Effects of Different Salinity Conditions on Regulation of ghrh-sst-gh-igf Axis in Nile Tilapia (Oreochromis niloticus): Insights from Transcriptional Signature
Abstract
1. Introduction
2. Results
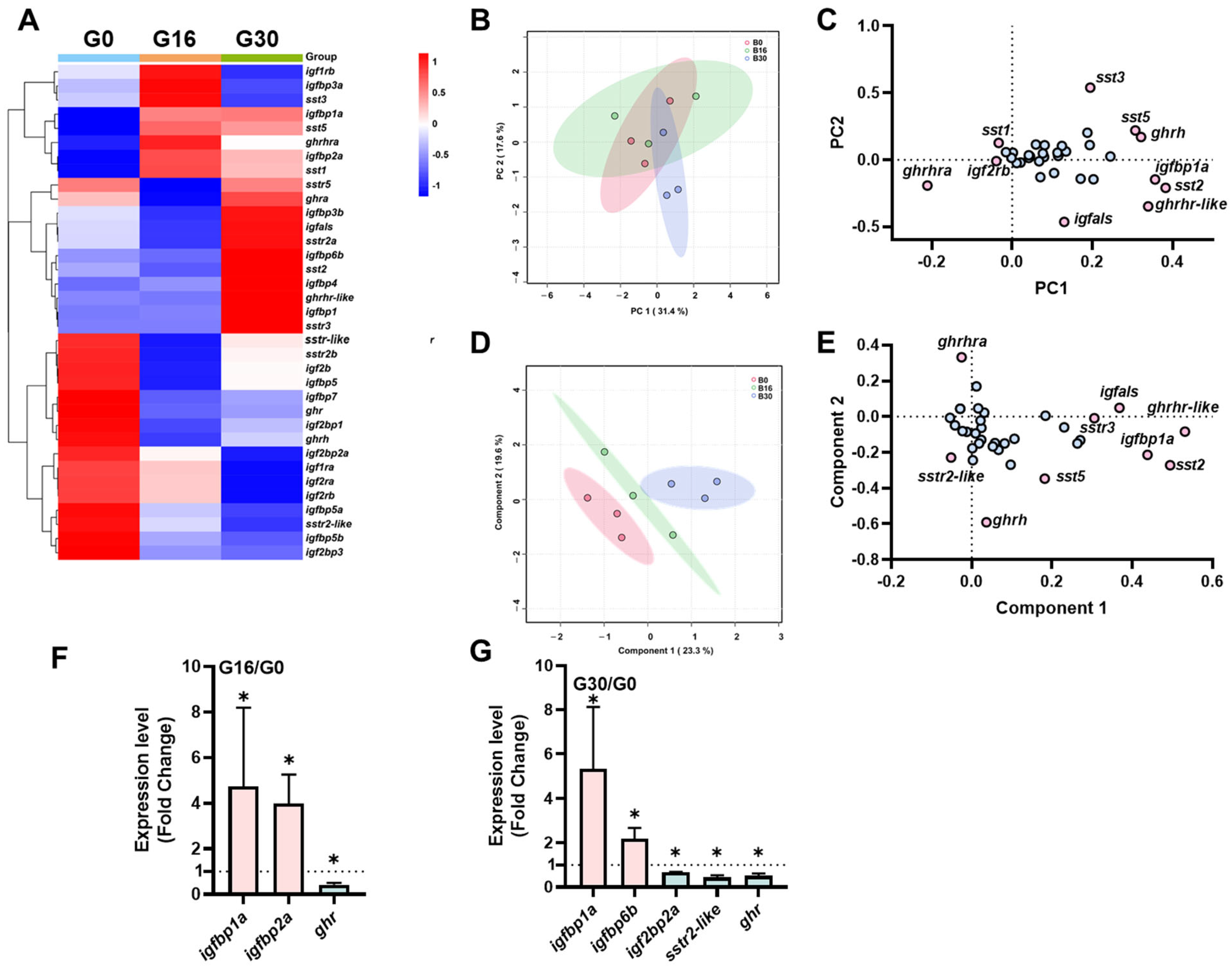
2.1. The Transcriptional Signature of the ghrh-sst-gh-igf Axis in the Brain Under Different Salinities
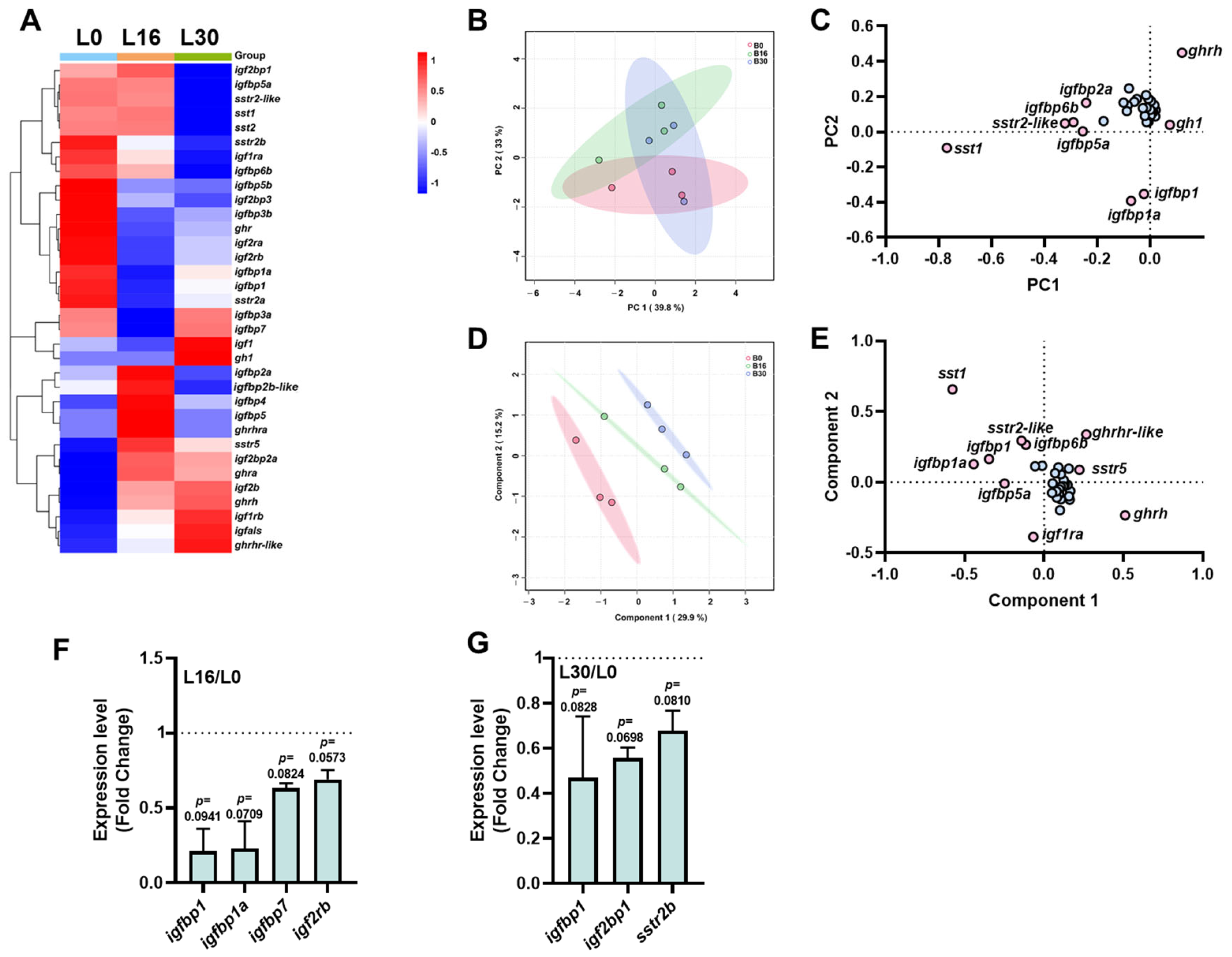
2.2. The Transcriptional Signature of the ghrh-sst-gh-igf Axis in the Intestine Under Different Salinities
2.3. The Transcriptional Signature of the ghrh-sst-gh-igf Axis in the Liver Under Different Salinities
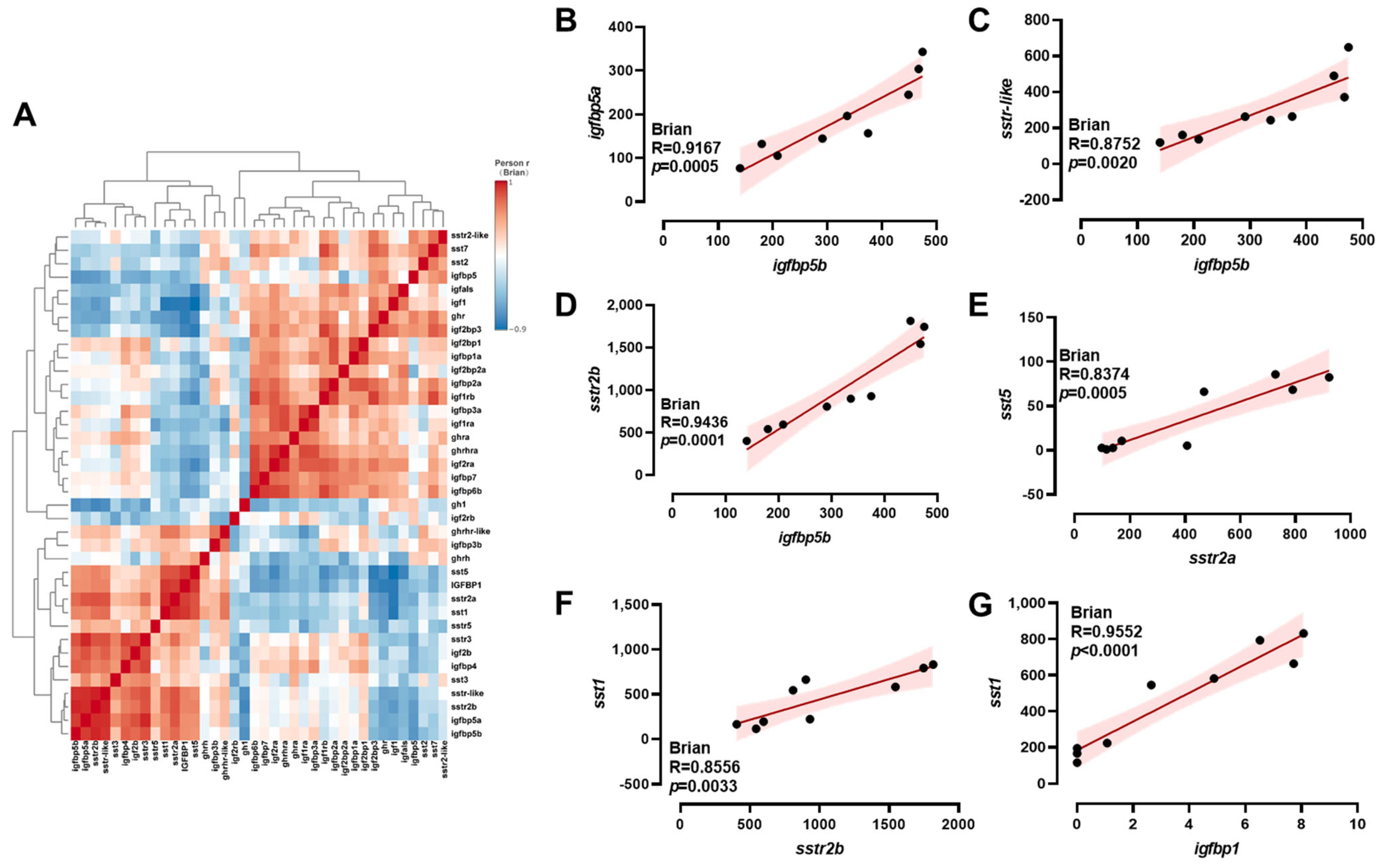
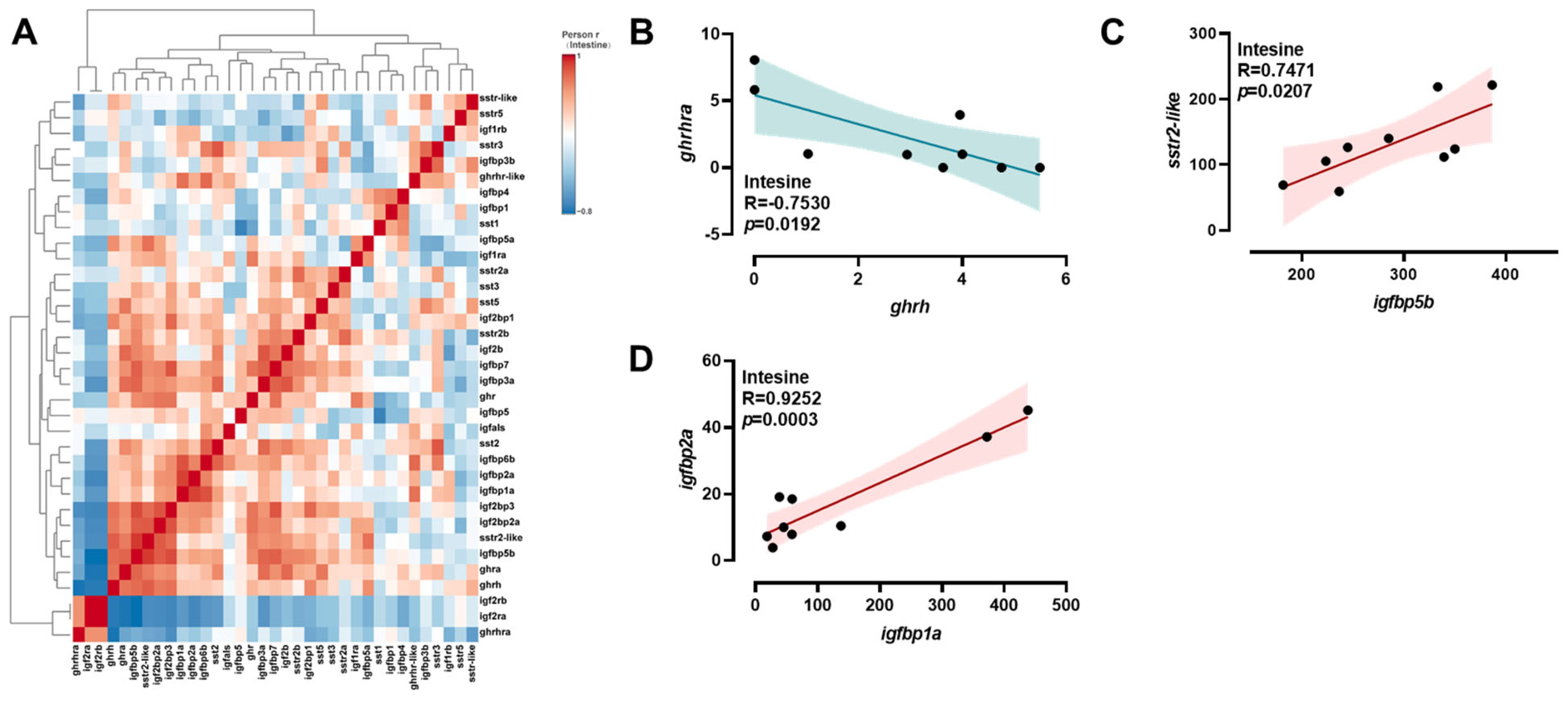
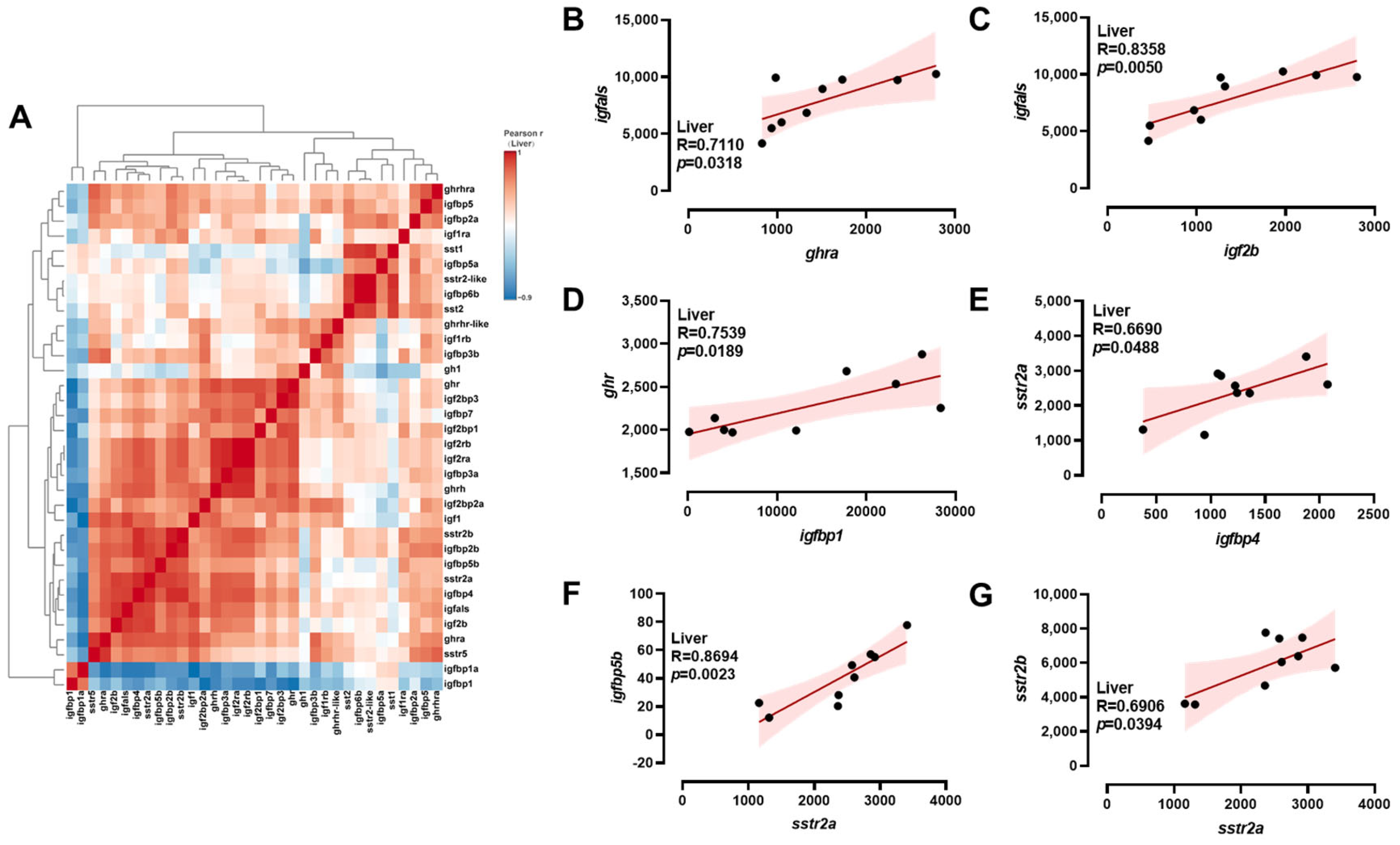
2.4. Correlation Analysis of ghrh-sst-gh-igf Axis Genes
3. Discussion
3.1. Brian Transcriptional Response of ghrh-sst-gh-igf Axis Genes to Salinity Stress
3.2. Divergent Roles of igfbp5a and igfbp5b in Salinity Adaptation
3.3. Intestinal Transcriptional Re-Programming Under Salinity Stress
3.4. Hepatic Energy Homeostasis and the ghrh-sst-gh-igf-Axis
4. Materials and Methods
4.1. Ethical Statement
4.2. Experimental Design and Sample Collection
4.3. RNA-Seq and Analysis
4.4. Quantitative Real-Time PCR (qPCR) for Verification of RNA-Seq
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boyd, C.E.; McNevin, A.A.; Davis, R.P. The contribution of fisheries and aquaculture to the global protein supply. Food Secur. 2022, 14, 805–827. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Doan, H.V. Tilapia Fish for Future Sustainable Aquaculture; Springer: Berlin/Heidelberg, Germany, 2023. [Google Scholar]
