Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions
Abstract
1. Introduction
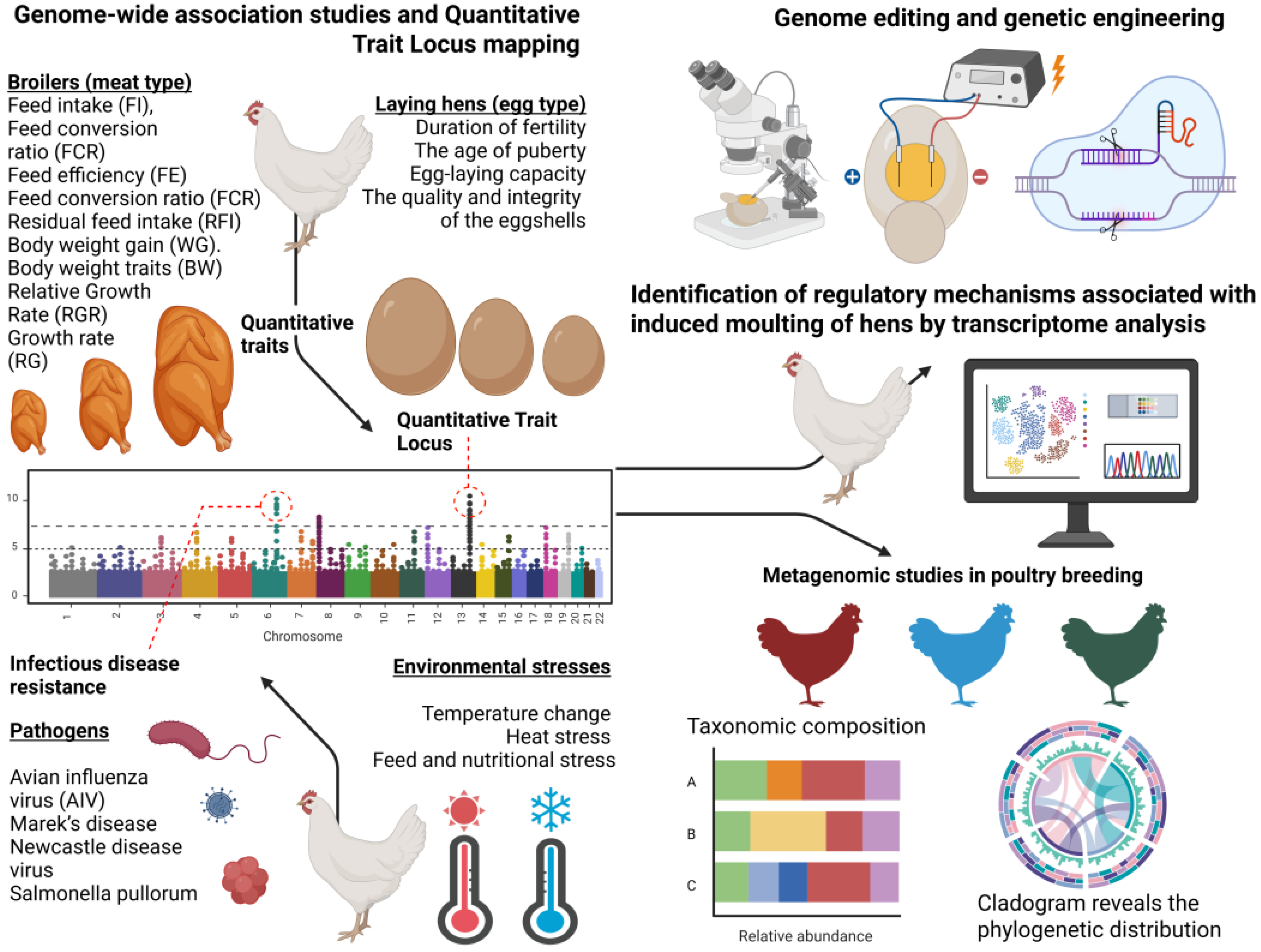
2. Whole-Genome and Transcriptome Studies of Association with Economically Important Traits in Chickens
2.1. Genome-Wide Association Studies with Quantitative Traits of Increased Productivity of Meat Chicken Breed
2.2. Genome-Wide Association Studies with Traits Related to Poultry Reproduction
2.2.1. Duration of Fertility
2.2.2. Age of Puberty
2.2.3. Egg-Laying Capacity
2.2.4. Quality and Integrity of Eggshells
3. Identification of Regulatory Mechanisms Associated with Artificial Molting of Chickens Using Transcriptome Analysis
4. Metagenomic Research in Poultry Production
5. Genetic Studies of Resistance to Infectious Diseases in Chickens
6. Studies on the Genetics of Climate Adaptability of Chickens
7. Prospects for Genetic Improvement of Chickens
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NGS | Next-generation sequencing |
| QTL | The quantitative trait locus |
| GWAS | Genome-wide association studies |
| FI | Feed intake |
| FC | Feed conversion ratio |
| FE | Feed utilization efficiency |
| WG | Body weight gain |
| BW | Body weight indices |
| RG | Growth rate |
| RGR | Relative growth rate |
| RFI | Residual feed intake |
| FCR | Feed conversion ratio |
| PGCs | Primordial germ cells |
| ICSI | Intracytoplasmic sperm injection |
References
- Bello, S.F.; Lawal, R.A.; Adeola, A.C.; Nie, Q. The Study of Selection Signature and Its Applications on Identification of Candidate Genes Using Whole Genome Sequencing Data in Chicken—A Review. Poult. Sci. 2023, 102, 102657. [Google Scholar] [CrossRef]
- International Chicken Genome Sequencing Consortium. Sequence and Comparative Analysis of the Chicken Genome Provide Unique Perspectives on Vertebrate Evolution. Nature 2004, 432, 695–716, Erratum in Nature 2005, 433, 777. [Google Scholar] [CrossRef]
- Sukhija, N.; Kanaka, K.K.; Goli, R.C.; Kapoor, P.; Sivalingam, J.; Verma, A.; Sharma, R.; Tripathi, S.B.; Malik, A.A. The Flight of Chicken Genomics and Allied Omics-a Mini Review. Ecol. Genet. Genom. 2023, 29, 100201. [Google Scholar] [CrossRef]
- Boschiero, C.; Moreira, G.C.M.; Gheyas, A.A.; Godoy, T.F.; Gasparin, G.; Mariani, P.D.S.C.; Paduan, M.; Cesar, A.S.M.; Ledur, M.C.; Coutinho, L.L. Genome-Wide Characterization of Genetic Variants and Putative Regions under Selection in Meat and Egg-Type Chicken Lines. BMC Genom. 2018, 19, 83. [Google Scholar] [CrossRef]
- Mebratie, W.; Reyer, H.; Wimmers, K.; Bovenhuis, H.; Jensen, J. Genome Wide Association Study of Body Weight and Feed Efficiency Traits in a Commercial Broiler Chicken Population, a Re-Visitation. Sci. Rep. 2019, 9, 922. [Google Scholar] [CrossRef] [PubMed]
- Chicken QTL Database. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/GG/traitmap?trait_ID=2151 (accessed on 11 July 2025).
