Seed Size Phenotype
3.4. TPR2 Affects Sucrose Levels in Seeds
3.5. TPR2 Proteins and Hormone Activity During Seed Development
3.6. TPR2-Mediated Effects on Cell Growth
4. Materials and Methods
4.1. Plant Growth Conditions and Seed Measurement
4.2. Cosmid and Single-Gene Cloning
4.3. Plant Transformation
4.4. Transcript Analysis
4.5. Carbohydrate Analysis
4.6. Protein Analysis
4.7. Lipid Analysis
4.8. Data Analysis and Presentation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| GA | Gibberellin |
| GWAS | Genome-wide association study |
| kb | Kilobase |
| PCR | Polymerase chain reaction |
| QTG QTL | Quantitative trait gene Quantitative trait locus |
| SSQ1 | Seed size QTL1 |
| TPR2 | Tetratricopeptide Repeat Protein 2 |
References
- Gnan, S.; Priest, A.; Kover, P.X. The Genetic Basis of Natural Variation in Seed Size and Seed Number and Their Trade-Off Using Arabidopsis thaliana MAGIC Lines. Genetics 2014, 198, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Shantharaj, D.; Kang, X.; Ni, M. Transcriptional and Hormonal Signaling Control of Arabidopsis Seed Development. Curr. Opin. Plant Biol. 2010, 13, 611–620. [Google Scholar] [CrossRef]
- Ando, A.; Kirkbride, R.C.; Qiao, H.; Chen, Z.J. Endosperm and Maternal-Specific Expression of EIN2 in the Endosperm Affects Endosperm Cellularization and Seed Size in Arabidopsis. Genetics 2022, 223, iyac161. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Zhao, X.Y.; Shao, X.X.; Wang, F.; Zhou, C.; Liu, Y.G.; Zhang, Y.; Zhang, X.S. Abscisic Acid Regulates Early Seed Development in Arabidopsis by ABI5-Mediated Transcription of Short Hypocotyl Under Blue1. Plant Cell 2014, 26, 1053–1068. [Google Scholar] [CrossRef]
- Gomez, M.D.; Cored, I.; Barro-Trastoy, D.; Sanchez-Matilla, J.; Tornero, P.; Perez-Amador, M.A. DELLA Proteins Positively Regulate Seed Size in Arabidopsis. Development 2023, 150, dev201853. [Google Scholar] [CrossRef]
- Hu, S.; Yang, H.; Gao, H.; Yan, J.; Xie, D. Control of Seed Size by Jasmonate. Sci. China Life Sci. 2021, 64, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid Regulates Seed Size and Shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and Transport-Mediated Dynamic Auxin Distribution during Seed Development Controls Seed Size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Baud, S.; Dubreucq, B.; Miquel, M.; Rochat, C.; Lepiniec, L. Storage Reserve Accumulation in Arabidopsis: Metabolic and Developmental Control of Seed Filling. Arab. Book 2008, 6, e0113. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Maternal Control of Seed Size in Plants. J. Exp. Bot. 2015, 66, 1087–1097. [Google Scholar] [CrossRef]
- Orozco-Arroyo, G.; Paolo, D.; Ezquer, I.; Colombo, L. Networks Controlling Seed Size in Arabidopsis. Plant Reprod. 2015, 28, 17–32. [Google Scholar] [CrossRef]
- Baud, S.; Boutin, J.-P.; Miquel, M.; Lepiniec, L.; Rochat, C. An Integrated Overview of Seed Development in Arabidopsis thaliana Ecotype WS. Plant Physiol. Biochem. 2002, 40, 151–160. [Google Scholar] [CrossRef]
- Liu, X.; Nakajima, K.P.; Adhikari, P.B.; Bradford, K.J.; Wu, X.; Zhu, K.; Kagenishi, T.; Kurotani, K.; Ishida, T.; Nakamura, M.; et al. Fertilization-Dependent Phloem End Gate Regulates Seed Size. Curr. Biol. 2025, 35, 2049–2063. [Google Scholar] [CrossRef]