- Zimmermann, S.; Kiessling, A.; Zhang, J. The future of intensive tilapia production and the circular bioeconomy without effluents: Biofloc technology, recirculation aquaculture systems, bio-RAS, partitioned aquaculture systems and integrated multitrophic aquaculture. Rev. Aquac. 2023, 15, 22–31. [Google Scholar] [CrossRef]
- Bal, A.; Panda, F.; Pati, S.G.; Das, K.; Agrawal, P.K.; Paital, B. Modulation of physiological oxidative stress and antioxidant status by abiotic factors especially salinity in aquatic organisms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 241, 108971. [Google Scholar] [CrossRef]
- Velasco, J.; Gutiérrez-Cánovas, C.; Botella-Cruz, M.; Sánchez-Fernández, D.; Arribas, P.; Carbonell, J.A.; Millán, A.; Pallarés, S. Effects of salinity changes on aquatic organisms in a multiple stressor context. Philos. Trans. R. Soc. B Biol. Sci. 2018, 374, 20180011. [Google Scholar] [CrossRef]
- Canosa, L.F.; Bertucci, J.I. The effect of environmental stressors on growth in fish and its endocrine control. Front. Endocrinol. 2023, 14, 1109461. [Google Scholar] [CrossRef]
- El-Leithy, A.A.A.; Hemeda, S.A.; Naby, W.S.H.A.E.; Nahas, A.F.E.; Helmy, Z.A. Optimum salinity for Nile tilapia (Oreochromis niloticus) growth and mRNA transcripts of ion-regulation, inflammatory, stress- and immune-related genes. Fish Physiol Biochem. 2019, 45, 1217–1232. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Canosa, L.F. Chapter 1 Neuroendocrine Systems of the Fish Brain. Fish Physiol. 2009, 28, 3–74. [Google Scholar]
- Canosa, L.F.; Chang, J.P.; Peter, R.E. Neuroendocrine control of growth hormone in fish. Gen. Comp. Endocrinol. 2007, 151, 1–26. [Google Scholar] [CrossRef]