- Hu, Z.-L.; Park, C.A.; Wu, X.-L.; Reecy, J.M. Animal QTLdb: An Improved Database Tool for Livestock Animal QTL/Association Data Dissemination in the Post-Genome Era. Nucleic Acids Res. 2013, 41, D871–D879. [Google Scholar] [CrossRef] [PubMed]
- Dementeva, N.V.; Mitrofanova, O.V.; Tyshchenko, V.I.; Terletskiy, V.P.; Yakovlev, A.F. The rate of weight gain and productivity of a chicken broiler cross with various polymorphic types of the myostatin gene. Russ. J. Genet. Appl. Res. 2017, 7, 1–5. [Google Scholar] [CrossRef]
- Moreira, G.C.M.; Godoy, T.F.; Boschiero, C.; Gheyas, A.; Gasparin, G.; Andrade, S.C.S.; Paduan, M.; Montenegro, H.; Burt, D.W.; Ledur, M.C.; et al. Variant Discovery in a QTL Region on Chromosome 3 Associated with Fatness in Chickens. Anim. Genet. 2015, 46, 141–147. [Google Scholar] [CrossRef]
- Moreira, G.C.M.; Poleti, M.D.; Pértille, F.; Boschiero, C.; Cesar, A.S.M.; Godoy, T.F.; Ledur, M.C.; Reecy, J.M.; Garrick, D.J.; Coutinho, L.L. Unraveling Genomic Associations with Feed Efficiency and Body Weight Traits in Chickens through an Integrative Approach. BMC Genet. 2019, 20, 83. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ben-Jemaa, S.; Perini, F.; Cendron, F.; Biscarini, F.; Lasagna, E.; Penasa, M.; Cassandro, M. Genome-Wide Mapping of Signatures of Selection Using a High-Density Array Identified Candidate Genes for Growth Traits and Local Adaptation in Chickens. Genet. Sel. Evol. 2023, 55, 20. [Google Scholar] [CrossRef]
- Cha, J.; Choo, H.; Srikanth, K.; Lee, S.-H.; Son, J.-W.; Park, M.-R.; Kim, N.; Jang, G.W.; Park, J.-E. Genome-Wide Association Study Identifies 12 Loci Associated with Body Weight at Age 8 Weeks in Korean Native Chickens. Genes 2021, 12, 1170. [Google Scholar] [CrossRef] [PubMed]
- Sell-Kubiak, E.; Wimmers, K.; Reyer, H.; Szwaczkowski, T. Genetic Aspects of Feed Efficiency and Reduction of Environmental Footprint in Broilers: A Review. J. Appl. Genet. 2017, 58, 487–498. [Google Scholar] [CrossRef]
- Yang, L.; He, T.; Xiong, F.; Chen, X.; Fan, X.; Jin, S.; Geng, Z. Identification of Key Genes and Pathways Associated with Feed Efficiency of Native Chickens Based on Transcriptome Data via Bioinformatics Analysis. BMC Genom. 2020, 21, 292. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Otecko, N.O.; Peng, M.; Weng, Z.; Li, W.; Chen, J.; Zhong, M.; Zhong, F.; Jin, S.; Geng, Z.; et al. Genome-Wide Genetic Structure and Selection Signatures for Color in 10 Traditional Chinese Yellow-Feathered Chicken Breeds. BMC Genom. 2020, 21, 316. [Google Scholar] [CrossRef]
- Moreira, G.C.M.; Boschiero, C.; Cesar, A.S.M.; Reecy, J.M.; Godoy, T.F.; Pértille, F.; Ledur, M.C.; Moura, A.S.A.M.T.; Garrick, D.J.; Coutinho, L.L. Integration of Genome Wide Association Studies and Whole Genome Sequencing Provides Novel Insights into Fat Deposition in Chicken. Sci. Rep. 2018, 8, 16222. [Google Scholar] [CrossRef]
- Zhang, S.; Li, H.; Shi, H. Single Marker and Haplotype Analysis of the Chicken Apolipoprotein B Gene T123G and D9500D9- Polymorphism Reveals Association with Body Growth and Obesity. Poult. Sci. 2006, 85, 178–184. [Google Scholar] [CrossRef]
- Sun, Y.N.; Gao, Y.; Qiao, S.P.; Wang, S.Z.; Duan, K.; Wang, Y.X.; Li, H.; Wang, N. Epigenetic DNA Methylation in the Promoters of Peroxisome Proliferator-Activated Receptor γ in Chicken Lines Divergently Selected for Fatness. J. Anim. Sci. 2014, 92, 48–53. [Google Scholar] [CrossRef]
- Wen, C.; Mai, C.; Cai, R.; Gou, Q.; Zhang, B.; Li, J.; Sun, C.; Yang, N. Inheritance of the Duration of Fertility in Chickens and Its Correlation with Laying Performance. Genet. Sel. Evol. 2022, 54, 41. [Google Scholar] [CrossRef]
- Azmal, S.A.; Nan, J.; Bhuiyan, A.A.; Elokil, A.A.; Ali, M.I.; Adetula, A.A.; Ma, S.; Sun, C.; Han, Z.; Yuan, J.; et al. A Genome-Wide Single Nucleotide Polymorphism Scan Reveals Genetic Markers Associated with Fertility Rate in Chinese Jing Hong Chicken. Poult. Sci. 2020, 99, 2873–2887. [Google Scholar] [CrossRef]
- Luo, W.; Huang, X.; Li, J.; Gu, L. Investigating the Genetic Determination of Duration-of-Fertility Trait in Breeding Hens. Sci. Rep. 2024, 14, 14819. [Google Scholar] [CrossRef]
- Liao, R.; Zhang, X.; Chen, Q.; Wang, Z.; Wang, Q.; Yang, C.; Pan, Y. Genome-wide association study reveals novel variants for growth and egg traits in Dongxiang blue-shelled and White Leghorn chickens. Anim. Genet. 2016, 47, 588–596. [Google Scholar] [CrossRef]
- Liu, L.; Cui, Z.; Xiao, Q.; Zhang, H.; Zhao, X.; Wang, Y.; Yin, H.; Li, D.; Zhu, Q. Polymorphisms in the Chicken Growth Differentiation Factor 9 Gene Associated with Reproductive Traits. Biomed. Res. Int. 2018, 2018, 9345473. [Google Scholar] [CrossRef]
- You, Z.; Yuan, J.; Wang, Y.; Sun, Y.; Ni, A.; Li, Y.; Ma, H.; Ma, T.; Chen, J. Integrated Transcriptomic Analysis on Chicken Ovary Reveals CYP21A1 Affects Follicle Granulosa Cell Development and Steroid Hormone Synthesis. Poult. Sci. 2024, 103, 103589. [Google Scholar] [CrossRef]
- Sun, C.; Lu, J.; Yi, G.; Yuan, J.; Duan, Z.; Qu, L.; Xu, G.; Wang, K.; Yang, N.; Veitia, R.A. Promising Loci and Genes for Yolk and Ovary Weight in Chickens Revealed by a Genome-Wide Association Study. PLoS ONE 2015, 10, e0137145. [Google Scholar] [CrossRef] [PubMed]
- Seminara, S.B.; Messager, S.; Chatzidaki, E.E.; Thresher, R.R.; Acierno, J.S.; Shagoury, J.K.; Bo-Abbas, Y.; Kuohung, W.; Schwinof, K.M.; Hendrick, A.G.; et al. The GPR54 Gene as a Regulator of Puberty. N. Engl. J. Med. 2003, 349, 1614–1627. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs Modulate microRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef]
- Han, W.; Zhu, Y.; Su, Y.; Li, G.; Qu, L.; Zhang, H.; Wang, K.; Zou, J.; Liu, H. High-Throughput Sequencing Reveals Circulating miRNAs as Potential Biomarkers for Measuring Puberty Onset in Chicken (Gallus Gallus). PLoS ONE 2016, 11, e0154958. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wu, Y.; Shen, J.; Pan, A.; Zhang, H.; Sun, J.; Liang, Z.; Huang, T.; Du, J.; Pi, J. Genome-Wide Association Study of Egg Production Traits in Shuanglian Chickens Using Whole Genome Sequencing. Genes 2023, 14, 2129. [Google Scholar] [CrossRef]
- Du, Y.; Liu, L.; He, Y.; Dou, T.; Jia, J.; Ge, C. Endocrine and Genetic Factors Affecting Egg Laying Performance in Chickens: A Review. Br. Poult. Sci. 2020, 61, 538–549. [Google Scholar] [CrossRef]
- Fedorova, Z.; Vachrameev, A. The influence of the mass of incubation eggs on the growth indicators of a live weight of the bred young chickens and the quality of their eggs. Genet. Breed. Anim. 2023, 3, 47–52. [Google Scholar] [CrossRef]
- Liu, W.; Li, D.; Liu, J.; Chen, S.; Qu, L.; Zheng, J.; Xu, G.; Yang, N. A Genome-Wide SNP Scan Reveals Novel Loci for Egg Production and Quality Traits in White Leghorn and Brown-Egg Dwarf Layers. PLoS ONE 2011, 6, e28600. [Google Scholar] [CrossRef]
- Zhang, G.X.; Fan, Q.C.; Wang, J.Y.; Zhang, T.; Xue, Q.; Shi, H.Q. Genome-Wide Association Study on Reproductive Traits in Jinghai Yellow Chicken. Anim. Reprod. Sci. 2015, 163, 30–34. [Google Scholar] [CrossRef]
- Kudinov, A.A.; Dementieva, N.V.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Larkina, T.A.; Mishina, A.I.; Plemyashov, K.V.; Griffin, D.K.; Romanov, M.N. Genome-Wide Association Studies Targeting the Yield of Extraembryonic Fluid and Production Traits in Russian White Chickens. BMC Genom. 2019, 20, 270. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Sun, C.; Dou, T.; Yi, G.; Qu, L.; Qu, L.; Wang, K.; Yang, N. Identification of Promising Mutants Associated with Egg Production Traits Revealed by Genome-Wide Association Study. PLoS ONE 2015, 10, e0140615. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, N.; Yan, Y.; Li, G.; Liu, A.; Wu, G.; Sun, C. Genome-Wide Association Analysis of Egg Production Performance in Chickens across the Whole Laying Period. BMC Genet. 2019, 20, 67. [Google Scholar] [CrossRef]
