Therapeutic Potential of Glutaminase Inhibition Targeting Metabolic Adaptations in Resistant Melanomas to Targeted Therapy
Abstract
1. Introduction
2. Results
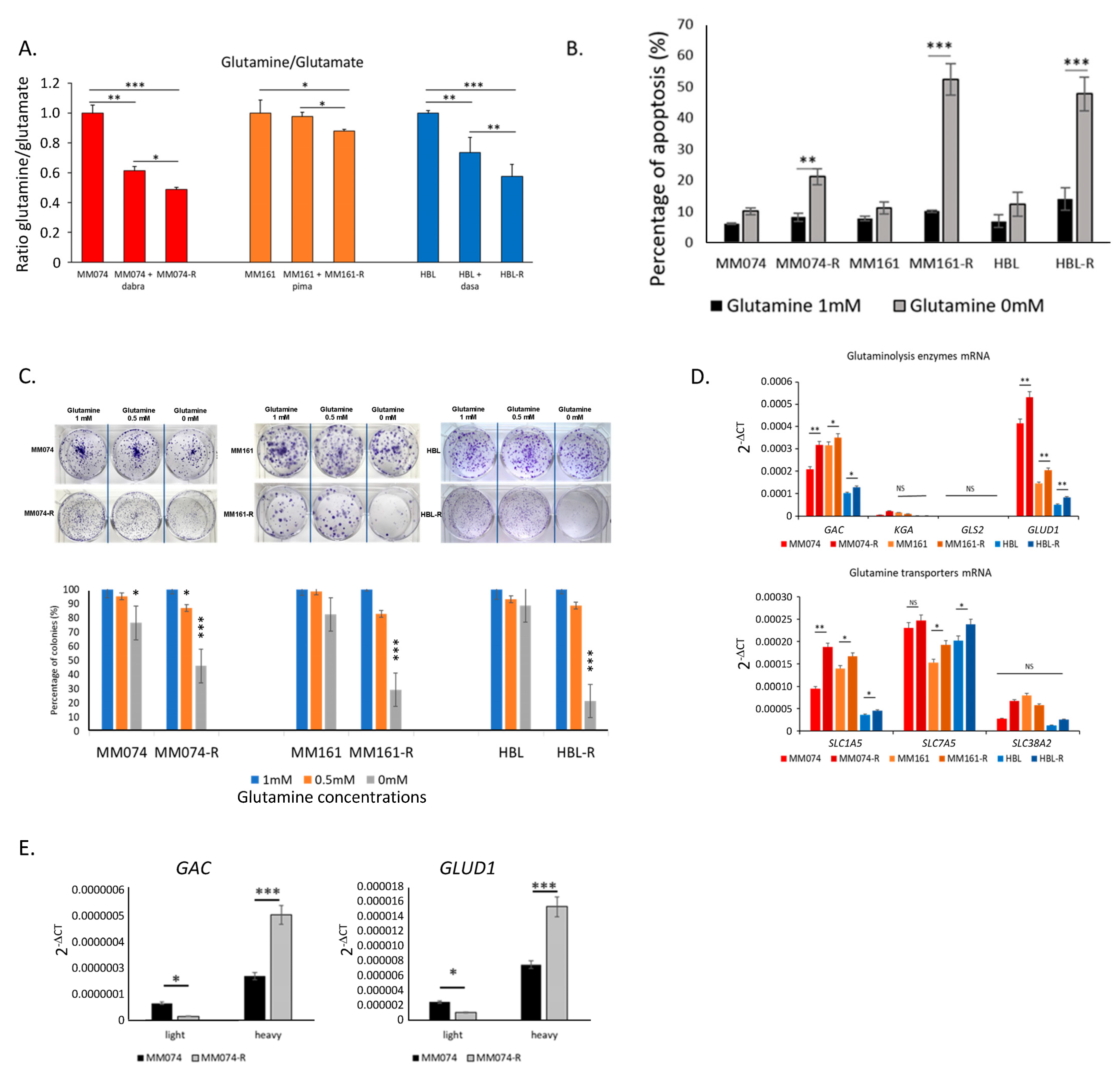
2.1. Melanoma Cells with Acquired Resistance Depend on Glutamine and Rely on Glutaminolysis for Their Survival
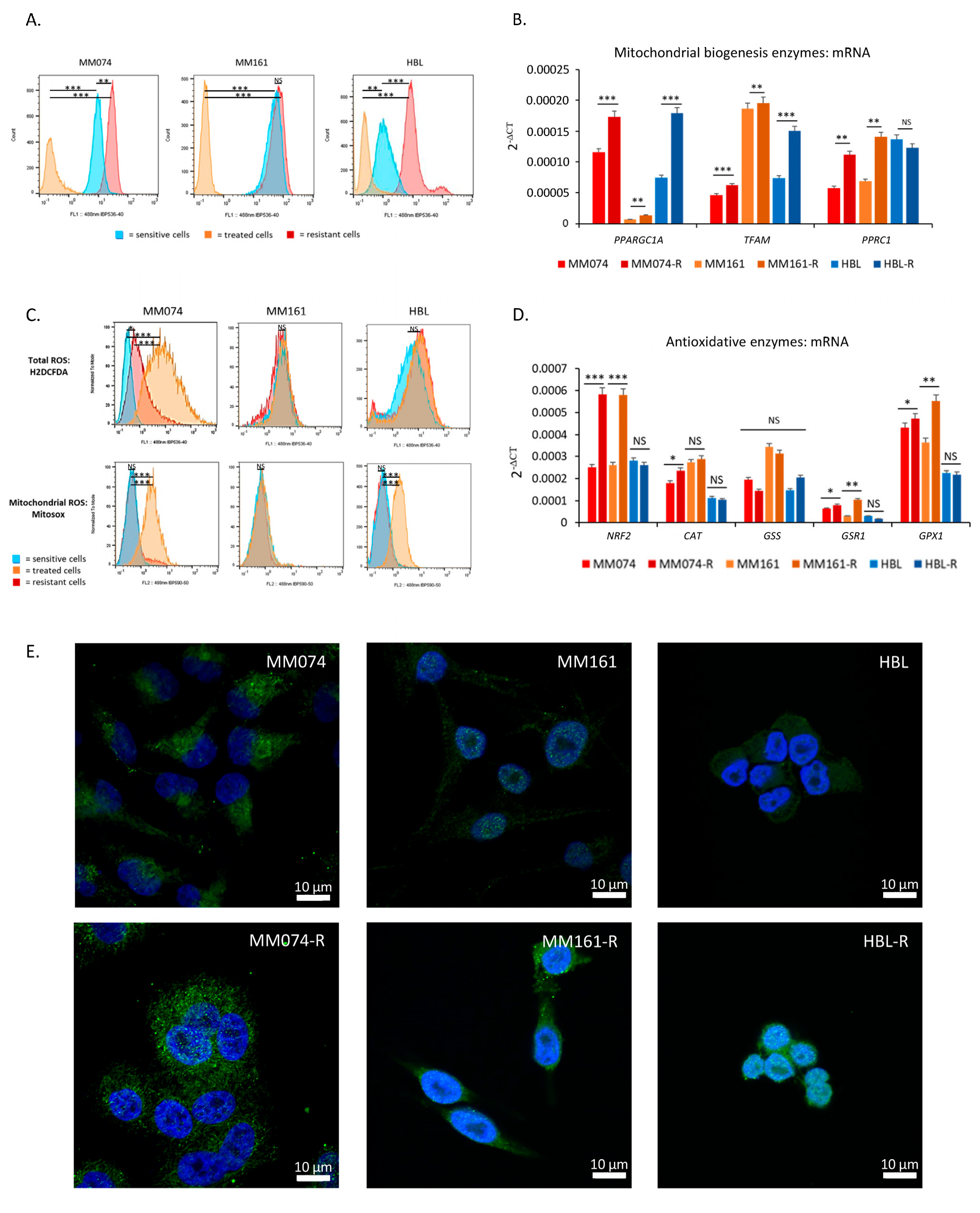
2.2. Resistant Cells Have Higher Mitochondrial Content and Better Antioxidative Defenses
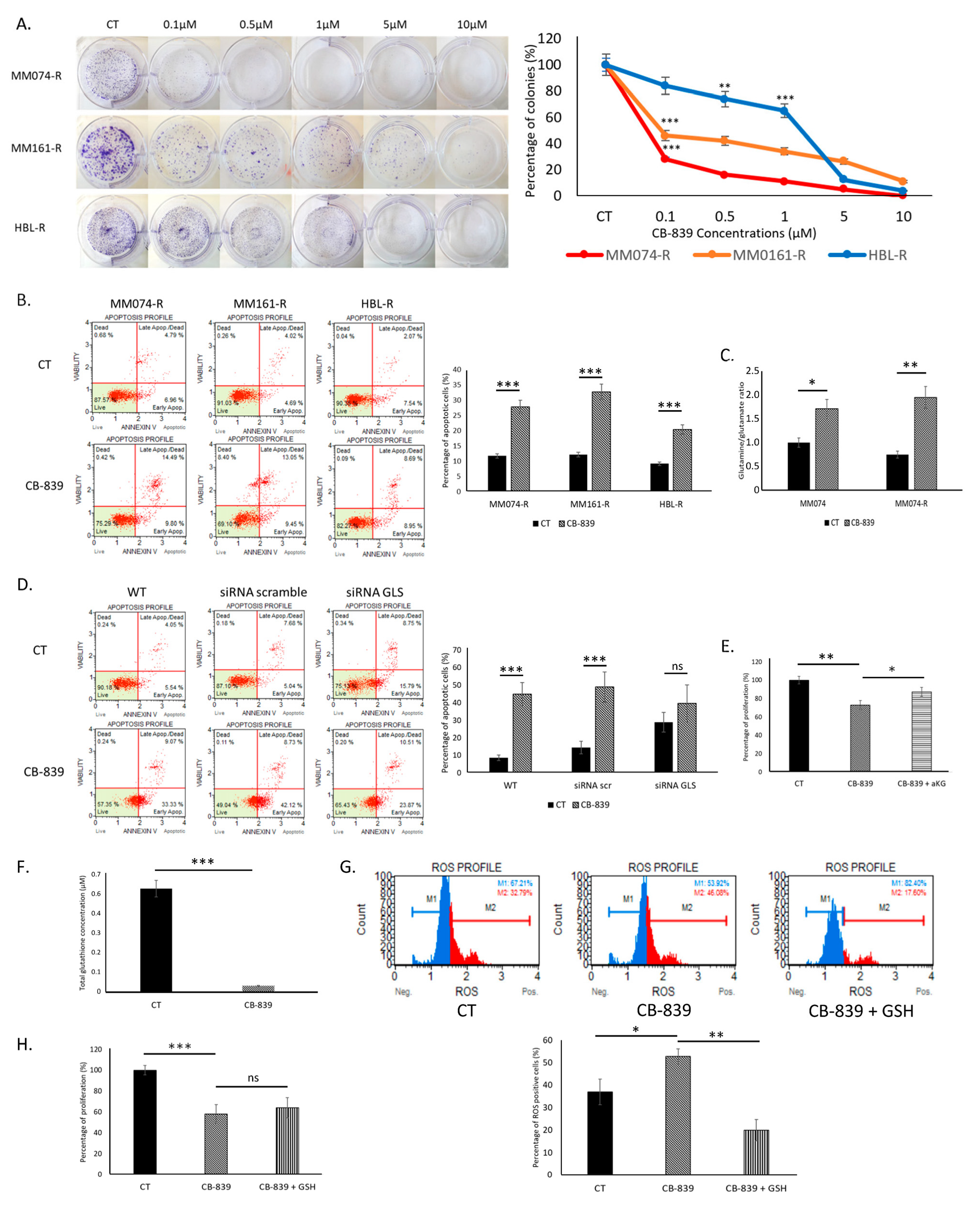
2.3. Glutaminase Inhibitor CB-839 Blocks Glutaminolysis and Decreases Survival of Resistant Cells
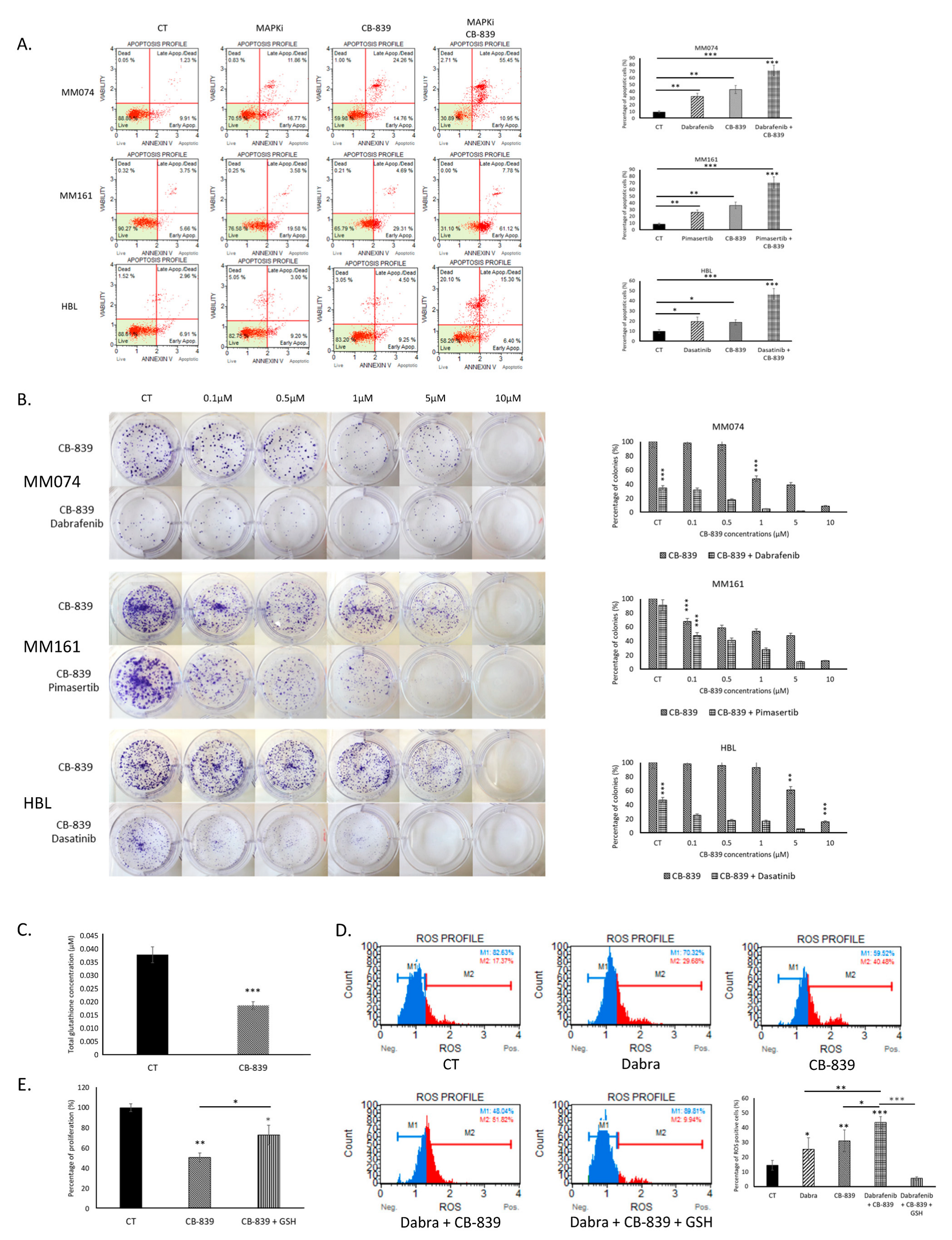
2.4. Glutaminolysis Inhibition Improves the Effectiveness of Targeted Therapy in Sensitive Cells by Increasing Oxidative Stress
