In Silico Identification of circPIM1/miR-16-5p/miR-195-5p/PIM1 Feed-Forward Loop in Recurrent Grade 2 Meningioma
Abstract
1. Introduction
2. Results
2.1. Identification of Candidate miRNAs Regulating the Expression of 34HR-MNG Transcripts
2.2. Identification of circRNAs Sponging Candidate MR-miRNAs
2.3. PIM1 Is Upregulated in Recurrent WHO Grade 2 MNGs
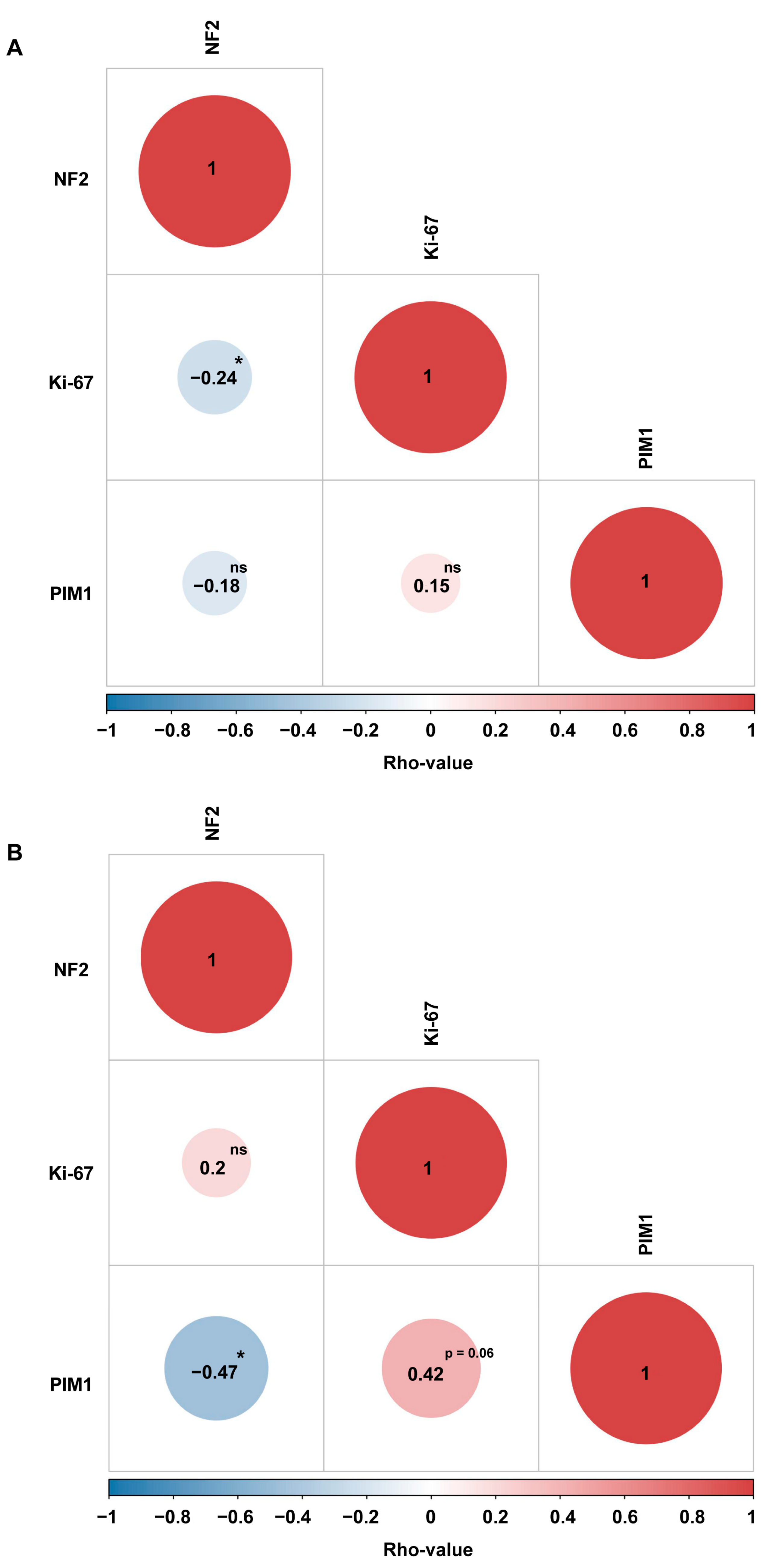
2.4. PIM1 Upregulation Is Associated with the Most Aggressive Molecular MNG Phenotype
2.5. MR-CircRNAs 0076215 and 0076216 Positively Correlates with PIM1
2.6. MR-miRNAs and PIM1 Are Expressed in Meningeal Tissue
3. Discussion
4. Materials and Methods
4.1. Identification of Candidate miRNAs Involved in MNG Recurrence
4.2. Identification of Candidate circRNAs Involved in MNG Recurrence
4.3. Correlation Between MR- circRNA Host Gene Expression and MNG Clinical and Molecular Data
4.4. Tissue and Single-Cell Expression Analysis
4.5. qRT-PCR and ceRNA Network
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016–2020. Neuro Oncol. 2023, 25, iv1–iv99. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Youngblood, M.W.; Duran, D.; Montejo, J.D.; Li, C.; Omay, S.B.; Özduman, K.; Sheth, A.H.; Zhao, A.Y.; Tyrtova, E.; Miyagishima, D.F.; et al. Correlations between Genomic Subgroup and Clinical Features in a Cohort of More than 3000 Meningiomas. J. Neurosurg. 2020, 133, 1345–1354. [Google Scholar] [CrossRef]
- Beylerli, O.; Ilyasova, T.; Shi, H.; Sufianov, A. MicroRNAs in Meningiomas: Potential Biomarkers and Therapeutic Targets. Noncoding RNA Res. 2024, 9, 641–648. [Google Scholar] [CrossRef]
- Wang, J.Z.; Nassiri, F.; Aldape, K.; von Deimling, A.; Sahm, F. The Epigenetic Landscape of Meningiomas. Adv. Exp. Med. Biol. 2023, 1416, 175–188. [Google Scholar] [CrossRef]
- Cirnigliaro, M.; Barbagallo, C.; Gulisano, M.; Domini, C.N.; Barone, R.; Barbagallo, D.; Ragusa, M.; Di Pietro, C.; Rizzo, R.; Purrello, M. Expression and Regulatory Network Analysis of MiR-140-3p, a New Potential Serum Biomarker for Autism Spectrum Disorder. Front. Mol. Neurosci. 2017, 10, 250. [Google Scholar] [CrossRef]
- Slavik, H.; Balik, V.; Vrbkova, J.; Rehulkova, A.; Vaverka, M.; Hrabalek, L.; Ehrmann, J.; Vidlarova, M.; Gurska, S.; Hajduch, M.; et al. Identification of Meningioma Patients at High Risk of Tumor Recurrence Using MicroRNA Profiling. Neurosurgery 2020, 87, 1055–1063. [Google Scholar] [CrossRef]
- Urbschat, S.; Landau, B.; Bewersdorf, N.-C.; Schuster, C.; Wagenpfeil, G.; Schulz-Schaeffer, W.J.; Oertel, J.; Ketter, R. MicroRNA 200a as a Histologically Independent Marker for Meningioma Recurrence: Results of a Four MicroRNA Panel Analysis in Meningiomas. Cancer Med. 2023, 12, 8433–8444. [Google Scholar] [CrossRef]
- Wang, M.; Deng, X.; Ying, Q.; Jin, T.; Li, M.; Liang, C. MicroRNA-224 Targets ERG2 and Contributes to Malignant Progressions of Meningioma. Biochem. Biophys. Res. Commun. 2015, 460, 354–361. [Google Scholar] [CrossRef]