- Yunker, W.K.; Smith, S.; Graves, C.; Davis, P.J.; Unniappan, S.; Rivier, J.E.; Peter, R.E.; Chang, J.P. Endogenous Hypothalamic Somatostatins Differentially Regulate Growth Hormone Secretion from Goldfish Pituitary Somatotropes in Vitro. Endocrinology 2003, 144, 4031–4041. [Google Scholar] [CrossRef]
- Caputo, M.; Pigni, S.; Agosti, E.; Daffara, T.; Ferrero, A.; Filigheddu, N.; Prodam, F. Regulation of GH and GH Signaling by Nutrients. Cells 2021, 10, 1376. [Google Scholar] [CrossRef]
- Montero-Hidalgo, A.J.; Rio-Moreno, M.d.; Pérez-Gómez, J.M.; Luque, R.M.; Kineman, R.D. Update on regulation of GHRH and its actions on GH secretion in health and disease. Rev. Endocr. Metab. Disord. 2025, 26, 305–320. [Google Scholar] [CrossRef]
- Lappin, F.M.; Shaw, R.L.; Macqueen, D.J. Targeted sequencing for high-resolution evolutionary analyses following genome duplication in salmonid fish: Proof of concept for key components of the insulin-like growth factor axis. Mar. Genom. 2016, 30, 15–26. [Google Scholar] [CrossRef]
- Hu, W.; Cao, Y.; Liu, Q.; Yuan, C.; Hu, Z. Effect of salinity on the physiological response and transcriptome of spotted seabass (Lateolabrax maculatus). Mar. Pollut. Bull. 2024, 203, 116432. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, C.; Qi, M.; Liu, Q.; Hu, Z. The Effect of Salinity Stress on Enzyme Activities, Histology, and Transcriptome of Silver Carp (Hypophthalmichthys molitrix). Biology 2022, 11, 1580. [Google Scholar] [CrossRef]
- Azodi, M.; Bahabadi, M.N.; Ghasemi, A.; Morshedi, V.; Mozanzadeh, M.T.; Shahraki, R.; Khademzadeh, O.; Hamedi, S.; Avizhgan, S. Effects of salinity on gills’ chloride cells, stress indices, and gene expression of Asian seabass (Lates calcarifer, Bloch, 1790). Fish Physiol. Biochem. 2021, 47, 2027–2039. [Google Scholar] [CrossRef]