- McLaughlin, J.E.; Boyer, J.S. Sugar-Responsive Gene Expression, Invertase Activity, and Senescence in Aborting Maize Ovaries at Low Water Potentials. Ann. Bot. 2004, 94, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jürgens, G.; Berger, F. Arabidopsis haiku Mutants Reveal New Controls of Seed Size by Endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of Seed Mass and Seed Yield by the Floral Homeotic Gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of Final Seed and Organ Size by the DA1 Gene Family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY Family Gene, and HAIKU2 (IKU2), a Leucine-Rich Repeat Kinase Gene, Are Regulators of Seed Size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Ren, D.; Wang, X.; Yang, M.; Yang, L.; He, G.; Deng, X.W. A New Regulator of Seed Size Control in Arabidopsis Identified by a Genome-Wide Association Study. New Phytol. 2018, 222, 895–906. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR2 Gene of Arabidopsis Links Auxin Signalling, Cell Division, and the Size of Seeds and Other Organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef]
- Garcia, D.; Fitz Gerald, J.N.; Berger, F. Maternal Control of Integument Cell Elongation and Zygotic Control of Endosperm Growth Are Coordinated to Determine Seed Size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef]
- Guo, J.; Fan, J.; Hauser, B.A.; Rhee, S.Y. Target Enrichment Improves Mapping of Complex Traits by Deep Sequencing. G3 Genes Genomes Genet. 2016, 6, 67–77. [Google Scholar] [CrossRef]
- Herridge, R.P.; Day, R.C.; Baldwin, S.; Macknight, R.C. Rapid Analysis of Seed Size in Arabidopsis for Mutant and QTL Discovery. Plant Methods 2011, 7, 3. [Google Scholar] [CrossRef]
- Wang, D.; Sun, W.; Yuan, Z.; Sun, Q.; Fan, K.; Zhang, C.; Yu, S. Identification of a Novel QTL and Candidate Gene Associated with Grain Size Using Chromosome Segment Substitution Lines in Rice. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.; Miao, H.; Chu, Y.; Cui, F.; Yang, W.; Wang, C.; Shen, Y.; Xu, T.; Zhao, L.; et al. QTL Identification for Seed Weight and Size Based on a High-Density SLAF-Seq Genetic Map in Peanut (Arachis hypogaea L.). BMC Plant Biol. 2019, 19, 537. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Vries, H.B.-D.; Hanhart, C.J.; Koornneef, M. Natural Allelic Variation at Seed Size Loci in Relation to Other Life History Traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 4710–4717. [Google Scholar] [CrossRef]
- Liang, S.; Duan, Z.; He, X.; Yang, X.; Yuan, Y.; Liang, Q.; Pan, Y.; Zhou, G.; Zhang, M.; Liu, S.; et al. Natural Variation in GmSW17 Controls Seed Size in Soybean. Nat. Commun. 2024, 15, 7417, Erratum in Nat. Commun. 2024, 15, 8250. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.X.; Paddock, K.J.; Zhang, Z.; Stacey, M.G. GmKIX8-1 Regulates Organ Size in Soybean and Is the Causative Gene for the Major Seed Weight QTL qSw17-1. New Phytol. 2020, 229, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Fan, J.; Rhee, S.Y. QTG-Finder: A Machine-Learning Based Algorithm to Prioritize Causal Genes of Quantitative Trait Loci in Arabidopsis and Rice. G3 Genes Genomes Genet. 2019, 9, 3129–3138. [Google Scholar] [CrossRef] [PubMed]
- Kvilekval, K.; Fedorov, D.; Obara, B.; Singh, A.; Manjunath, B.S. Bisque: A Platform for Bioimage Analysis and Management. Bioinformatics 2009, 26, 544–552. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Weigel, D.; Mott, R. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 2009, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.T.; Soanes, B.K.; Kusakina, J.; Larrieu, A.; Knop, K.; Joy, N.; Breidenbach, F.; Sherwood, A.V.; Barton, G.J.; Fica, S.M.; et al. m6A Modification of U6 snRNA Modulates Usage of Two Major Classes of Pre-mRNA 5′ Splice Site. eLife 2022, 11, e78808. [Google Scholar] [CrossRef]