- Hergalant, S.; Casse, J.-M.; Oussalah, A.; Houlgatte, R.; Helle, D.; Rech, F.; Vallar, L.; Guéant, J.-L.; Vignaud, J.-M.; Battaglia-Hsu, S.-F.; et al. MicroRNAs MiR-16 and MiR-519 Control Meningioma Cell Proliferation via Overlapping Transcriptomic Programs Shared with the RNA-Binding Protein HuR. Front. Oncol. 2023, 13, 1158773. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhong, Y.; Pan, S.; Zhi, S.; Li, Y.; Xiu, Z.; Wei, C.; Luo, J. Construction and Investigation of CircRNA-Associated CeRNA Regulatory Network in Molecular Subtypes of Breast Cancer. Curr. Comput. Aided Drug Des. 2022, 18, 185–195. [Google Scholar] [CrossRef]
- Barbagallo, D.; Palermo, C.I.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Spina, V.; Ragusa, M.; Di Pietro, C.; Scalia, G.; Purrello, M. Competing Endogenous RNA Network Mediated by Circ_3205 in SARS-CoV-2 Infected Cells. Cell Mol. Life Sci. 2022, 79, 75. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The Emerging Roles of CircRNAs in Cancer and Oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Merulla, A.E.; Stella, M.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Broggi, G.; Altieri, R.; Certo, F.; Caltabiano, R.; Ragusa, M.; et al. CircSMARCA5 Is an Upstream Regulator of the Expression of MiR-126-3p, MiR-515-5p, and Their MRNA Targets, Insulin-like Growth Factor Binding Protein 2 (IGFBP2) and NRAS Proto-Oncogene, GTPase (NRAS) in Glioblastoma. Int. J. Mol. Sci. 2022, 23, 13676. [Google Scholar] [CrossRef]
- Chen, K.; Huang, Z.; Liu, C.; Ouyang, Q.; Yan, Q.; Zheng, W.; Huang, Y. Hsa_circ_0004872 Mitigates Proliferation, Metastasis and Immune Escape of Meningioma Cells by Suppressing PD-L1. Metab. Brain Dis. 2024, 39, 895–907. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, Z.; Peng, Z.; Liu, A.; Yuan, W.; Han, D.; Peng, J. Hsa_circ_0004872 Alleviates Meningioma Progression by Sponging MiR-190a-3p/PTEN Signaling. BMC Cancer 2024, 24, 345. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, R.; Li, Q.; Li, Y.; Xuan, T.; Cao, S.; Zheng, J. SNHG1/MiR-556-5p/TCF12 Feedback Loop Enhances the Tumorigenesis of Meningioma through Wnt Signaling Pathway. J. Cell Biochem. 2020, 121, 1880–1889. [Google Scholar] [CrossRef]
- Zheng, J.; Pang, C.-H.; Du, W.; Wang, L.; Sun, L.-G.; Xing, Z.-Y. An Allele of Rs619586 Polymorphism in MALAT1 Alters the Invasiveness of Meningioma via Modulating the Expression of Collagen Type V Alpha (COL5A1). J. Cell Mol. Med. 2020, 24, 10223–10232. [Google Scholar] [CrossRef]
- Li, T.; Ren, J.; Ma, J.; Wu, J.; Zhang, R.; Yuan, H.; Han, X. LINC00702/MiR-4652-3p/ZEB1 Axis Promotes the Progression of Malignant Meningioma through Activating Wnt/β-Catenin Pathway. Biomed. Pharmacother. 2019, 113, 108718. [Google Scholar] [CrossRef]
- Ding, Y.; Ge, Y.; Wang, D.; Liu, Q.; Sun, S.; Hua, L.; Deng, J.; Luan, S.; Cheng, H.; Xie, Q.; et al. LncRNA-IMAT1 Promotes Invasion of Meningiomas by Suppressing KLF4/Hsa-MiR22-3p/Snai1 Pathway. Mol. Cells 2022, 45, 388–402. [Google Scholar] [CrossRef]