- Qu, L.; Shen, M.M.; Dou, T.C.; Ma, M.; Lu, J.; Wang, X.G.; Guo, J.; Hu, Y.P.; Li, Y.F.; Wang, K.H. Genome-Wide Association Studies for Mottled Eggs in Chickens Using a High-Density Single-Nucleotide Polymorphism Array. Animal 2021, 15, 100051. [Google Scholar] [CrossRef]
- Zhao, X.; Nie, C.; Zhang, J.; Li, X.; Zhu, T.; Guan, Z.; Chen, Y.; Wang, L.; Lv, X.Z.; Yang, W.; et al. Identification of Candidate Genomic Regions for Chicken Egg Number Traits Based on Genome-Wide Association Study. BMC Genom. 2021, 22, 610. [Google Scholar] [CrossRef]
- Gao, J.; Xu, W.; Zeng, T.; Tian, Y.; Wu, C.; Liu, S.; Zhao, Y.; Zhou, S.; Lin, X.; Cao, H.; et al. Genome-Wide Association Study of Egg-Laying Traits and Egg Quality in LingKun Chickens. Front. Vet. Sci. 2022, 9, 877739. [Google Scholar] [CrossRef]
- Kang, H.; Lu, Y.; Zhang, W.; Hua, G.; Gan, J.; Deng, X.; Zhang, Z.; Li, H. Genome-Wide Association Study Identifies a Novel Candidate Gene for Egg Production Traits in Chickens. Anim. Genet. 2024, 55, 480–483. [Google Scholar] [CrossRef]
- Cai, D.; Wang, Z.; Zhou, Z.; Lin, D.; Ju, X.; Nie, Q. Integration of Transcriptome Sequencing and Whole Genome Resequencing Reveal Candidate Genes in Egg Production of Upright and Pendulous-Comb Chickens. Poult. Sci. 2023, 102, 102504. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Cao, D.; Liu, J.; Li, F.; Han, H.; Han, H.; Lei, Q.; Liu, W.; Li, D.; et al. Elucidation of the genetic determination of clutch traits in Chinese local chickens of the Laiwu Black breed. BMC Genom. 2023, 24, 686. [Google Scholar] [CrossRef]
- Li, Q.; Duan, Z.; Sun, C.; Zheng, J.; Xu, G.; Yang, N. Genetic Variations for the Eggshell Crystal Structure Revealed by Genome-Wide Association Study in Chickens. BMC Genom. 2021, 22, 786. [Google Scholar] [CrossRef]
- Sun, C.; Qu, L.; Yi, G.; Yuan, J.; Duan, Z.; Shen, M.; Qu, L.; Xu, G.; Wang, K.; Yang, N. Genome-Wide Association Study Revealed a Promising Region and Candidate Genes for Eggshell Quality in an F2 Resource Population. BMC Genom. 2015, 16, 565. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, F.; Liu, L.; Zheng, C.W.; Wang, D.H.; Hou, Z.C.; Ning, Z.H.; Leung, F.C.C. Integrating Transcriptome and Genome Re-Sequencing Data to Identify Key Genes and Mutations Affecting Chicken Eggshell Qualities. PLoS ONE 2015, 10, e0125890. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Zhong, C.; Jiang, X.; Wu, G.; Li, G.; Yan, Y.; Yang, N.; Sun, C. Genetic pat-terns and genome-wide association analysis of eggshell quality traits of egg-type chicken across an extended laying period. Poult. Sci. 2024, 103, 103458. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Chen, Z.-T.; Zheng, R.; Diao, S.; Teng, J.; Yuan, X.; Zhang, H.; Chen, Z.; Zhang, X.; Li, J.; et al. New Insights From Imputed Whole-Genome Sequence-Based Genome-Wide Association Analysis and Transcriptome Analysis: The Genetic Mechanisms Underlying Residual Feed Intake in Chickens. Front. Genet. 2020, 11, 243. [Google Scholar] [CrossRef]
- Reyer, H.; Hawken, R.; Murani, E.; Ponsuksili, S.; Wimmers, K. The Genetics of Feed Conversion Efficiency Traits in a Commercial Broiler Line. Sci. Rep. 2015, 5, 16387. [Google Scholar] [CrossRef]
- Yang, R.; Xu, Z.; Wang, Q.; Zhu, D.; Bian, C.; Ren, J.; Huang, Z.; Zhu, X.; Tian, Z.; Wang, Y.; et al. Genome-wide Association Study and Genomic Prediction for Growth Traits in Yellow-Plumage Chicken Using Genotyping-by-Sequencing. Genet. Sel. Evol. 2021, 53, 82. [Google Scholar] [CrossRef]
- Fan, Q.C.; Wu, P.F.; Dai, G.J.; Zhang, G.X.; Zhang, T.; Xue, Q.; Shi, H.Q.; Wang, J.Y. Identification of 19 Loci for Reproductive Traits in a Local Chinese Chicken by Genome-Wide Study. Genet. Mol. Res. 2017, 16, 16019431. [Google Scholar] [CrossRef]
- Darwish, H.Y.A.; Dalirsefat, S.B.; Dong, X.; Hua, G.; Chen, J.; Zhang, Y.; Li, J.; Xu, J.; Li, J.; Deng, X.; et al. Genome-Wide Association Study and a Post Replication Analysis Revealed a Promising Genomic Region and Candidate Genes for Chicken Eggshell Blueness. PLoS ONE 2019, 14, e0209181. [Google Scholar] [CrossRef]
- Tan, X.; Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Large-Scale Whole Genome Sequencing Study Reveals Genetic Architecture and Key Variants for Breast Muscle Weight in Native Chickens. Genes 2021, 13, 3. [Google Scholar] [CrossRef]
- Dadousis, C.; Somavilla, A.; Ilska, J.J.; Johnsson, M.; Batista, L.; Mellanby, R.J.; Headon, D.; Gottardo, P.; Whalen, A.; Wilson, D.; et al. A Genome-Wide Association Analysis for Body Weight at 35 Days Measured on 137,343 Broiler Chickens. Genet. Sel. Evol. 2021, 53, 70. [Google Scholar] [CrossRef]
- Psifidi, A.; Banos, G.; Matika, O.; Desta, T.T.; Bettridge, J.; Hume, D.A.; Dessie, T.; Christley, R.; Wigley, P.; Hanotte, O.; et al. Genome-Wide Association Studies of Immune, Disease and Production Traits in Indigenous Chicken Ecotypes. Genet. Sel. Evol. 2016, 48, 74. [Google Scholar] [CrossRef]
- Zhang, J.; Geng, X.; Zhang, Y.; Zhao, X.; Zhang, P.; Sun, G.; Li, W.; Li, D.; Han, R.; Li, G.; et al. Interaction Between Cecal Metabolites and Liver Lipid Metabolism Pathways During Induced Molting in Laying Hens. Front. Physiol. 2022, 13, 862721. [Google Scholar] [CrossRef]
- Hassanien, H.H.M. Effect of Force Molting Programs on Egg Production and Quality of Laying Hens. Asian J. Poult. Sci. 2011, 5, 13–20. [Google Scholar] [CrossRef]
- Sun, M.; Wang, H.; Zhu, X.; Zhang, X.; Min, Y.; Ge, M.; Jiang, X.; Yu, W. The Mechanism of Egg Production Improvement in Laying Hens before and after Molting Revealed by Transcriptome and Metabolome Integration. Poult. Sci. 2025, 104, 105125. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, Y.; Wen, J.; Jia, Y.; Wang, L.; Lv, X.; Yang, W.; Qu, C.; Li, H.; Wang, H.; et al. Transcriptomic Analysis of Laying Hens Revealed the Role of Aging-Related Genes during Forced Molting. Genes 2021, 12, 1767. [Google Scholar] [CrossRef] [PubMed]
- Onbaşılar, E.E.; Erol, H. Effects of Different Forced Molting Methods on Postmolt Production, Corticosterone Level, and Immune Response to Sheep Red Blood Cells in Laying Hens. J. Appl. Poult. Res. 2007, 16, 529–536. [Google Scholar] [CrossRef]
- Zhang, Z.; Du, H.; Yang, C.; Li, Q.; Qiu, M.; Song, X.; Yu, C.; Jiang, X.; Liu, L.; Hu, C.; et al. Comparative Transcriptome Analysis Reveals Regulators Mediating Breast Muscle Growth and Development in Three Chicken Breeds. Anim. Biotechnol. 2019, 30, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Febel, H.; Mezes, M.; Horn, P.; Balogh, K.; Romvari, R. Differential Utilization of Hepatic and Myocardial Fatty Acids during Forced Molt of Laying Hens. Poult. Sci. 2005, 84, 106–112. [Google Scholar] [CrossRef]
- Kubena, L.F.; Byrd, J.A.; Moore, R.W.; Ricke, S.C.; Nisbet, D.J. Effects of Drinking Water Treatment on Susceptibility of Laying Hens to Salmonella Enteritidis during Forced Molt. Poult. Sci. 2005, 84, 204–211. [Google Scholar] [CrossRef]
- Mpango, M.M.; Madekurozwa, M.-C. Comparative Histomorphological and Ultrastructural Study of the Luminal Epithelium of the Isthmus in Laying and Moulting Domestic Fowls (Gallus Domesticus). Anat. Histol. Embryol. 2018, 47, 444–455. [Google Scholar] [CrossRef]