- Prasad, B.D.; Goel, S.; Krishna, P. In Silico Identification of Carboxylate Clamp Type Tetratricopeptide Repeat Proteins in Arabidopsis and Rice as Putative Co-Chaperones of HSP90/HSP70. PLoS ONE 2010, 5, e12761. [Google Scholar] [CrossRef]
- Angeles-Núñez, J.G.; Tiessen, A. Mutation of the Transcription Factor LEAFY COTYLEDON2 Alters the Chemical Composition of Arabidopsis Seeds, Decreasing Oil and Protein Content, While Maintaining High Levels of Starch and Sucrose in Mature Seeds. J. Plant Physiol. 2011, 168, 1891–1900. [Google Scholar] [CrossRef]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon Source–Sink Relationship in Arabidopsis thaliana: The Role of Sucrose Transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Sugar Import and Metabolism during Seed Development. Trends Plant Sci. 1997, 2, 169–174. [Google Scholar] [CrossRef]
- Angeles-Núñez, J.G.; Tiessen, A. Arabidopsis Sucrose Synthase 2 and 3 Modulate Metabolic Homeostasis and Direct Carbon Towards Starch Synthesis in Developing Seeds. Planta 2010, 232, 701–718. [Google Scholar] [CrossRef]
- Rosado, A.; Schapire, A.L.; Bressan, R.A.; Harfouche, A.L.; Hasegawa, P.M.; Valpuesta, V.; Botella, M.A. The Arabidopsis Tetratricopeptide Repeat-Containing Protein TTL1 Is Required for Osmotic Stress Responses and Abscisic Acid Sensitivity. Plant Physiol. 2006, 142, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, X.; Chen, S.; Liu, C.; Yang, L.; Li, K.; Liao, J.; Zheng, X.; Li, H.; Li, Y.; et al. ABI5–FLZ13 Module Transcriptionally Represses Growth-Related Genes to Delay Seed Germination in Response to ABA. Plant Commun. 2023, 4, 100636. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Ho, C.W.; Grierson, D. AtTRP1 Encodes a Novel TPR Protein That Interacts with the Ethylene Receptor ERS1 and Modulates Development in Arabidopsis. J. Exp. Bot. 2009, 60, 3697–3714. [Google Scholar] [CrossRef]
- Lin, Z.; Arciga-Reyes, L.; Zhong, S.; Alexander, L.; Hackett, R.; Wilson, I.; Grierson, D. SlTPR1, a Tomato Tetratricopeptide Repeat Protein, Interacts with the Ethylene Receptors NR and LeETR1, Modulating Ethylene and Auxin Responses and Development. J. Exp. Bot. 2008, 59, 4271–4287. [Google Scholar] [CrossRef]
- Filardo, F.; Robertson, M.; Singh, D.P.; Parish, R.W.; Swain, S.M. Functional Analysis of HvSPY, a Negative Regulator of GA Response, in Barley Aleurone Cells and Arabidopsis. Planta 2008, 229, 523–537. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Olszewski, N.E. Mutations at the SPINDLY Locus of Arabidopsis Alter Gibberellin Signal Transduction. Plant Cell 1993, 5, 887–896. [Google Scholar]
- Tseng, T.-S.; Swain, S.M.; Olszewski, N.E. Ectopic Expression of the Tetratricopeptide Repeat Domain of SPINDLY Causes Defects in Gibberellin Response. Plant Physiol. 2001, 126, 1250–1258. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in Fruit Development and Maturation. Plant J. 2020, 105, 446–458. [Google Scholar] [CrossRef]