- Xing, H.; Wang, S.; Li, Q.; Ma, Y.; Sun, P. Long Noncoding RNA LINC00460 Targets MiR-539/MMP-9 to Promote Meningioma Progression and Metastasis. Biomed. Pharmacother. 2018, 105, 677–682. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Xu, J.; Huang, Z.; Bu, X.; Wang, D.; Ge, H.; Zhang, G.; Gu, J.; Kang, D.; et al. Long Non-Coding RNA MEG3 Modifies Cell-Cycle, Migration, Invasion, and Proliferation Through AKAP12 by Sponging MiR-29c in Meningioma Cells. Front. Oncol. 2020, 10, 537763. [Google Scholar] [CrossRef]
- Nyalundja, A.D.; Mugisha, F.; Karekezi, C. The Natural History and Treatment of Meningiomas: An Update. Semin. Neurol. 2024, 44, 1–15. [Google Scholar] [CrossRef]
- Huntoon, K.; Toland, A.M.S.; Dahiya, S. Meningioma: A Review of Clinicopathological and Molecular Aspects. Front. Oncol. 2020, 10, 579599. [Google Scholar] [CrossRef]
- Corniola, M.V.; Lemée, J.-M.; Meling, T.R. Histological Transformation in Recurrent WHO Grade I Meningiomas. Sci. Rep. 2020, 10, 11220. [Google Scholar] [CrossRef]
- Li, Z.; Gao, Y.; Zhang, J.; Han, L.; Zhao, H. DNA Methylation Meningioma Biomarkers: Attributes and Limitations. Front. Mol. Neurosci. 2023, 16, 1182759. [Google Scholar] [CrossRef]
- Biswas, D.; Halder, A.; Barpanda, A.; Ghosh, S.; Chauhan, A.; Bhat, L.; Epari, S.; Shetty, P.; Moiyadi, A.; Ball, G.R.; et al. Integrated Meta-Omics Analysis Unveils the Pathways Modulating Tumorigenesis and Proliferation in High-Grade Meningioma. Cells 2023, 12, 2483. [Google Scholar] [CrossRef]
- Nassiri, F.; Liu, J.; Patil, V.; Mamatjan, Y.; Wang, J.Z.; Hugh-White, R.; Macklin, A.M.; Khan, S.; Singh, O.; Karimi, S.; et al. A Clinically Applicable Integrative Molecular Classification of Meningiomas. Nature 2021, 597, 119–125. [Google Scholar] [CrossRef]
- Choudhury, A.; Magill, S.T.; Eaton, C.D.; Prager, B.C.; Chen, W.C.; Cady, M.A.; Seo, K.; Lucas, C.-H.G.; Casey-Clyde, T.J.; Vasudevan, H.N.; et al. Meningioma DNA Methylation Groups Identify Biological Drivers and Therapeutic Vulnerabilities. Nat. Genet. 2022, 54, 649–659. [Google Scholar] [CrossRef]
- Thirimanne, H.N.; Almiron-Bonnin, D.; Nuechterlein, N.; Arora, S.; Jensen, M.; Parada, C.A.; Qiu, C.; Szulzewsky, F.; English, C.W.; Chen, W.C.; et al. Meningioma Transcriptomic Landscape Demonstrates Novel Subtypes with Regional Associated Biology and Patient Outcome. Cell Genom. 2024, 4, 100566. [Google Scholar] [CrossRef]
- Bayley, J.C.; Hadley, C.C.; Harmanci, A.O.; Harmanci, A.S.; Klisch, T.J.; Patel, A.J. Multiple Approaches Converge on Three Biological Subtypes of Meningioma and Extract New Insights from Published Studies. Sci. Adv. 2022, 8, eabm6247. [Google Scholar] [CrossRef]
- Bender, L.; Somme, F.; Lhermitte, B.; Ahle, G.; Boone, M.; Blonski, M.; Pouget, C.; Truc, G.; Cebula, H.; Noël, G. High Risk of Recurrence for Grade II Meningioma: A 10-Year Multicenter Analysis of Prognosis Factors. Chin. Clin. Oncol. 2021, 10, 26. [Google Scholar] [CrossRef]
- Aghi, M.K.; Carter, B.S.; Cosgrove, G.R.; Ojemann, R.G.; Amin-Hanjani, S.; Martuza, R.L.; Curry, W.T.; Barker, F.G. Long-Term Recurrence Rates of Atypical Meningiomas after Gross Total Resection with or without Postoperative Adjuvant Radiation. Neurosurgery 2009, 64, 56–60; discussion 60. [Google Scholar] [CrossRef]