- Laporta, L.; Micera, E.; Surdo, N.C.; Moramarco, A.M.; Di Modugno, G.; Zarrilli, A. A Functional Study on L-Type Calcium Channels in Granulosa Cells of Small Follicles in Laying and Forced Molt Hens. Anim. Reprod. Sci. 2011, 126, 265–270. [Google Scholar] [CrossRef]
- Kakhki, R.A.M.; Mousavi, Z.; Anderson, K.E. An Appraisal of Moulting on Post-Moult Egg Production and Egg Weight Distribution in White Layer Hens; Meta-Analysis. Br. Poult. Sci. 2018, 59, 278–285. [Google Scholar] [CrossRef]
- Huo, S.; Li, Y.; Guo, Y.; Zhang, S.; Li, P.; Gao, P. Improving Effects of Epimedium Flavonoids on the Selected Reproductive Features in Layer Hens after Forced Molting. Poult. Sci. 2020, 99, 2757–2765. [Google Scholar] [CrossRef] [PubMed]
- Han, G.P.; Lee, K.-C.; Kang, H.K.; Oh, H.N.; Sul, W.J.; Kil, D.Y. Analysis of Excreta Bacterial Community after Forced Molting in Aged Laying Hens. Asian-Australas. J. Anim. Sci. 2019, 32, 1715–1724. [Google Scholar] [CrossRef]
- Onbaşılar, E.E.; Kahraman, M.; Güngör, Ö.F.; Kocakaya, A.; Karakan, T.; Pirpanahi, M.; Doğan, B.; Metin, D.; Akan, M.; Şehu, A.; et al. Effects of Cage Type on Performance, Welfare, and Microbiological Properties of Laying Hens during the Molting Period and the Second Production Cycle. Trop. Anim. Health Prod. 2020, 52, 3713–3724. [Google Scholar] [CrossRef]
- Piórkowska, K.; Żukowski, K.; Połtowicz, K.; Nowak, J.; Wojtysiak, D.; Derebecka, N.; Wesoły, J.; Ropka-Molik, K. Transcriptomic Changes in Broiler Chicken Hypothalamus during Growth and Development. Int. J. Genom. 2018, 2018, 6049469. [Google Scholar] [CrossRef]
- Xing, S.; Liu, R.; Zhao, G.; Liu, L.; Groenen, M.A.M.; Madsen, O.; Zheng, M.; Yang, X.; Crooijmans, R.P.M.A.; Wen, J. RNA-Seq Analysis Reveals Hub Genes Involved in Chicken Intramuscular Fat and Abdominal Fat Deposition During Development. Front. Genet. 2020, 11, 1009. [Google Scholar] [CrossRef]
- Guo, J.; Ito, S.; Nguyen, H.T.; Yamamoto, K.; Iwata, H. Effects on the Hepatic Transcriptome of Chicken Embryos in Ovo Exposed to Phenobarbital. Ecotoxicol. Environ. Saf. 2018, 160, 94–103. [Google Scholar] [CrossRef]
- Yin, Z.; Lian, L.; Zhu, F.; Zhang, Z.-H.; Hincke, M.; Yang, N.; Hou, Z.-C. The Transcriptome Landscapes of Ovary and Three Oviduct Segments during Chicken (Gallus Gallus) Egg Formation. Genomics 2020, 112, 243–251. [Google Scholar] [CrossRef]
- Wang, X.; Jia, Y.; Ren, J.; Liu, H.; Adam, F.A.; Wang, X.; Yang, Z. Insights into the Chicken Bursa of Fabricius Response to Newcastle Disease Virus at 48 and 72 Hours Post-Infection through RNA-Seq. Vet. Microbiol. 2019, 236, 108389. [Google Scholar] [CrossRef]
- Guo, Y.; Balasubramanian, B.; Zhao, Z.-H.; Liu, W.-C. Marine Algal Polysaccharides Alleviate Aflatoxin B1-Induced Bursa of Fabricius Injury by Regulating Redox and Apoptotic Signaling Pathway in Broilers. Poult. Sci. 2020, 100, 844–857. [Google Scholar] [CrossRef]
- Wang, C.; Shan, H.; Chen, H.; Bai, X.; Ding, J.; Ye, D.; Adam, F.E.A.; Yang, Y.; Wang, J.; Yang, Z. Probiotics and Vitamins Modulate the Cecal Microbiota of Laying Hens Submitted to Induced Molting. Front. Microbiol. 2023, 14, 1180838. [Google Scholar] [CrossRef]
- Ma, P.; Xue, F.; Chen, J.; Zhang, X.; Xu, X.; Ma, Z.; Zhang, H.; Wu, Y.; Li, L.; Qu, Y.; et al. Transcriptomic Insight into the Underlying Mechanism of Induced Molting on Reproductive Remodeling, Performance and Egg Quality in Laying Hen. Poult. Sci. 2025, 104, 104692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, C.; Liu, X.; Zhang, X.; An, Z.; Zhou, E.; Li, J.; Li, Z.; Li, W.; Sun, G.; et al. Multi-Stage Transcriptome Analysis Revealed the Growth Mechanism of Feathers and Hair Follicles during Induction Molting by Fasting in the Late Stage of Egg Laying. Biology 2023, 12, 1345. [Google Scholar] [CrossRef]
- Stanley, D.; Denman, S.E.; Hughes, R.J.; Geier, M.S.; Crowley, T.M.; Chen, H.; Haring, V.R.; Moore, R.J. Intestinal Microbiota Associated with Differential Feed Conversion Efficiency in Chickens. Appl. Microbiol. Biotechnol. 2012, 96, 1361–1369. [Google Scholar] [CrossRef]
- Shetty, S.A.; Marathe, N.P.; Shouche, Y.S. Opportunities and Challenges for Gut Microbiome Studies in the Indian Population. Microbiome 2013, 1, 24. [Google Scholar] [CrossRef]
- Deusch, S.; Tilocca, B.; Camarinha-Silva, A.; Seifert, J. News in Livestock Research—Use of Omics-Technologies to Study the Microbiota in the Gastrointestinal Tract of Farm Animals. Comput. Struct. Biotechnol. J. 2015, 13, 55–63. [Google Scholar] [CrossRef]
- Brugman, S.; Ikeda-Ohtsubo, W.; Braber, S.; Folkerts, G.; Pieterse, C.M.J.; Bakker, P.A.H.M. A Comparative Review on Microbiota Manipulation: Lessons From Fish, Plants, Livestock, and Human Research. Front. Nutr. 2018, 5, 80. [Google Scholar] [CrossRef]
- Wen, C.; Yan, W.; Sun, C.; Ji, C.; Zhou, Q.; Zhang, D.; Zheng, J.; Yang, N. The Gut Microbiota Is Largely Independent of Host Genetics in Regulating Fat Deposition in Chickens. ISME J. 2019, 13, 1422–1436. [Google Scholar] [CrossRef]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A Comparison of the Microbiome and the Metabolome of Different Regions of the Equine Hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef]
- Edwards, J.E.; Shetty, S.A.; van den Berg, P.; Burden, F.; van Doorn, D.A.; Pellikaan, W.F.; Dijkstra, J.; Smidt, H. Multi-Kingdom Characterization of the Core Equine Fecal Microbiota Based on Multiple Equine (Sub)Species. Anim. Microbiome 2020, 2, 6. [Google Scholar] [CrossRef]
- Segura-Wang, M.; Grabner, N.; Koestelbauer, A.; Klose, V.; Ghanbari, M. Genome-Resolved Metagenomics of the Chicken Gut Microbiome. Front. Microbiol. 2021, 12, 726923. [Google Scholar] [CrossRef]
- Carrasco, J.M.D.; Casanova, N.A.; Miyakawa, M.E.F. Microbiota, Gut Health and Chicken Productivity: What Is the Connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Cheled-Shoval, S.L.; Gamage, N.S.W.; Amit-Romach, E.; Forder, R.; Marshal, J.; Van Kessel, A.; Uni, Z. Differences in Intestinal Mucin Dynamics between Germ-Free and Conventionally Reared Chickens after Mannan-Oligosaccharide Supplementation. Poult. Sci. 2014, 93, 636–644. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal Microbiome of Poultry and Its Interaction with Host and Diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the Chicken Gastrointestinal Tract: Influence on Health, Productivity and Disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken Gut Microbiota: Importance and Detection Technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef]
- Rovira, P.; McAllister, T.; Lakin, S.M.; Cook, S.R.; Doster, E.; Noyes, N.R.; Weinroth, M.D.; Yang, X.; Parker, J.K.; Boucher, C.; et al. Characterization of the Microbial Resistome in Conventional and “Raised Without Antibiotics” Beef and Dairy Production Systems. Front. Microbiol. 2019, 10, 1980. [Google Scholar] [CrossRef]
- Chen, Y.; Ni, J.; Li, H. Effect of Green Tea and Mulberry Leaf Powders on the Gut Microbiota of Chicken. BMC Vet. Res. 2019, 15, 77. [Google Scholar] [CrossRef]
- Lee, S.-H.; Bang, S.; Jang, H.-H.; Lee, E.-B.; Kim, B.-S.; Kim, S.-H.; Kang, S.-H.; Lee, K.-W.; Kim, D.-W.; Kim, J.-B.; et al. Effects of Allium Hookeri on Gut Microbiome Related to Growth Performance in Young Broiler Chickens. PLoS ONE 2020, 15, e0226833. [Google Scholar] [CrossRef]
- Shi, S.; Qi, Z.; Jiang, W.; Quan, S.; Sheng, T.; Tu, J.; Shao, Y.; Qi, K. Effects of Probiotics on Cecal Microbiome Profile Altered by Duck Escherichia Coli 17 Infection in Cherry Valley Ducks. Microb. Pathog. 2020, 138, 103849. [Google Scholar] [CrossRef]