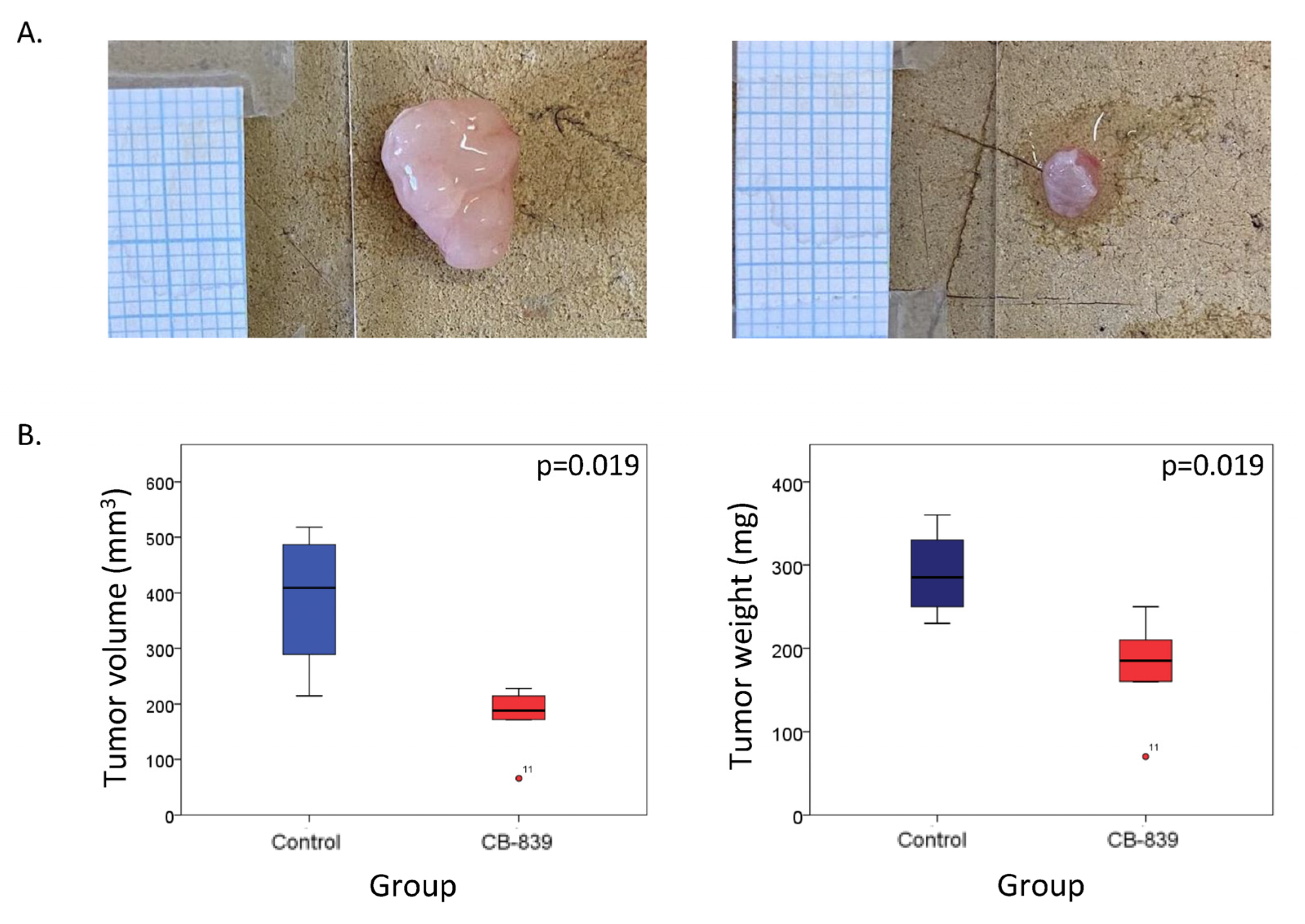
2.5. Glutaminolysis Inhibition Decreases Tumor Development in Mice Bearing BRAFi Resistant Xenografts
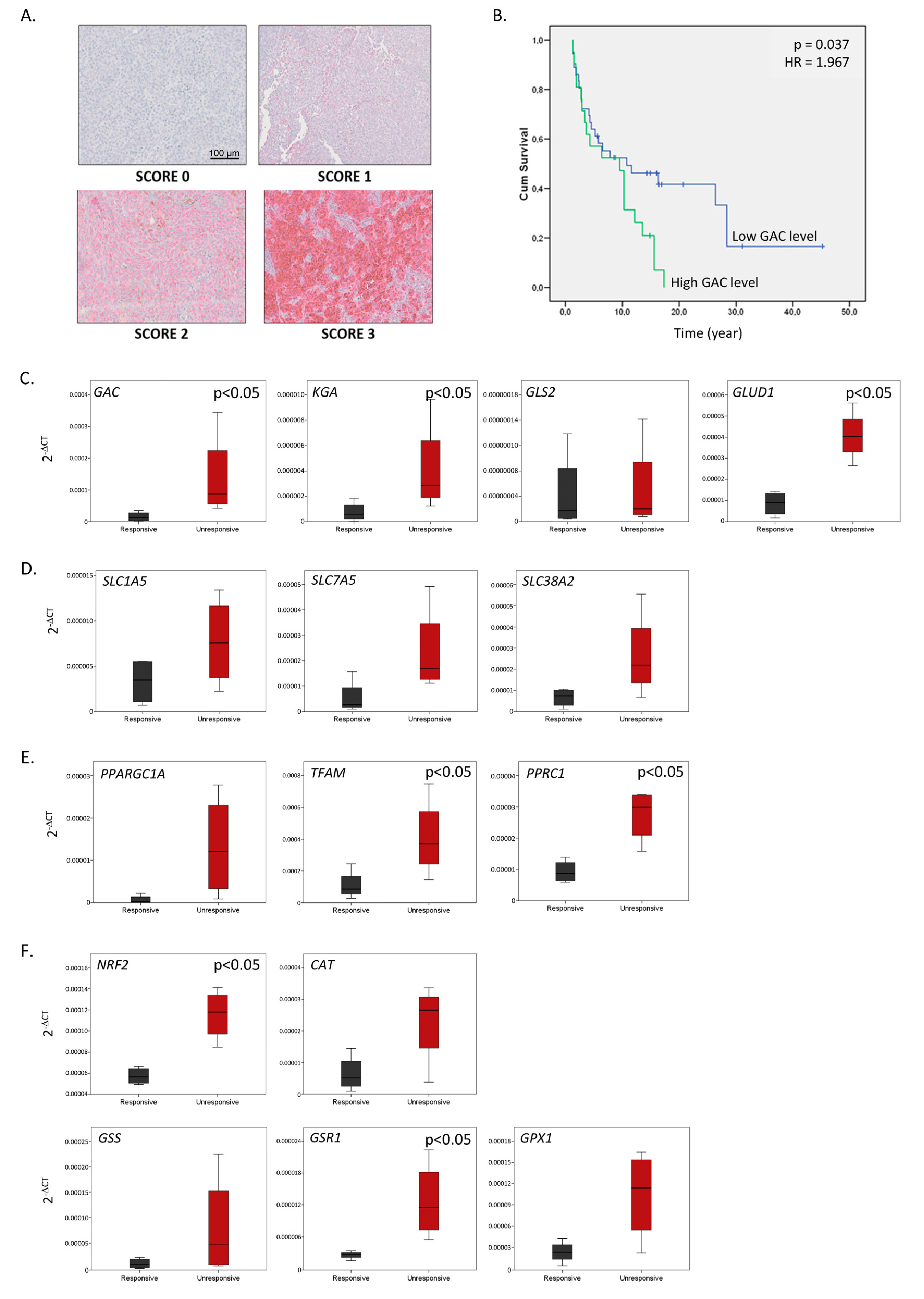
2.6. The Glutaminolysis Enzyme GAC Is a Prognostic Marker in Metastatic Melanoma and a Signature Comprising GAC and Other Glutamine Metabolism-Related Enzymes May Predict Resistance to BRAFi
3. Discussion
4. Materials and Methods
4.1. Inhibitors
4.2. Cell Culture
4.3. Glutamine/Glutamate Dosage
4.4. Clonogenic Assay
4.5. Cell Proliferation
4.6. RT-qPCR
4.7. Polysome Profiling
4.8. Flow Cytometry Assessment of Apoptosis, Oxidative Stress, Mitochondrial Levels, and ROS Level
4.9. Immunofluorescence
4.10. Glutathione Measurement