- Chen, W.C.; Choudhury, A.; Youngblood, M.W.; Polley, M.-Y.C.; Lucas, C.-H.G.; Mirchia, K.; Maas, S.L.N.; Suwala, A.K.; Won, M.; Bayley, J.C.; et al. Targeted Gene Expression Profiling Predicts Meningioma Outcomes and Radiotherapy Responses. Nat. Med. 2023, 29, 3067–3076. [Google Scholar] [CrossRef]
- Maleki, E.H.; Bahrami, A.R.; Matin, M.M. Cancer Cell Cycle Heterogeneity as a Critical Determinant of Therapeutic Resistance. Genes. Dis. 2024, 11, 189–204. [Google Scholar] [CrossRef]
- Li, P.-C.; Tu, M.-J.; Ho, P.Y.; Batra, N.; Tran, M.M.L.; Qiu, J.-X.; Wun, T.; Lara, P.N.; Hu, X.; Yu, A.-X.; et al. In Vivo Fermentation Production of Humanized Noncoding RNAs Carrying Payload MiRNAs for Targeted Anticancer Therapy. Theranostics 2021, 11, 4858–4871. [Google Scholar] [CrossRef]
- Cai, S.; Shi, C.-J.; Lu, J.-X.; Wang, Y.-P.; Yuan, T.; Wang, X.-P. MiR-124-3p Inhibits the Viability and Motility of Glioblastoma Multiforme by Targeting RhoG. Int. J. Mol. Med. 2021, 47, 69. [Google Scholar] [CrossRef]
- Lv, Z.; Yang, L. MiR-124 Inhibits the Growth of Glioblastoma through the Downregulation of SOS1. Mol. Med. Rep. 2013, 8, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.-H.; Wu, S.-Q.; Zhang, Y.-D. Downregulation of MiR-124 Promotes the Growth and Invasiveness of Glioblastoma Cells Involving Upregulation of PPP1R13L. Int. J. Mol. Med. 2013, 32, 101–107. [Google Scholar] [CrossRef]
- Qiao, W.; Guo, B.; Zhou, H.; Xu, W.; Chen, Y.; Liang, Y.; Dong, B. MiR-124 Suppresses Glioblastoma Growth and Potentiates Chemosensitivity by Inhibiting AURKA. Biochem. Biophys. Res. Commun. 2017, 486, 43–48. [Google Scholar] [CrossRef]
- Lin, J.; Wen, X.; Zhang, X.; Sun, X.; Yunzhi, L.; Peng, R.; Zhu, M.; Wang, M.; Zhang, Y.; Luo, W.; et al. MiR-135a-5p and MiR-124-3p Inhibit Malignancy of Glioblastoma by Downregulation of Syndecan Binding Protein. J. Biomed. Nanotechnol. 2018, 14, 1317–1329. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, Y. Circular RNA HIPK3 Promotes Glioma Progression by Binding to MiR-124-3p. Gene 2019, 690, 81–89. [Google Scholar] [CrossRef]
- Wei, J.; Wang, F.; Kong, L.-Y.; Xu, S.; Doucette, T.; Ferguson, S.D.; Yang, Y.; McEnery, K.; Jethwa, K.; Gjyshi, O.; et al. MiR-124 Inhibits STAT3 Signaling to Enhance T Cell-Mediated Immune Clearance of Glioma. Cancer Res. 2013, 73, 3913–3926. [Google Scholar] [CrossRef]
- Deng, D.; Wang, L.; Chen, Y.; Li, B.; Xue, L.; Shao, N.; Wang, Q.; Xia, X.; Yang, Y.; Zhi, F. MicroRNA-124-3p Regulates Cell Proliferation, Invasion, Apoptosis, and Bioenergetics by Targeting PIM1 in Astrocytoma. Cancer Sci. 2016, 107, 899–907. [Google Scholar] [CrossRef]
- Lv, X.-B.; Jiao, Y.; Qing, Y.; Hu, H.; Cui, X.; Lin, T.; Song, E.; Yu, F. MiR-124 Suppresses Multiple Steps of Breast Cancer Metastasis by Targeting a Cohort of pro-Metastatic Genes in Vitro. Chin. J. Cancer 2011, 30, 821–830. [Google Scholar] [CrossRef]
- Ando, T.; Yoshida, T.; Enomoto, S.; Asada, K.; Tatematsu, M.; Ichinose, M.; Sugiyama, T.; Ushijima, T. DNA Methylation of MicroRNA Genes in Gastric Mucosae of Gastric Cancer Patients: Its Possible Involvement in the Formation of Epigenetic Field Defect. Int. J. Cancer 2009, 124, 2367–2374. [Google Scholar] [CrossRef]