- Xiang, K.; Yang, Q.; Liu, M.; Yang, X.; Li, J.; Hou, Z.; Wen, H. Crosstalk between Growth and Osmoregulation of GHRH-SST-GH-IGF Axis in Triploid Rainbow Trout (Oncorhynchus mykiss). Int. J. Mol. Sci. 2022, 23, 8691. [Google Scholar] [CrossRef]
- Hou, Z.-S.; Xin, Y.-R.; Zeng, C.; Zhao, H.-K.; Tian, Y.; Li, J.-F.; Wen, H. GHRH-SST-GH-IGF axis regulates crosstalk between growth and immunity in rainbow trout (Oncorhynchus mykiss) infected with Vibrio anguillarum. Fish Shellfish. Immunol. 2020, 106, 887–897. [Google Scholar] [CrossRef]
- Seale, A.P.; Cao, K.; Chang, R.J.A.; Goodearly, T.R.; Malintha, G.H.T.; Merlo, R.S.; Peterson, T.L.; Reighard, J.R. Salinity tolerance of fishes: Experimental approaches and implications for aquaculture production. Rev. Aquac. 2024, 16, 1351–1373. [Google Scholar] [CrossRef]
- Chen, J.; Cai, M.; Zhan, C. Neuronal Regulation of Feeding and Energy Metabolism: A Focus on the Hypothalamus and Brainstem. Neurosci. Bull. 2024, 41, 665–675. [Google Scholar] [CrossRef]
- Verburg-van Kemenade, B.M.L.; Cohen, N.; Chadzinska, M. Neuroendocrine-immune interaction: Evolutionarily conserved mechanisms that maintain allostasis in an ever-changing environment. Dev. Comp. Immunol. 2017, 66, 2–23. [Google Scholar] [CrossRef]
- Veldhuis, J.D.; Iranmanesh, A.; Erickson, D.; Roelfsema, F.; Bowers, C.Y. Lifetime Regulation of Growth Hormone (GH) Secretion. In Handbook of Neuroendocrinology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 237–257. [Google Scholar]
- Yan, J.-J.; Lee, Y.-C.; Tsou, Y.-L.; Tseng, Y.-C.; Hwang, P.-P. Insulin-like growth factor 1 triggers salt secretion machinery in fish under acute salinity stress. J. Endocrinol. 2020, 246, 277–288. [Google Scholar] [CrossRef]