- Joo, S.S.; Yoon, J.H.; Jung, J.Y.; Joo, S.Y.; An, S.H.; Ban, B.C.; Kong, C.; Kim, M. The Modulatory Effects of Lacticaseibacillus Paracasei Strain NSMJ56 on Gut Immunity and Microbiome in Early-Age Broiler Chickens. Animals 2022, 12, 3413. [Google Scholar] [CrossRef]
- Sayed, Y.; Hassan, M.; Salem, H.M.; Al-Amry, K.; Eid, G.E. Prophylactic Influences of Prebiotics on Gut Microbiome and Immune Response of Heat-Stressed Broiler Chickens. Sci. Rep. 2023, 13, 13991. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, T.; Dong, W.; Sun, G.; Liu, J.; Peng, N.; Zhao, S. Metagenome-Assembled Genome Reveals Species and Functional Composition of Jianghan Chicken Gut Microbiota and Isolation of Pediococcus Acidilactic with Probiotic Properties. Microbiome 2024, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Qi, G.; Wang, J.; Zhang, H.; Qiu, K.; Wu, S. Intestinal Microbiota of Layer Hens and Its Association with Egg Quality and Safety. Poult. Sci. 2022, 101, 102008. [Google Scholar] [CrossRef] [PubMed]
- Chousalkar, K.K.; Khan, S.; McWhorter, A.R. Microbial quality, safety and storage of eggs. Curr. Opin. Food Sci. 2021, 38, 91–95. [Google Scholar] [CrossRef]
- Ricke, S.C.; Dittoe, D.K.; Olson, E.G. Microbiome Applications for Laying Hen Performance and Egg Production. Poult. Sci. 2022, 101, 101784. [Google Scholar] [CrossRef]
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive Microbial and Functional Diversity within the Chicken Cecal Microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The Chicken Gut Metagenome and the Modulatory Effects of Plant-Derived Benzylisoquinoline Alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef]
- Nene, M.; Kunene, N.W.; Pierneef, R.; Hadebe, K. Profiling the Diversity of the Village Chicken Faecal Microbiota Using 16S rRNA Gene and Metagenomic Sequencing Data to Reveal Patterns of Gut Microbiome Signatures. Front. Microbiol. 2025, 15, 1487595. [Google Scholar] [CrossRef]
- Glendinning, L.; Stewart, R.D.; Pallen, M.J.; Watson, K.A.; Watson, M. Assembly of Hundreds of Novel Bacterial Genomes from the Chicken Caecum. Genome Biol. 2020, 21, 34. [Google Scholar] [CrossRef]
- Gilroy, R.; Ravi, A.; Getino, M.; Pursley, I.; Horton, D.L.; Alikhan, N.-F.; Baker, D.; Gharbi, K.; Hall, N.; Watson, M.; et al. Extensive Microbial Diversity within the Chicken Gut Microbiome Revealed by Metagenomics and Culture. PeerJ 2021, 9, e10941. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Zhu, B.; Gao, G.F.; Guo, Y.; Hu, Y. Author Correction: Metagenome-Assembled Genomes and Gene Catalog from the Chicken Gut Microbiome Aid in Deciphering Antibiotic Resistomes. Commun. Biol. 2022, 5, 1289. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The Abundance and Variety of Carbohydrate-Active Enzymes in the Human Gut Microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Aarestrup, F.M. The Livestock Reservoir for Antimicrobial Resistance: A Personal View on Changing Patterns of Risks, Effects of Interventions and the Way Forward. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140085. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.C.; Wu, N.; Weimer, B.C.; Gao, G.F.; et al. The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Cao, J.; Bi, Y.; Lv, N.; Liu, F.; Liang, S.; Shi, Y.; Jiao, X.; Gao, G.F.; et al. Antibiotic Resistance Gene Reservoir in Live Poultry Markets. J. Infect. 2019, 78, 445–453. [Google Scholar] [CrossRef]
- Bai, X.; Zhong, H.; Cui, X.; Wang, T.; Gu, Y.; Li, M.; Miao, X.; Li, J.; Lu, L.; Xu, W.; et al. Metagenomic Profiling Uncovers Microbiota and Antibiotic Resistance Patterns across Human, Chicken, Pig Fecal, and Soil Environments. Sci. Total Environ. 2024, 947, 174734. [Google Scholar] [CrossRef]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes among Foodborne Pathogens of the Enterobacteriaceae Family—A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef]
- Oladeinde, A.; Cook, K.; Lakin, S.M.; Woyda, R.; Abdo, Z.; Looft, T.; Herrington, K.; Zock, G.; Lawrence, J.P.; Thomas, J.C.; et al. Horizontal Gene Transfer and Acquired Antibiotic Resistance in Salmonella Enterica Serovar Heidelberg Following In Vitro Incubation in Broiler Ceca. Appl. Environ. Microbiol. 2019, 85, e01903–e01919. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human Gut Microbiome Viewed across Age and Geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Meehan, C.J.; Koenig, J.E.; Dhanani, A.S.; Rose, R.A.; Howlett, S.E.; Beiko, R.G. Microbial Shifts in the Aging Mouse Gut. Microbiome 2014, 2, 50. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and Succession of the Intestinal Bacterial Community of the Maturing Broiler Chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef]
- Shaufi, M.A.M.; Sieo, C.C.; Chong, C.W.; Gan, H.M.; Ho, Y.W. Deciphering Chicken Gut Microbial Dynamics Based on High-Throughput 16S rRNA Metagenomics Analyses. Gut Pathog. 2015, 7, 4. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut Microbiota Composition Correlates with Diet and Health in the Elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Awad, W.A.; Mann, E.; Dzieciol, M.; Hess, C.; Schmitz-Esser, S.; Wagner, M.; Hess, M. Age-Related Differences in the Luminal and Mucosa-Associated Gut Microbiome of Broiler Chickens and Shifts Associated with Campylobacter Jejuni Infection. Front. Cell. Infect. Microbiol. 2016, 6, 154. [Google Scholar] [CrossRef]
- Kretzschmar-McCluskey, V.; Curtis, P.A.; Anderson, K.E.; Kerth, L.K.; Berry, W.D. Influence of Hen Age and Molting Treatments on Shell Egg Exterior, Interior, and Contents Microflora and Salmonella Prevalence During a Second Production Cycle. Poult. Sci. 2008, 87, 2146–2151. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Gong, Y.; Zhang, J.; Li, D.; Tian, Y.; Han, R.; Guo, Y.; Sun, G.; Li, W.; et al. Fasting-Induced Molting Impacts the Intestinal Health by Altering the Gut Microbiota. Animals 2024, 14, 1640. [Google Scholar] [CrossRef]
- Zekarias, B.; Ter Huurne, A.A.H.M.; Landman, W.J.M.; Rebel, J.M.J.; Pol, J.M.A.; Gruys, E. Immunological Basis of Differences in Disease Resistance in the Chicken. Vet. Res. 2002, 33, 109–125. [Google Scholar] [CrossRef]
- Gul, H.; Habib, G.; Khan, I.M.; Rahman, S.U.; Khan, N.M.; Wang, H.; Khan, N.U.; Liu, Y. Genetic Resilience in Chickens against Bacterial, Viral and Protozoal Pathogens. Front. Vet. Sci. 2022, 9, 1032983. [Google Scholar] [CrossRef]
- Heward, J.A.; Lindsay, M.A. Long Non-Coding RNAs in the Regulation of the Immune Response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef]
- Dar, M.A.; Mumtaz, P.T.; AhmadBhat, S.; Nabi, M.; Taban, Q.; Shah, R.A.; Khan, H.M.; Ahmad, S.M.; Dar, M.A.; Mumtaz, P.T.; et al. Genetics of Disease Resistance in Chicken. In Application of Genetics and Genomics in Poultry Science; IntechOpen: London, UK, 2018; ISBN 978-1-78923-631-6. [Google Scholar]
- Yu, H.; Mi, C.; Wang, Q.; Dai, G.; Zhang, T.; Zhang, G.; Xie, K.; Zhao, Z. Long Noncoding RNA Profiling Reveals That LncRNA BTN3A2 Inhibits the Host Inflammatory Response to Eimeria Tenella Infection in Chickens. Front. Immunol. 2022, 13, 891001. [Google Scholar] [CrossRef]
- Chen, M. Genetic Resolution of Disease Resistance in Poultry: A Genome-Wide Association Study Perspective. Anim. Mol. Breed. 2024, 14, 10–18. [Google Scholar] [CrossRef]
- Sulimova, G.E.; Oyun, N.Y.; Sevastianova, A.; Alexandrov, A.V.; Vakhrameev, A.B.; Kuzevanova, A.Y.; Alimov, A.A. Evaluation of polymorphism loci associated with viral diseases in spangled Orloff chicken breed. Russ. J. Genet. 2017, 53, 1119–1125. [Google Scholar] [CrossRef]