- Agirre, X.; Vilas-Zornoza, A.; Jiménez-Velasco, A.; Martin-Subero, J.I.; Cordeu, L.; Gárate, L.; San José-Eneriz, E.; Abizanda, G.; Rodríguez-Otero, P.; Fortes, P.; et al. Epigenetic Silencing of the Tumor Suppressor MicroRNA Hsa-MiR-124a Regulates CDK6 Expression and Confers a Poor Prognosis in Acute Lymphoblastic Leukemia. Cancer Res. 2009, 69, 4443–4453. [Google Scholar] [CrossRef]
- El-Gewely, M.R.; Andreassen, M.; Walquist, M.; Ursvik, A.; Knutsen, E.; Nystad, M.; Coucheron, D.H.; Myrmel, K.S.; Hennig, R.; Johansen, S.D. Differentially Expressed MicroRNAs in Meningiomas Grades I and II Suggest Shared Biomarkers with Malignant Tumors. Cancers 2016, 8, 31. [Google Scholar] [CrossRef]
- Ludwig, N.; Kim, Y.-J.; Mueller, S.C.; Backes, C.; Werner, T.V.; Galata, V.; Sartorius, E.; Bohle, R.M.; Keller, A.; Meese, E. Posttranscriptional Deregulation of Signaling Pathways in Meningioma Subtypes by Differential Expression of MiRNAs. Neuro Oncol. 2015, 17, 1250–1260. [Google Scholar] [CrossRef]
- Herzog, S.; Fink, M.A.; Weitmann, K.; Friedel, C.; Hadlich, S.; Langner, S.; Kindermann, K.; Holm, T.; Böhm, A.; Eskilsson, E.; et al. Pim1 Kinase Is Upregulated in Glioblastoma Multiforme and Mediates Tumor Cell Survival. Neuro Oncol. 2015, 17, 223–242. [Google Scholar] [CrossRef]
- Tursynbay, Y.; Zhang, J.; Li, Z.; Tokay, T.; Zhumadilov, Z.; Wu, D.; Xie, Y. Pim-1 Kinase as Cancer Drug Target: An Update. Biomed. Rep. 2016, 4, 140–146. [Google Scholar] [CrossRef]
- Torres-Ayuso, P.; Katerji, M.; Mehlich, D.; Lookingbill, S.A.; Sabbasani, V.R.; Liou, H.; Casillas, A.L.; Chauhan, S.S.; Serwa, R.; Rubin, M.R.; et al. PIM1 Targeted Degradation Prevents the Emergence of Chemoresistance in Prostate Cancer. Cell Chem. Biol. 2024, 31, 326–337.e11. [Google Scholar] [CrossRef]
- Lee, K.D.; DePowell, J.J.; Air, E.L.; Dwivedi, A.K.; Kendler, A.; McPherson, C.M. Atypical Meningiomas: Is Postoperative Radiotherapy Indicated? Neurosurg. Focus. 2013, 35, E15. [Google Scholar] [CrossRef]
- Ukai, R.; Wanibuchi, M.; Komatsu, K.; Kimura, Y.; Akiyama, Y.; Mikami, T.; Mikuni, N. Recurrence Interval Within 1 Year Leads to Death in Patients with Grade 2 Meningioma. World Neurosurg. 2020, 142, e58–e65. [Google Scholar] [CrossRef]
- Youngblood, M.W.; Miyagishima, D.F.; Jin, L.; Gupte, T.; Li, C.; Duran, D.; Montejo, J.D.; Zhao, A.; Sheth, A.; Tyrtova, E.; et al. Associations of Meningioma Molecular Subgroup and Tumor Recurrence. Neuro Oncol. 2021, 23, 783–794. [Google Scholar] [CrossRef]
- DeSisto, J.; O’Rourke, R.; Jones, H.E.; Pawlikowski, B.; Malek, A.D.; Bonney, S.; Guimiot, F.; Jones, K.L.; Siegenthaler, J.A. Single-Cell Transcriptomic Analyses of the Developing Meninges Reveal Meningeal Fibroblast Diversity and Function. Dev. Cell 2020, 54, 43–59.e4. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liao, X.; Hu, D.; Guan, D.; Tian, M. Back to the Origin: Mechanisms of CircRNA-Directed Regulation of Host Genes in Human Disease. Noncoding RNA 2024, 10, 49. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, S.; Chen, Z.; He, Z.; Xu, Y.; Li, Z. CircRNA-ENO1 Promoted Glycolysis and Tumor Progression in Lung Adenocarcinoma through Upregulating Its Host Gene ENO1. Cell Death Dis. 2019, 10, 885. [Google Scholar] [CrossRef]