- Mahmoudi, R.; Naderi, M.; Dopeikar, H.; Khorasaninasab, S.A.; Seyedalhosseini, S.H. Interactive effects of triploidy induction and culture densities on growth performance and stress, immune, and metabolic responses in juvenile rainbow trout (Oncorhynchus mykiss). Aquac. Rep. 2025, 40, 102540. [Google Scholar] [CrossRef]
- Waters, J.A.; Urbano, I.; Robinson, M.; House, C.D. Insulin-like growth factor binding protein 5: Diverse roles in cancer. Front. Oncol. 2022, 12, 1052457. [Google Scholar] [CrossRef]
- Dai, W.; Bai, Y.; Hebda, L.; Zhong, X.; Liu, J.; Kao, J.; Duan, C. Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ. 2013, 21, 568–581. [Google Scholar] [CrossRef]
- Garcia de la Serrana, D.; Macqueen, D.J. Insulin-Like Growth Factor-Binding Proteins of Teleost Fishes. Front. Endocrinol. 2018, 9, 80. [Google Scholar] [CrossRef]
- Allard, J.B.; Duan, C. IGF-Binding Proteins: Why Do They Exist and Why Are There So Many? Front. Endocrinol. 2018, 9, 117. [Google Scholar] [CrossRef]
- Edwards, S.L.; Marshall, W.S. Principles and Patterns of Osmoregulation and Euryhalinity in Fishes. In Fish Physiology; Academic Press: Cambridge, MA, USA, 2012; pp. 1–44. [Google Scholar]
- Reinecke, M.; Björnsson, B.T.; Dickhoff, W.W.; McCormick, S.D.; Navarro, I.; Power, D.M.; Gutiérrez, J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 2005, 142, 20–24. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Zuloaga, R.; Mercado, L.; Einarsdottir, I.E.; Björnsson, B.T.; Valdés, J.A.; Molina, A. Chronic stress inhibits growth and induces proteolytic mechanisms through two different nonoverlapping pathways in the skeletal muscle of a teleost fish. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2018, 314, R102–R113. [Google Scholar] [CrossRef]