- Xu, L.; He, Y.; Ding, Y.; Liu, G.E.; Zhang, H.; Cheng, H.H.; Taylor, R.L.; Song, J. Genetic Assessment of Inbred Chicken Lines Indicates Genomic Signatures of Resistance to Marek’s Disease. J. Anim. Sci. Biotechnol. 2018, 9, 65. [Google Scholar] [CrossRef]
- Lin, X.; Chen, H. Genetic Strategies in Poultry to Combat Environmental Stress: An Analysis Based on GWAS. Anim. Mol. Breed. 2024, 14, 62–71. [Google Scholar] [CrossRef]
- Smith, J.; Lipkin, E.; Soller, M.; Fulton, J.E.; Burt, D.W. Mapping QTL Associated with Resistance to Avian Oncogenic Marek’s Disease Virus (MDV) Reveals Major Candidate Genes and Variants. Genes 2020, 11, 1019. [Google Scholar] [CrossRef]
- Li, X.; Nie, C.; Liu, Y.; Chen, Y.; Lv, X.; Wang, L.; Zhang, J.; Li, K.; Jia, Y.; Ban, L.; et al. A Genome-Wide Association Study Explores the Genetic Determinism of Host Resistance to Salmonella Pullorum Infection in Chickens. Genet. Sel. Evol. 2019, 51, 51. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, Q.; Hu, Y.; Sun, Y.; Liu, R.; Zheng, M.; Wen, J.; Li, P.; Liu, L.; Zhao, G. Genomewide Association Study of Immune Traits in Chicken F2 Resource Population. J. Anim. Breed. Genet. 2016, 133, 197–206. [Google Scholar] [CrossRef]
- Luo, C.; Qu, H.; Ma, J.; Wang, J.; Hu, X.; Li, N.; Shu, D. A Genome-Wide Association Study Identifies Major Loci Affecting the Immune Response against Infectious Bronchitis Virus in Chicken. Infect. Genet. Evol. 2014, 21, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Walugembe, M.; Mushi, J.R.; Amuzu-Aweh, E.N.; Chiwanga, G.H.; Msoffe, P.L.; Wang, Y.; Saelao, P.; Kelly, T.; Gallardo, R.A.; Zhou, H.; et al. Genetic Analyses of Tanzanian Local Chicken Ecotypes Challenged with Newcastle Disease Virus. Genes 2019, 10, 546. [Google Scholar] [CrossRef]
- Ali, A.; Ponnampalam, E.N.; Pushpakumara, G.; Cottrell, J.J.; Suleria, H.A.R.; Dunshea, F.R. Cinnamon: A Natural Feed Additive for Poultry Health and Production—A Review. Animals 2021, 11, 2026. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.C.; Garcia Junior, A.A.P.; Gromboni, J.G.G.; Filho, R.V.F.; do Nascimento, C.S.; Wenceslau, A.A. Influence of Heat Stress, Sex and Genetic Groups on Reference Genes Stability in Muscle Tissue of Chicken. PLoS ONE 2017, 12, e0176402. [Google Scholar] [CrossRef]
- Al-Zghoul, M.B.; Saleh, K.M.; Ababneh, M.M.K. Effects of Pre-Hatch Thermal Manipulation and Post-Hatch Acute Heat Stress on the mRNA Expression of Interleukin-6 and Genes Involved in Its Induction Pathways in 2 Broiler Chicken Breeds. Poult. Sci. 2019, 98, 1805–1819. [Google Scholar] [CrossRef]
- Galal, A.; Radwan, L.M.; Rezik, H.H.; Ayoub, H. Expression Levels of HSP70 and CPT-1 in Three Local Breeds of Chickens Reared under Normal or Heat Stress Conditions after the Introduction of the Naked Neck Gene. J. Therm. Biol. 2019, 80, 113–118. [Google Scholar] [CrossRef]
- Radwan, L.M. Genetic Improvement of Egg Laying Traits in Fayoumi Chickens Bred under Conditions of Heat Stress through Selection and Gene Expression Studies. J. Therm. Biol. 2020, 89, 102546. [Google Scholar] [CrossRef]
- Irivboje, Y.I.; Sanni, M.T.; Fafiolu, A.O.; Olowofeso, O.; Ikeobi, C.O.N. Genetic Polymorphisms in Part of Intron 7 and Exon 8 of HSP90AA1 Gene and Its Association with Heat Tolerance Traits in Two Exotic Layer Chicken Strains. Trop. Anim. Health Prod. 2020, 52, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Ncho, C.M.; Choi, Y.-H. Regulation of Gene Expression in Chickens by Heat Stress. Anim. Sci. Biotechnol. 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Adu-Asiamah, P.; Zhang, Y.; Amoah, K.; Leng, Q.Y.; Zheng, J.H.; Yang, H.; Zhang, W.L.; Zhang, L. Evaluation of Physiological and Molecular Responses to Acute Heat Stress in Two Chicken Breeds. Animal 2021, 15, 100106. [Google Scholar] [CrossRef]
- Fedorova, E.S.; Dementieva, N.V.; Shcherbakov, Y.S.; Stanishevskaya, O.I. Identification of Key Candidate Genes in Runs of Homozygosity of the Genome of Two Chicken Breeds, Associated with Cold Adaptation. Biology 2022, 11, 547. [Google Scholar] [CrossRef]
- Xu, N.-Y.; Si, W.; Li, M.; Gong, M.; Larivière, J.-M.; Nanaei, H.A.; Bian, P.-P.; Jiang, Y.; Zhao, X. Genome-Wide Scan for Selective Footprints and Genes Related to Cold Tolerance in Chantecler Chickens. Zool. Res. 2021, 42, 710–720. [Google Scholar] [CrossRef]
- Yevshin, I.S.; Shagimardanova, E.I.; Ryabova, A.S.; Pintus, S.S.; Kolpakov, F.A.; Gusev, O.A. Genome of Russian Snow-White Chicken Reveals Genetic Features Associated with Adaptations to Cold and Diseases. Int. J. Mol. Sci. 2024, 25, 11066. [Google Scholar] [CrossRef]
- Romanov, M.N.; Abdelmanova, A.S.; Fisinin, V.I.; Gladyr, E.A.; Volkova, N.A.; Koshkina, O.A.; Rodionov, A.N.; Vetokh, A.N.; Gusev, I.V.; Anshakov, D.V.; et al. Selective Footprints and Genes Relevant to Cold Adaptation and Other Phenotypic Traits Are Unscrambled in the Genomes of Divergently Selected Chicken Breeds. J. Anim. Sci. Biotechnol. 2023, 14, 35. [Google Scholar] [CrossRef]
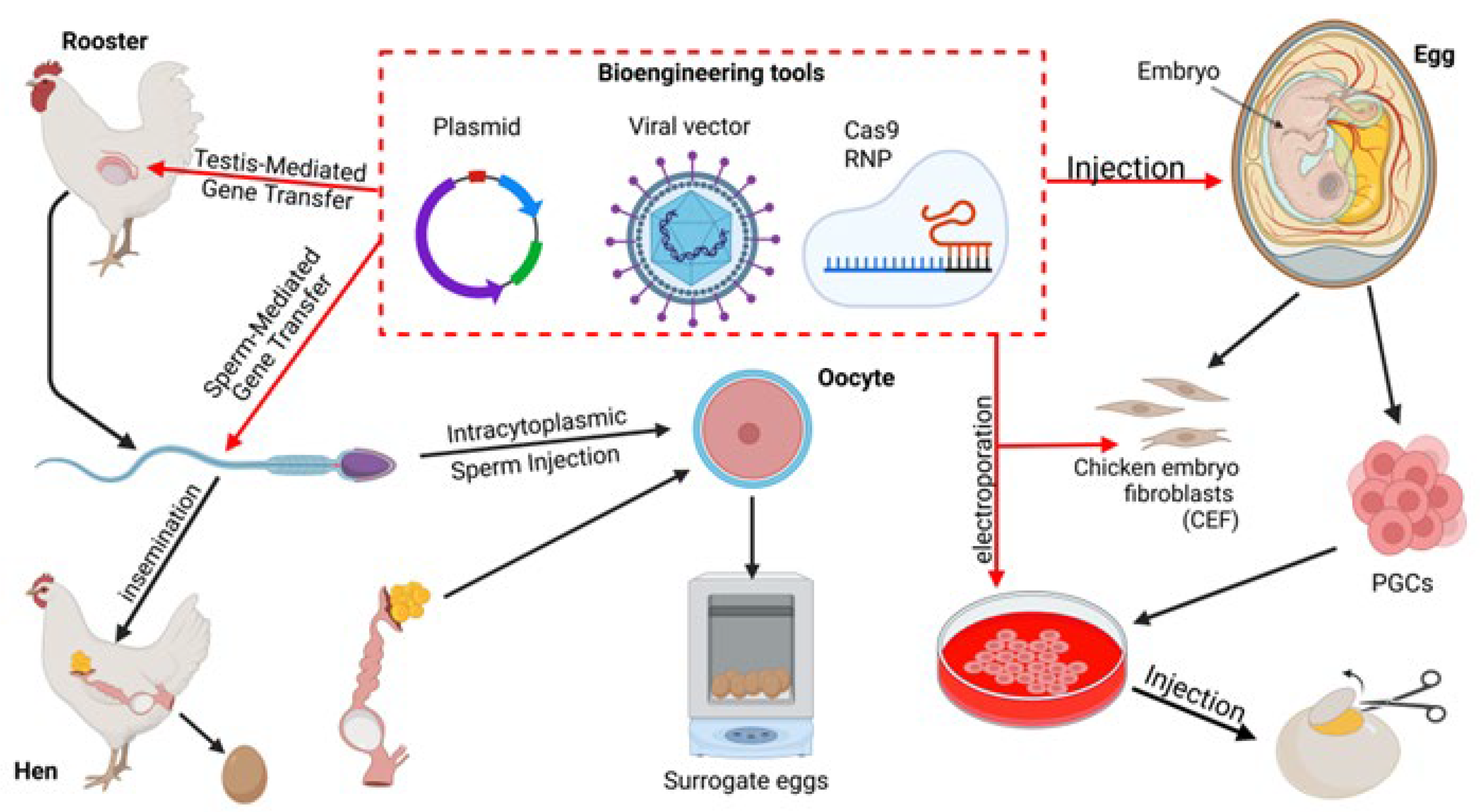
- Sid, H.; Schusser, B. Applications of Gene Editing in Chickens: A New Era Is on the Horizon. Front. Genet. 2018, 9, 456. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kagami, H.; Tagami, T. Development, Differentiation and Manipulation of Chicken Germ Cells. Dev. Growth Differ. 2013, 55, 20–40. [Google Scholar] [CrossRef]
- Collarini, E.J.; Leighton, P.A.; Van de Lavoir, M.-C. Production of Transgenic Chickens Using Cultured Primordial Germ Cells and Gonocytes. Methods Mol. Biol. 2019, 1874, 403–430. [Google Scholar] [CrossRef]
- Mathan; Zaib, G.; Jin, K.; Zuo, Q.; Habib, M.; Zhang, Y.; Li, B. Formation, Application, and Significance of Chicken Primordial Germ Cells: A Review. Animals 2023, 13, 1096. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y. Germ Cells and Transgenesis in Chickens. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 61–80. [Google Scholar] [CrossRef]
- Dehdilani, N.; Taemeh, S.Y.; Goshayeshi, L.; Dehghani, H. Genetically Engineered Birds; Pre-CRISPR and CRISPR Era†. Biol. Reprod. 2022, 106, 24–46. [Google Scholar] [CrossRef]
- Kalina, J.; Senigl, F.; Micáková, A.; Mucksová, J.; Blazková, J.; Yan, H.; Poplstein, M.; Hejnar, J.; Trefil, P. Retrovirus-Mediated in Vitro Gene Transfer into Chicken Male Germ Line Cells. Reproduction 2007, 134, 445–453. [Google Scholar] [CrossRef]
- Mizushima, S.; Sasanami, T.; Ono, T.; Kuroiwa, A. Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing. Genes 2023, 14, 757. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.; Han, J.H.; Lee, H.J.; Ruud, I.; Kim, T.H. Targeted Modulation of Chicken Genes In Vitro Using CRISPRa and CRISPRi Toolkit. Genes 2023, 14, 906. [Google Scholar] [CrossRef]
- Schaefer-Klein, J.; Givol, I.; Barsov, E.V.; Whitcomb, J.M.; VanBrocklin, M.; Foster, D.N.; Federspiel, M.J.; Hughes, S.H. The EV-O-Derived Cell Line DF-1 Supports the Efficient Replication of Avian Leukosis-Sarcoma Viruses and Vectors. Virology 1998, 248, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Miao, A.; Wu, S.; Du, X.; Gao, F. Genetically Modified Chickens as Bioreactors for Protein-Based Drugs. Front. Genome Ed. 2024, 6, 1522837. [Google Scholar] [CrossRef]
- Lee, H.J.; Yoon, J.W.; Jung, K.M.; Kim, Y.M.; Park, J.S.; Lee, K.Y.; Park, K.J.; Hwang, Y.S.; Park, Y.H.; Rengaraj, D.; et al. Targeted Gene Insertion into Z Chromosome of Chicken Primordial Germ Cells for Avian Sexing Model Development. FASEB J. 2019, 33, 8519–8529. [Google Scholar] [CrossRef]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.-S.; Han, J.Y. Targeted Gene Knockout in Chickens Mediated by TALENs. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [PubMed]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted Mutagenesis in Chicken Using CRISPR/Cas9 System. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef]