- Ou, R.; Lv, J.; Zhang, Q.; Lin, F.; Zhu, L.; Huang, F.; Li, X.; Li, T.; Zhao, L.; Ren, Y.; et al. CircAMOTL1 Motivates AMOTL1 Expression to Facilitate Cervical Cancer Growth. Mol. Ther. Nucleic Acids 2020, 19, 50–60. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhou, W.; Duan, L.; Zhang, J.; Lu, X.; Jin, L.; Yu, Y. Circular RNA Circ-VANGL1 as a Competing Endogenous RNA Contributes to Bladder Cancer Progression by Regulating MiR-605-3p/VANGL1 Pathway. J. Cell Physiol. 2019, 234, 3887–3896. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, M.; Xue, C.; Chen, S.; Zheng, L.; Deng, H.; Tang, F.; Li, G.; Xiong, W.; Zeng, Z.; et al. Understanding the Roles and Regulation Patterns of CircRNA on Its Host Gene in Tumorigenesis and Tumor Progression. J. Exp. Clin. Cancer Res. 2023, 42, 86. [Google Scholar] [CrossRef]
- Zhou, T.; Xie, X.; Li, M.; Shi, J.; Zhou, J.J.; Knox, K.S.; Wang, T.; Chen, Q.; Gu, W. Rat BodyMap Transcriptomes Reveal Unique Circular RNA Features across Tissue Types and Developmental Stages. RNA 2018, 24, 1443–1456. [Google Scholar] [CrossRef]
- Bai, N.; Peng, E.; Qiu, X.; Lyu, N.; Zhang, Z.; Tao, Y.; Li, X.; Wang, Z. CircFBLIM1 Act as a CeRNA to Promote Hepatocellular Cancer Progression by Sponging MiR-346. J. Exp. Clin. Cancer Res. 2018, 37, 172. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Abdullah, S.T.; Taheri, M.; Samadian, M. A Review on the Role of Mir-16-5p in the Carcinogenesis. Cancer Cell Int. 2022, 22, 342. [Google Scholar] [CrossRef]
- Wang, H.; Pan, J.; Yu, L.; Meng, L.; Liu, Y.; Chen, X. MicroRNA-16 Inhibits Glioblastoma Growth in Orthotopic Model by Targeting Cyclin D1 and WIP1. Onco Targets Ther. 2020, 13, 10807–10816. [Google Scholar] [CrossRef]
- Krell, A.; Wolter, M.; Stojcheva, N.; Hertler, C.; Liesenberg, F.; Zapatka, M.; Weller, M.; Malzkorn, B.; Reifenberger, G. MiR-16-5p Is Frequently down-Regulated in Astrocytic Gliomas and Modulates Glioma Cell Proliferation, Apoptosis and Response to Cytotoxic Therapy. Neuropathol. Appl. Neurobiol. 2019, 45, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Bonafé, G.A.; Dos Santos, J.S.; Ziegler, J.V.; Umezawa, K.; Ribeiro, M.L.; Rocha, T.; Ortega, M.M. Growth Inhibitory Effects of Dipotassium Glycyrrhizinate in Glioblastoma Cell Lines by Targeting MicroRNAs Through the NF-ΚB Signaling Pathway. Front. Cell Neurosci. 2019, 13, 216. [Google Scholar] [CrossRef]
- Huang, X.; Hou, Y.; Weng, X.; Pang, W.; Hou, L.; Liang, Y.; Wang, Y.; Du, L.; Wu, T.; Yao, M.; et al. Diethyldithiocarbamate-Copper Complex (CuET) Inhibits Colorectal Cancer Progression via MiR-16-5p and 15b-5p/ALDH1A3/PKM2 Axis-Mediated Aerobic Glycolysis Pathway. Oncogenesis 2021, 10, 4. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, W.; Du, Y.; Guo, Q.; Mao, Y.; Zhou, X.; Hua, D. Serum MiR-16 as a Potential Biomarker for Human Cancer Diagnosis: Results from a Large-Scale Population. J. Cancer Res. Clin. Oncol. 2019, 145, 787–796. [Google Scholar] [CrossRef]