- Martin, S.A.M.; Macqueen, D.J.; Boudinot, P.; Secombes, C.J.; Wang, T.; Castro, R.; Alzaid, A. Cross Talk Between Growth and Immunity: Coupling of the IGF Axis to Conserved Cytokine Pathways in Rainbow Trout. Endocrinology 2016, 157, 1942–1955. [Google Scholar] [CrossRef]
- Peterson, B.C.; Small, B.C. Effects of fasting on circulating IGF-binding proteins, glucose, and cortisol in channel catfish (Ictalurus punctatus). Domest. Anim. Endocrinol. 2004, 26, 231–240. [Google Scholar] [CrossRef]
- Baxter, R.C. Signaling Pathways of the Insulin-like Growth Factor Binding Proteins. Endocr. Rev. 2023, 44, 753–778. [Google Scholar] [CrossRef]
- Dai, N.; Zhao, L.; Wrighting, D.; Krämer, D.; Majithia, A.; Wang, Y.; Cracan, V.; Borges-Rivera, D.; Mootha Vamsi, K.; Nahrendorf, M.; et al. IGF2BP2/IMP2-Deficient Mice Resist Obesity through Enhanced Translation of Ucp1 mRNA and Other mRNAs Encoding Mitochondrial Proteins. Cell Metab. 2015, 21, 609–621. [Google Scholar] [CrossRef]
- Wang, X.; Lu, L.; Li, Y.; Li, M.; Chen, C.; Feng, Q.; Zhang, C.; Duan, C. Molecular and functional characterization of two distinct IGF binding protein-6 genes in zebrafish. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2009, 296, R1348–R1357. [Google Scholar] [CrossRef]
- Li, W.; Lin, H. The endocrine regulation network of growth hormone synthesis and secretion in fish: Emphasis on the signal integration in somatotropes. Sci. China Life Sci. 2010, 53, 462–470. [Google Scholar] [CrossRef]
- Robinson, M.W.; Harmon, C.; O’Farrelly, C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016, 13, 267–276. [Google Scholar] [CrossRef]