- Chojnacka-Puchta, L.; Sawicka, D. CRISPR/Cas9 Gene Editing in a Chicken Model: Current Approaches and Applications. J. Appl. Genet. 2020, 61, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Lee, K.Y.; Han, J.Y. Precise Genome Editing in Poultry and Its Application to Industries. Genes 2020, 11, 1182. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.-H.; Gim, G.-M.; Yum, S.-Y.; Jang, G. Current Status and Future of Gene Engineering in Livestock. BMB Rep. 2024, 57, 50–59. [Google Scholar] [CrossRef]
- Cooper, C.A.; Doran, T.J.; Challagulla, A.; Tizard, M.L.V.; Jenkins, K.A. Innovative Approaches to Genome Editing in Avian Species. J. Anim. Sci. Biotechnol. 2018, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Mukae, T.; Yoshii, K.; Watanobe, T.; Tagami, T.; Oishi, I. Production and Characterization of Eggs from Hens with Ovomucoid Gene Mutation. Poult. Sci. 2021, 100, 452–460. [Google Scholar] [CrossRef]
- Kim, G.-D.; Lee, J.H.; Song, S.; Kim, S.W.; Han, J.S.; Shin, S.P.; Park, B.-C.; Park, T.S. Generation of Myostatin-Knockout Chickens Mediated by D10A-Cas9 Nickase. FASEB J. 2020, 34, 5688–5696. [Google Scholar] [CrossRef]
- Park, T.S.; Park, J.; Lee, J.H.; Park, J.-W.; Park, B.-C. Disruption of G0/G1 Switch Gene 2 (G0S2) Reduced Abdominal Fat Deposition and Altered Fatty Acid Composition in Chicken. FASEB J. 2019, 33, 1188–1198. [Google Scholar] [CrossRef]
- Koslová, A.; Trefil, P.; Mucksová, J.; Reinišová, M.; Plachý, J.; Kalina, J.; Kučerová, D.; Geryk, J.; Krchlíková, V.; Lejčková, B.; et al. Precise CRISPR/Cas9 Editing of the NHE1 Gene Renders Chickens Resistant to the J Subgroup of Avian Leukosis Virus. Proc. Natl. Acad. Sci. USA 2020, 117, 2108–2112. [Google Scholar] [CrossRef]
- Idoko-Akoh, A.; Goldhill, D.H.; Sheppard, C.M.; Bialy, D.; Quantrill, J.L.; Sukhova, K.; Brown, J.C.; Richardson, S.; Campbell, C.; Taylor, L.; et al. Creating Resistance to Avian Influenza Infection through Genome Editing of the ANP32 Gene Family. Nat. Commun. 2023, 14, 6136. [Google Scholar] [CrossRef]
- Zuo, Q.; Wang, Y.; Cheng, S.; Lian, C.; Tang, B.; Wang, F.; Lu, Z.; Ji, Y.; Zhao, R.; Zhang, W.; et al. Site-Directed Genome Knockout in Chicken Cell Line and Embryos Can Use CRISPR/Cas Gene Editing Technology. G3 2016, 6, 1787–1792. [Google Scholar] [CrossRef]
- Cooper, C.A.; Challagulla, A.; Jenkins, K.A.; Wise, T.G.; O’Neil, T.E.; Morris, K.R.; Tizard, M.L.; Doran, T.J. Generation of Gene Edited Birds in One Generation Using Sperm Transfection Assisted Gene Editing (STAGE). Transgenic Res. 2017, 26, 331–347. [Google Scholar] [CrossRef]
| Trait | SNP ID | RefSNP | Gene | Genomic Location | Alleles | p-Value | Reference |
|---|---|---|---|---|---|---|---|
| residual feed intake (RFI) | gga27299173 | rs794348453 | ENSGALG00000005551 (DKK3) | chr5:8134988 | T/A | 8.62 × 10−7 | [43] |
| gga19252996 | rs735238610 | ENSGALG00010025920 (GTSF1) | chr27:1220239 | A/G | 1.55 × 10−7 | [47] | |
| gga10101246 | rs314351418 | ENSGALG00000004834 (COPS3) | chr14:4782740 | A/G | 4.53 × 10−7 | [47] | |
| gga10101220 | rs741733192 | ENSGALG00010017495 (COPS3) | chr14:4782376 | A/G | 2.33 × 10−7 | [47] | |
| feed intake (FI) | gga13915072 | rs14175506 | ENSGALG00010019808 | chr2:45414140 | T/C | 1.24 × 10−5 | [43] |
| gga40794011 | - | - | chr5:6549732 | C/T | 4.17 × 10−5 | [48] | |
| gga27235132 | rs315684001 | ENSGALG00010024055 | chr5:6628773 | A/G | 9.12 × 10−5 | [48] | |
| gga28669902 | rs15718055 | ENSGALG00010016935 | chr5:45362683 | C/A | 7.24 × 10−5 | [48] | |
| gga11900846 | rs313913143 | ENSGALG00010029371 | chr19:1878406 | A/C | 2.45 × 10−5 | [48] | |
| gga19012391 | rs15467593 | ENSGALG00000038948 (KDM5B) | chr26:539371 | C/T | 3.16 × 10−5 | [48] | |
| gga16547347 | rs15150566 | - | chr2:122907241 | A/G | 1.1 × 10−5 | [5] | |
| feed conversion ratio (FCR) | gga26455105 | rs738402168 | ENSGALG00000041854 | chr4:76507176 | A/C | 9.77 × 10−42 | [49] |
| gga26459075 | rs80762609 | ENSGALG00010011096 | chr4:76588703 | A/G | 8.52 × 10−14 | [49] | |
| gga26453154 | rs316119236 | ENSGALG00000014421 (LCORL) | chr4:76440302 | A/T | 6.62 × 10−21 | [49] | |
| gga26456559 | rs740878900 | ENSGALG00010010997 | chr4:76541002 | C/G | 6.51 × 10−21 | [49] | |
| gga26457701 | rs14739738 | ENSGALG00000030070 (QDPR) | chr4:76563623 | A/G | 4.24 × 10−15 | [49] | |
| gga26420206 | rs740848166 | ENSGALG00010010298 | chr4:75224686 | A/T | 3.7 × 10−13 | [49] | |
| gga26463849 | rs80564409 | ENSGALG00000014485 (LDB2) | chr4:76748107 | G/A | 3.74 × 10−16 | [49] | |
| gga8697511 | rs313167401 | ENSGALG00010028273 | chr12:5990423 | T/G | 3.74 × 10−10 | [49] | |
| gga26455535 | rs738469542 | ENSGALG00010010979 | chr4:76516142 | A/G | 3.5 × 10−43 | [49] | |
| gga26408980 | rs316896826 | - | chr4:74916879 | T/A | 3.5 × 10−19 | [49] | |
| gga26432816 | rs16436155 | ENSGALG00000041121 (SLIT2) | chr4:75683233 | T/G | 3.3 × 10−15 | [49] | |
| gga26470496 | rs316070373 | ENSGALG00000014485 (LDB2) | chr4:76923106 | C/T | 2.85 × 10−13 | [49] | |
| gga26408556 | rs738916134 | ENSGALG00010010027 | chr4:74884976 | C/A | 2.34 × 10−10 | [49] | |
| gga26456458 | - | ENSGALG00000006334 (LAP3) | chr4:76538559 | C/T | 2.21 × 10−13 | [49] | |
| egg number | gga40794121 | - | - | chr5:48969832 | A/ | 1.4 × 10−18 | [38] |
| gga40794155 | - | ENSGALG00000037911 | chr5:48997447 | T/ | 5.54 × 10−13 | [38] | |
| gga40794267 | - | ENSGALG00000037911 | chr5:49015358 | T/ | 2.48 × 10−12 | [38] | |
| gga40794301 | - | ENSGALG00000011244 (DLK1) | chr5:49016281 | G/ | 5.76 × 10−12 | [38] | |
| gga40794078 | - | ENSGALG00000042132 (SUGP1) | chr28:3540022 | T/ | 1.4 × 10−10 | [38] | |
| gga40794259 | - | ENSGALG00000037911 | chr5:48988619 | C/ | 9.02 × 10−10 | [38] | |
| gga40794420 | - | ENSGALG00000021598 | chr21:5262861 | G/ | 1.55 × 10−9 | [38] | |
| egg shell thickness | gga19003886 | rs736368342 | ENSGALG00000000606 (ARL8A) | chr26:348743 | A/G | 3.76 × 10−7 | [22] |
| gga19003889 | rs735278795 | ENSGALG00000000606 (ARL8A) | chr26:348810 | G/A | 3.76 × 10−7 | [22] | |
| gga5865222 | rs13968878 | ENSGALG00000016967 (ENOX1) | chr1:166941530 | G/A | 2.81 × 10−8 | [32] | |
| gga19003886 | rs736368342 | ENSGALG00000000606 (ARL8A) | chr26:348743 | A/G | 3.76 × 10−7 | [22] | |
| egg shell weight | gga15205102 | rs13636444 | ENSGALG00010008389 (GALNT1) | chr2:84108965 | G/A | 5.85 × 10−9 | [32] |
| gga23503286 | rs14411624 | ENSGALG00010011743 | chr3:107750850 | C/T | 1.41 × 10−7 | [32] | |
| gga8069820 | rs14022717 | ENSGALG00010024544 | chr11:9050275 | G/T, A | 8.62 × 10−7 | [32] | |
| age at first egg | gga9401651 | rs318240317 | ENSGALG00000001768 (TENM2) | chr13:5042404 | T/C | 1.42 × 10−6 | [32] |
| egg shell percentage | gga11229394 | rs793955278 | ENSGALG00010027632 | chr17:6527642 | C/A | 2.98 × 10−7 | [22] |
| gga14284065 | rs317955040 | - | chr2:57645767 | C/T | 5.53 × 10−10 | [22] | |
| gga20208557 | rs793960563 | ENSGALT00010040903 (SLC8A1) | chr3:16388607 | A/G | 1.52 × 10−9 | [22] | |
| gga20618399 | rs15305641 | - | chr3:27338039 | G/A | 1.43 × 10−8 | [22] | |
| gga2247149 | rs14834812 | ENSGALG00000013037 (BCL2L13) | chr1:61905467 | T/C | 1.74 × 10−7 | [22] | |
| gga7655209 | rs740613354 | ENSGALG00010018848 (MEF2A) | chr10:17024031 | T/G | 5.56 × 10−8 | [22] | |