- Reis, P.P.; Drigo, S.A.; Carvalho, R.F.; Lopez Lapa, R.M.; Felix, T.F.; Patel, D.; Cheng, D.; Pintilie, M.; Liu, G.; Tsao, M.-S. Circulating MiR-16-5p, MiR-92a-3p, and MiR-451a in Plasma from Lung Cancer Patients: Potential Application in Early Detection and a Regulatory Role in Tumorigenesis Pathways. Cancers 2020, 12, 2071. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, X.; Hua, L.; Huang, L. Melatonin Inhibits the Malignant Progression of Glioblastoma via Regulating MiR-16-5p/PIM1. Curr. Neurovasc Res. 2022, 19, 92–99. [Google Scholar] [CrossRef]
- Joshi, R.; Sharma, A.; Kulshreshtha, R. Noncoding RNA Landscape and Their Emerging Roles as Biomarkers and Therapeutic Targets in Meningioma. Mol. Ther. Oncol. 2024, 32, 200782. [Google Scholar] [CrossRef]
- Song, L.-R.; Li, D.; Weng, J.-C.; Li, C.-B.; Wang, L.; Wu, Z.; Zhang, J.-T. MicroRNA-195 Functions as a Tumor Suppressor by Directly Targeting Fatty Acid Synthase in Malignant Meningioma. World Neurosurg. 2020, 136, e355–e364. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.S.; Hong, Y.G.; Kang, C.S. Significance of COX-2 and VEGF Expression in Histopathologic Grading and Invasiveness of Meningiomas. APMIS 2014, 122, 16–24. [Google Scholar] [CrossRef]
- Yamasaki, F.; Yoshioka, H.; Hama, S.; Sugiyama, K.; Arita, K.; Kurisu, K. Recurrence of Meningiomas. Cancer 2000, 89, 1102–1110. [Google Scholar] [CrossRef]
- Yang, K.; Zou, Z.; Wu, Y.; Hu, G. MiR-195 Suppression Alleviates Apoptosis and Oxidative Stress in CCl4-Induced ALI in Mice by Targeting Pim-1. Exp. Mol. Pathol. 2020, 115, 104438. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Yu, Y.; Li, M.; Wang, G.; Chen, R.; Fan, G.-C.; Martin, C.; Xiong, S.; Peng, T. Inhibition of MicroRNA 195 Prevents Apoptosis and Multiple-Organ Injury in Mouse Models of Sepsis. J. Infect. Dis. 2016, 213, 1661–1670. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Fromm, B.; Høye, E.; Domanska, D.; Zhong, X.; Aparicio-Puerta, E.; Ovchinnikov, V.; Umu, S.U.; Chabot, P.J.; Kang, W.; Aslanzadeh, M.; et al. MirGeneDB 2.1: Toward a Complete Sampling of All Major Animal Phyla. Nucleic Acids Res. 2022, 50, D204–D210. [Google Scholar] [CrossRef]
- Tastsoglou, S.; Skoufos, G.; Miliotis, M.; Karagkouni, D.; Koutsoukos, I.; Karavangeli, A.; Kardaras, F.S.; Hatzigeorgiou, A.G. DIANA-MiRPath v4.0: Expanding Target-Based MiRNA Functional Analysis in Cell-Type and Tissue Contexts. Nucleic Acids Res. 2023, 51, W154–W159. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. StarBase v2.0: Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Sun, Z.-Y.; Yang, C.-L.; Huang, L.-J.; Mo, Z.-C.; Zhang, K.-N.; Fan, W.-H.; Wang, K.-Y.; Wu, F.; Wang, J.-G.; Meng, F.-L.; et al. CircRNADisease v2.0: An Updated Resource for High-Quality Experimentally Supported CircRNA-Disease Associations. Nucleic Acids Res. 2024, 52, D1193–D1200. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A Web Tool for Exploring Circular RNAs and Their Interacting Proteins and MicroRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Rishik, S.; Hirsch, P.; Grandke, F.; Fehlmann, T.; Keller, A. MiRNATissueAtlas 2025: An Update to the Uniformly Processed and Annotated Human and Mouse Non-Coding RNA Tissue Atlas. Nucleic Acids Res. 2025, 53, D129–D137. [Google Scholar] [CrossRef]