4.11. Animal Study
4.12. Patient Samples
4.13. Immunohistochemistry
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| α-KG | Alpha-ketoglutarate |
| 5-FU | 5-Fluorouracil |
| 18S | 18S ribosomal RNA |
| ANOVA | Analysis of Variance |
| ATP | Adenosine Triphosphate |
| BRAFi | BRAF inhibitor |
| BSA | Bovine Serum Albumin |
| cKITi | cKIT inhibitor |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DTT | Dithiothreitol |
| GAC | glutaminase C |
| GSH | glutathione |
| HE | hematoxylin and eosin |
| HR | hazard ratio |
| IHC | immunohistochemistry |
| MAPK | Mitogen-Activated Protein Kinase |
| MEKi | MEK inhibitor |
| NS | Not Significant |
| OS | overall survival |
| PBS | Phosphate Buffered Saline |
| PCR | Polymerase Chain Reaction |
| PERCIST | PET Response Criteria in Solid Tumors |
| RTKi | Receptor Tyrosine Kinase (RTK) inhibitor |
| ROS | Reactive Oxygen Species |
| RPM | Revolutions Per Minute |
| RT-qPCR | Reverse Transcription Quantitative PCR |
| TCA | Tricarboxylic Acid |
| TFAM | Mitochondrial Transcription Factor A |
References
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.B.; Arachchi, H.; Arora, J.A.; Auman, T.; Ayala, B. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, P.; Trinh, A.; Khamari, R.; Kluza, J. Melanoma metabolism contributes to the cellular responses to MAPK/ERK pathway inhibitors. Biochim. Biophys. Acta. (BBA)—Gen. Subj. 2018, 1862, 999–1005. [Google Scholar] [CrossRef]