- Gabillard, J.-C.; Kamangar, B.B.; Montserrat, N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss). J. Endocrinol. 2006, 191, 15–24. [Google Scholar] [CrossRef]
- Reindl, K.M.; Sheridan, M.A. Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2012, 163, 231–245. [Google Scholar] [CrossRef]
- Delafontaine, P.; Song, Y.-H.; Li, Y. Expression, Regulation, and Function of IGF-1, IGF-1R, and IGF-1 Binding Proteins in Blood Vessels. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 435–444. [Google Scholar] [CrossRef]
- Melmed, S.; Liu, N.-A.; Ren, S.-G.; Chesnokova, V.; Zhou, C.; Khalafi, R.; Pichurin, O.; Ben-Shlomo, A. Constitutive Somatostatin Receptor Subtype 2 Activity Attenuates GH Synthesis. Endocrinology 2013, 154, 2399–2409. [Google Scholar] [CrossRef]
- Su, M.; Feng, Z.; Zhong, Y.; Ye, Z.; Zhang, J. Insights into salinity effect on growth of the spotted scat (Scatophagus argus): Exploring the optimum salinity for its culture. Aquac. Rep. 2025, 41, 102667. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhao, H.; Soufan, O.; Xia, J.; Tang, R.; Li, L.; Li, D. Transcriptome and physiological analysis reveal alterations in muscle metabolisms and immune responses of grass carp (Ctenopharyngodon idellus) cultured at different stocking densities. Aquaculture 2019, 503, 186–197. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Mager, S.; Ludewig, U. Massive Loss of DNA Methylation in Nitrogen-, but Not in Phosphorus-Deficient Zea mays Roots Is Poorly Correlated With Gene Expression Differences. Front. Plant Sci. 2018, 9, 497. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J.; Stegle, O. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Liu, Z.; Koid, A.E.; Terrado, R.; Campbell, V.; Caron, D.A.; Heidelberg, K.B. Changes in gene expression of Prymnesium parvum induced by nitrogen and phosphorus limitation. Front. Microbiol. 