- Hirano, T.; Kinoshita, N.; Morikawa, K.; Yanagida, M. Snap Helix with Knob and Hole: Essential Repeats in S. pombe Nuclear Protein nuc2 +. Cell 1990, 60, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, R.S.; Boguski, M.S.; Goebl, M.; Hieter, P. A Repeating Amino Acid Motif in CDC23 Defines a Family of Proteins and a New Relationship among Genes Required for Mitosis and RNA Synthesis. Cell 1990, 60, 307–317. [Google Scholar] [CrossRef]
- Debeaujon, I.; Lepiniec, L.; Pourcel, L.; Routaboul, J. Seed Coat Development and Dormancy. In Annual Plant Reviews; Blackwell: Oxford, UK, 2007; p. 392. [Google Scholar]
- Braun, D.M. Phloem Loading and Unloading of Sucrose: What a Long, Strange Trip from Source to Sink. Annu. Rev. Plant Biol. 2022, 73, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Park, S.O.; Hwang, S.; Hauser, B.A. The Phenotype of Arabidopsis Ovule Mutants Mimics the Morphology of Primitive Seed Plants. Proc. Biol. Sci. 2004, 271, 311–316. [Google Scholar] [CrossRef]
- Weis, J.H.; Quertermous, T. Size Fractionation Using Sucrose Gradients. Curr. Protoc. Mol. Biol. 1987, 1, 5.3.1–5.3.8. [Google Scholar] [CrossRef]
- Olszewski, N.E.; Martin, F.B.; Ausubel, F.M. Specialized Binary Vector for Plant Transformation: Expression of the Arabidopsis thaliana AHAS Gene in Nicotiana tabacum. Nucleic Acids Res. 1988, 16, 10765–10782. [Google Scholar] [CrossRef]
- Fraley, R.T.; Rogers, S.G.; Horsch, R.B.; Eichholtz, D.A.; Flick, J.S.; Fink, C.L.; Hoffmann, N.L.; Sanders, P.R. The SEV System: A New Disarmed Ti Plasmid Vector System for Plant Transformation. Nat. Biotechnol. 1985, 3, 629–635. [Google Scholar] [CrossRef]
- Gleave, A.P. A Versatile Binary Vector System with a T-DNA Organisational Structure Conducive to Efficient Integration of Cloned DNA into the Plant Genome. Plant Mol. Biol. 1992, 20, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Harder, A.; Wildgruber, R.; Nawrocki, A.; Fey, S.J.; Larsen, P.M.; Görg, A. Comparison of Yeast Cell Protein Solubilization Procedures for Two-Dimensional Electrophoresis. Electrophoresis 1999, 20, 826–829. [Google Scholar] [CrossRef]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein–Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hara, A.; Radin, N.S. Lipid Extraction of Tissues with a Low-Toxicity Solvent. Anal. Biochem. 1978, 90, 420–426. [Google Scholar] [CrossRef]
- Li, Y.; Beisson, F.; Pollard, M.; Ohlrogge, J. Oil Content of Arabidopsis Seeds: The Influence of Seed Anatomy, Light and Plant-to-Plant Variation. Phytochemistry 2006, 67, 904–915. [Google Scholar] [CrossRef]
- Cheng, C.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Cara, S.; Rhee, S.Y.; Hauser, B.A. Tetratricopeptide Repeat 2 Is a Quantitative Trait Locus That Controls Seed Size. Int. J. Mol. Sci. 2025, 26, 8310. https://doi.org/10.3390/ijms26178310
Wang Z, Cara S, Rhee SY, Hauser BA. Tetratricopeptide Repeat 2 Is a Quantitative Trait Locus That Controls Seed Size. International Journal of Molecular Sciences. 2025; 26(17):8310. https://doi.org/10.3390/ijms26178310
Chicago/Turabian StyleWang, Zhuolun, Stephanie Cara, Seung Y. Rhee, and Bernard A. Hauser. 2025. "Tetratricopeptide Repeat 2 Is a Quantitative Trait Locus That Controls Seed Size" International Journal of Molecular Sciences 26, no. 17: 8310. https://doi.org/10.3390/ijms26178310
APA StyleWang, Z., Cara, S., Rhee, S. Y., & Hauser, B. A. (2025). Tetratricopeptide Repeat 2 Is a Quantitative Trait Locus That Controls Seed Size. International Journal of Molecular Sciences, 26(17), 8310. https://doi.org/10.3390/ijms26178310