| MR-miRNA | Function in MNG or Other Cancers | References | Candidate MR Host Linear Transcripts | Candidate MR-circRNAs (Biogenerated by MR Host Linear Transcripts and Acting as a Sponge for MR-miRNAs) | AGO-CLIP Region p-Value |
|---|---|---|---|---|---|
| hsa-miR-16-5p | Tumor suppressor in MNG | [10] | CHEK1 | circCHEK1 (hsa_circ_0024791; hsa_circ_0024793; hsa_circ_0024794) | ≤10−6 |
| hsa-miR-16-5p | Tumor suppressor in MNG | [10] | PIM1 | circPIM1 (hsa_circ_0076213; hsa_circ_0076214; hsa_circ_0076215; hsa_circ_0076216) | ≤10−12 |
| hsa-miR-124-3p | Tumor suppressor in several cancers | [38,39,40,41,42,43,44,45,46,47,48,49] * | PIM1 | circPIM1 (hsa_circ_0076213; hsa_circ_0076214) | ≤10−12 |
| hsa-miR-193b-3p | Tumor suppressor in MNG | [50] | CHEK1 | circCHEK1 (hsa_circ_0024793) | ≤10−8 |
| hsa-miR-195-5p | Tumor suppressor in MNG | [51] | CHEK1 | circCHEK1 (hsa_circ_0024793) | ≤10−6 |
| hsa-miR-195-5p | Tumor suppressor in MNG | [51] | PIM1 | circPIM1 (hsa_circ_0076213; hsa_circ_0076214; hsa_circ_0076215; hsa_circ_0076216) | ≤10−12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotera, G.; Forte, C.; D’Urso, D.G.; Reina, D.; Zuccaro, N.; Toscano, A.G.; Caponnetto, A.; Barbagallo, C.; Broggi, G.; Certo, F.; et al. In Silico Identification of circPIM1/miR-16-5p/miR-195-5p/PIM1 Feed-Forward Loop in Recurrent Grade 2 Meningioma. Int. J. Mol. Sci. 2025, 26, 8263. https://doi.org/10.3390/ijms26178263
Sotera G, Forte C, D’Urso DG, Reina D, Zuccaro N, Toscano AG, Caponnetto A, Barbagallo C, Broggi G, Certo F, et al. In Silico Identification of circPIM1/miR-16-5p/miR-195-5p/PIM1 Feed-Forward Loop in Recurrent Grade 2 Meningioma. International Journal of Molecular Sciences. 2025; 26(17):8263. https://doi.org/10.3390/ijms26178263
Chicago/Turabian StyleSotera, Giuseppe, Carla Forte, Daniele Giuseppe D’Urso, Domenica Reina, Noemi Zuccaro, Andrea Giuseppe Toscano, Angela Caponnetto, Cristina Barbagallo, Giuseppe Broggi, Francesco Certo, and et al. 2025. "In Silico Identification of circPIM1/miR-16-5p/miR-195-5p/PIM1 Feed-Forward Loop in Recurrent Grade 2 Meningioma" International Journal of Molecular Sciences 26, no. 17: 8263. https://doi.org/10.3390/ijms26178263
APA StyleSotera, G., Forte, C., D’Urso, D. G., Reina, D., Zuccaro, N., Toscano, A. G., Caponnetto, A., Barbagallo, C., Broggi, G., Certo, F., Ragusa, M., Caltabiano, R., Di Pietro, C., Barbagallo, G. M. V., Purrello, M., & Barbagallo, D. (2025). In Silico Identification of circPIM1/miR-16-5p/miR-195-5p/PIM1 Feed-Forward Loop in Recurrent Grade 2 Meningioma. International Journal of Molecular Sciences, 26(17), 8263. https://doi.org/10.3390/ijms26178263