2015, 6, 631. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 2014, 34, 11929–11947, Erratum in J. Neurosci. 2015, 35, 846. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Meekuan, P.; Wang, Y.-X.; Feng, Z.-H.; Luo, S.-Y.; Zhang, Z.-X.; Xiao, J.; Yu, F.; Hou, Z.-S. Effects of Different Salinity Conditions on Regulation of ghrh-sst-gh-igf Axis in Nile Tilapia (Oreochromis niloticus): Insights from Transcriptional Signature. Int. J. Mol. Sci. 2025, 26, 8261. https://doi.org/10.3390/ijms26178261
Li Z, Meekuan P, Wang Y-X, Feng Z-H, Luo S-Y, Zhang Z-X, Xiao J, Yu F, Hou Z-S. Effects of Different Salinity Conditions on Regulation of ghrh-sst-gh-igf Axis in Nile Tilapia (Oreochromis niloticus): Insights from Transcriptional Signature. International Journal of Molecular Sciences. 2025; 26(17):8261. https://doi.org/10.3390/ijms26178261
Chicago/Turabian StyleLi, Zhao, Pichayapa Meekuan, Ya-Xin Wang, Zhuo-Hang Feng, Shuang-Yue Luo, Zheng-Xiang Zhang, Jun Xiao, Fan Yu, and Zhi-Shuai Hou. 2025. "Effects of Different Salinity Conditions on Regulation of ghrh-sst-gh-igf Axis in Nile Tilapia (Oreochromis niloticus): Insights from Transcriptional Signature" International Journal of Molecular Sciences 26, no. 17: 8261. https://doi.org/10.3390/ijms26178261
APA StyleLi, Z., Meekuan, P., Wang, Y.-X., Feng, Z.-H., Luo, S.-Y., Zhang, Z.-X., Xiao, J., Yu, F., & Hou, Z.-S. (2025). Effects of Different Salinity Conditions on Regulation of ghrh-sst-gh-igf Axis in Nile Tilapia (Oreochromis niloticus): Insights from Transcriptional Signature. International Journal of Molecular Sciences, 26(17), 8261. https://doi.org/10.3390/ijms26178261