| body weight at first egg | gga26454149 | rs16023603 | ENSGALG00000041854 | chr4:76488977 | G/T | 9.75 × 10−8 | [50] |
| egg shell color | gga10971513 | rs731126327 | ENSGALG00000037735 (CENPA) | chr16:58557 | C/G | 2.57 × 10−9 | [22] |
| gga18068242 | rs315477097 | ENSGALP00000043161 | chr21:803620 | G/C | 3.83 × 10−8 | [51] | |
| gga17077724 | rs15168063 | ENSGALP00000045123 | chr2:137478073 | T/A | 4.63 × 10−8 | [22] | |
| gga11908141 | rs14117102 | ENSGALG00010029870 | chr19:2048452 | T/C | 2.01 × 10−8 | [22] | |
| gga36013348 | rs793971423 | ENSGALG00010009886 | chrZ:73194176 | C/T | 8.98 × 10−9 | [22] | |
| gga6512746 | rs315232554 | ENSGALP00000051533 | chr1:183114617 | T/C | 1.5 × 10−9 | [22] | |
| gga18068999 | rs313199923 | ENSGALG00010019630 | chr21:822838 | T/C | 3.29 × 10−6 | [51] | |
| gga13707557 | rs316634461 | ENSGALP00000030515 | chr2:40713376 | G/A | 2.77 × 10−6 | [51] | |
| gga18072343 | rs16177221 | ENSGALG00000000978 (CEP104) | chr21:912580 | G/A | 2.84 × 10−6 | [51] | |
| breast muscle weight | gga19367973 | - | ENSGALG00000041204 (IGF2BP1) | chr27:3929034 | A/C | 3.09 × 10−8 | [52] |
| gga19367739 | rs741713216 | ENSGALG00000041204 (IGF2BP1) | chr27:3923534 | T/C | 1.5 × 10−8 | [52] | |
| gga19360817 | rs733674119 | ENSGALG00000001315 (UBE2Z) | chr27:3696784 | T/C | 1.89 × 10−8 | [52] | |
| gga19361902 | rs740150938 | ENSGALG00000041204 (IGF2BP1) | chr27:3727891 | A/T | 1.38 × 10−8 | [52] | |
| gga19366244 | rs739298135 | ENSGALG00000041204 (IGF2BP1) | chr27:3883310 | A/G | 6.21 × 10−9 | [52] | |
| gga19360904 | rs14303799 | ENSGALG00000041204 (IGF2BP1) | chr27:3699719 | T/C | 5.17 × 10−9 | [52] | |
| gga19361431 | rs737533546 | ENSGALG00000001525 (CALCOCO2) | chr27:3713052 | C/T | 4.16 × 10−9 | [52] | |
| gga19361079 | rs736156149 | ENSGALG00000001330 (ATP5G1) | chr27:3705603 | G/A | 2.39 × 10−9 | [52] | |
| gga26142808 | rs313870616 | ENSGALG00010017374 | chr4:66917344 | T/C | 2.79 × 10−11 | [53] | |
| body weight | gga2548108 | rs315749637 | - | chr1:69658667 | G/A | 9.86 × 10−9 | [53] |
| gga26144149 | rs315209025 | ENSGALG00000014124 (TEC) | chr4:66946846 | C/A | 1.38 × 10−10 | [53] | |
| gga26144434 | rs315209025 | ENSGALG00000014124 (TEC) | chr4:66952728 | C/A | 3.67 × 10−12 | [53] | |
| gga32875195 | rs313957679 | ENSGALG00000010543 (EPS15) | chr8:24178278 | G/A | 6.09 × 10−9 | [53] | |
| gga33041294 | rs14657331 | ENSGALG00010024151 (LEPROT) | chr8:28421453 | C/T | 2.64 × 10−14 | [53] | |
| gga33080862 | rs312436211 | ENSGALG00000011350 (NEGR1) | chr8:29394332 | C/T | 9.16 × 10−11 | [53] | |
| gga10122855 | rs14073523 | ENSGALG00000005215 (CACNA1H) | chr14:5337950 | A/G | 3.8 × 10−5 | [48] | |
| body weight gain | gga11364204 | rs312843163 | ENSGALG00000001094 (ADGRD2) | chr17:9904101 | A/G | 5.13 × 10−6 | [48] |
| gga16591237 | rs15151359 | - | chr2:124071457 | G/A | 3.72 × 10−5 | [48] | |
| gga28701462 | rs316866456 | ENSGALG00010018389 | chr5:46165348 | C/T | 8.13 × 10−5 | [48] | |
| gga26455105 | rs738402168 | ENSGALG00000041854 | chr4:76507176 | A/C | 1.39 × 10−62 | [49] | |
| average daily gain | gga26454915 | rs315397301 | ENSGALG00000041854 | chr4:76503015 | T/C | 2.21 × 10−60 | [49] |
| gga26408556 | rs738916134 | ENSGALG00000040208 | chr4:74884976 | C/A | 4.86 × 10−31 | [49] | |
| gga26450824 | rs80691090 | - | chr4:76367855 | G/C | 2.75 × 10−29 | [49] | |
| gga26459318 | rs16756269 | ENSGALG00010011096 | chr4:76593451 | G/A | 2.98 × 10−29 | [49] | |
| gga26455535 | rs738469542 | ENSGALG00010010979 | chr4:76516142 | A/G | 5.11 × 10−28 | [49] | |
| gga26430969 | rs315846457 | ENSGALG00000041121 (SLIT2) | chr4:75622029 | A/G | 1.25 × 10−25 | [49] | |
| gga11356458 | rs316227600 | ENSGALG00000001157 (DENND1A) | chr17:9636765 | T/C | 2.21 × 10−6 | [54] |
| Trait | SNP ID | RefSNP | Gene | Genomic Location | Alleles | p-Value | Reference |
|---|---|---|---|---|---|---|---|
| salmonella enterica serovan gallinarum antibody titre | gga11356458 | rs316227600 | ENSGALG00000001157 (DENND1A) | chr17:9636765 | T/C | 2.21 × 10−6 | [54] |
| gga14606249 | rs313247175 | ENSGALG00000027339 (LYRM4) | chr2:65707454 | T/C | 5.9 × 10−5 | [135] | |
| gga11356599 | - | ENSGALG00000001157 | chr17:9642868-9642868 | A/C | 1.21 × 10−6 | [54] | |
| avian influenza virus antibody titer | gga29091675 | rs14554319 | ENSGALG00000012137 (KTN1) | chr5:56259597 | T/C | 5.44 × 10−5 | [135] |
| gga28738351 | rs16505398 | - | chr5:47167537 | C/T | 2.69 × 10−14 | [136] | |
| antibody level against infectious bronchitis virus | gga4586819 | rs13623466 | ENSGALG00000016681 (DHRSX) | chr1:128713658 | C/T | 9.98 × 10−14 | [136] |
| gga32379046 | rs314472262 | ENSGALG00000004620 (LAMC1) | chr8:7544464 | T/C | 1.8 × 10−13 | [136] | |
| gga4604674 | rs313566132 | ENSGALG00000016689 (ASMTL) | chr1:129172183 | C/T | 2.19 × 10−13 | [136] | |
| gga11638399 | rs14112036 | ENSGALG00000003105 (ANKFN1) | chr18:6199395 | T/C | 7.22 × 10−6 | [54] | |
| mareks disease virus antibody titre | gga21222152 | rs14346868 | ENSGALG00000011473 (RPS6KA2) | chr3:42937011 | T/C | 1.05 × 10−6 | [54] |
| gga18354446 | rs15998498 | ENSGALG00000001608 (UNC5D) | chr22:1854894 | C/T | 6.75 × 10−6 | [137] | |
| pre-infection growth rate | gga21954944 | rs317939411 | ENSGALG00000014902 | chr3:63366122 | C/T | 5.42 × 10−6 | [137] |
| post-infection growth rate | gga11889533 | rs314290710 | ENSGALG00000001153 (AUTS2) | chr19:1607256 | G/T | 3.65 × 10−6 | [137] |
| immune response to newcastle disease | gga1757454 | rs314284996 | ENSGALG00000011894 (F1NJG4) | chr1:49441152 | T/C | 1.55 × 10−7 | PMID 34745207 |
| gga1822646 | rs737774287 | ENSGALG00000012291 (POLR2F) | chr1:51104995 | T/C | 4.0 × 10−10 | PMID 34745207 | |
| gga40793944 | - | ENSGALG00000041823 | chr1:51056044 | A/C | 6.42 × 10−8 | PMID 34745207 | |
| resistance to cestodes parasitism | gga31494288 | - | ENSGALG00000011318.8 (DNPEP) | chr7:22288125 | T/C | 1.8 × 10−11 | [54] |
| gga10144957 | - | ENSGALG00000044187 (LMF1) | chr14:5838224 | T/C | 1.06 × 10−8 | [54] | |
| resistance to eimeria parasitism | gga21878395 | - | ENSGALG00000014848 (TRDN) | chr3:61008720 | C/T | 8.09 × 10−7 | [54] |
| gga40793947 | - | ENSGALG00000030397 | chr16:161178-161178 | C/A | 1.0 × 10−5 | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilyazova, I.; Korytina, G.; Kochetova, O.; Savelieva, O.; Mikhaylova, E.; Vershinina, Z.; Chumakova, A.; Markelov, V.; Abdeeva, G.; Karunas, A.; et al. Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions. Int. J. Mol. Sci. 2025, 26, 8285. https://doi.org/10.3390/ijms26178285
Gilyazova I, Korytina G, Kochetova O, Savelieva O, Mikhaylova E, Vershinina Z, Chumakova A, Markelov V, Abdeeva G, Karunas A, et al. Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions. International Journal of Molecular Sciences. 2025; 26(17):8285. https://doi.org/10.3390/ijms26178285
Chicago/Turabian StyleGilyazova, Irina, Gulnaz Korytina, Olga Kochetova, Olga Savelieva, Elena Mikhaylova, Zilya Vershinina, Anna Chumakova, Vitaliy Markelov, Gulshat Abdeeva, Alexandra Karunas, and et al. 2025. "Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions" International Journal of Molecular Sciences 26, no. 17: 8285. https://doi.org/10.3390/ijms26178285
APA StyleGilyazova, I., Korytina, G., Kochetova, O., Savelieva, O., Mikhaylova, E., Vershinina, Z., Chumakova, A., Markelov, V., Abdeeva, G., Karunas, A., Khusnutdinova, E., & Gusev, O. (2025). Advances in Genomics and Postgenomics in Poultry Science: Current Achievements and Future Directions. International Journal of Molecular Sciences, 26(17), 8285. https://doi.org/10.3390/ijms26178285