- Theodosakis, N.; Held, M.A.; Marzuka-Alcala, A.; Meeth, K.M.; Micevic, G.; Long, G.V.; Scolyer, R.A.; Stern, D.F.; Bosenberg, M.W. BRAF Inhibition Decreases Cellular Glucose Uptake in Melanoma in Association with Reduction in Cell Volume. Mol. Cancer Ther. 2015, 14, 1680–1692. [Google Scholar] [CrossRef] [PubMed]
- Falck Miniotis, M.; Arunan, V.; Eykyn, T.R.; Marais, R.; Workman, P.; Leach, M.O.; Beloueche-Babari, M. MEK1/2 inhibition decreases lactate in BRAF-driven human cancer cells. Cancer Res. 2013, 73, 4039–4049. [Google Scholar] [CrossRef] [PubMed]
- Hardeman, K.N.; Peng, C.; Paudel, B.B.; Meyer, C.T.; Luong, T.; Tyson, D.R.; Young, J.D.; Quaranta, V.; Fessel, J.P. Dependence On Glycolysis Sensitizes BRAF-mutated Melanomas For Increased Response To Targeted BRAF Inhibition. Sci. Rep. 2017, 7, 42604. [Google Scholar] [CrossRef]
- Zecena, H.; Tveit, D.; Wang, Z.; Farhat, A.; Panchal, P.; Liu, J.; Singh, S.J.; Sanghera, A.; Bainiwal, A.; Teo, S.Y.; et al. Systems biology analysis of mitogen activated protein kinase inhibitor resistance in malignant melanoma. BMC Syst. Biol. 2018, 12, 33. [Google Scholar] [CrossRef]
- Baenke, F.; Chaneton, B.; Smith, M.; Van Den Broek, N.; Hogan, K.; Tang, H.; Viros, A.; Martin, M.; Galbraith, L.; Girotti, M.R.; et al. Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Mol. Oncol. 2016, 10, 73–84. [Google Scholar] [CrossRef]
- Hernandez-Davies, J.E.; Tran, T.Q.; Reid, M.A.; Rosales, K.R.; Lowman, X.H.; Pan, M.; Moriceau, G.; Yang, Y.; Wu, J.; Lo, R.S.; et al. Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. J. Transl. Med. 2015, 13, 210. [Google Scholar] [CrossRef]
- Soumoy, L.; Schepkens, C.; Krayem, M.; Najem, A.; Tagliatti, V.; Ghanem, G.E.; Saussez, S.; Colet, J.-M.; Journe, F. Metabolic Reprogramming in Metastatic Melanoma with Acquired Resistance to Targeted Therapies: Integrative Metabolomic and Proteomic Analysis. Cancers 2020, 12, 1323. [Google Scholar] [CrossRef]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef]
- Halama, A.; Suhre, K. Advancing Cancer Treatment by Targeting Glutamine Metabolism—A Roadmap. Cancers 2022, 14, 553. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Chen, S. Metabolic Signaling Cascades Prompted by Glutaminolysis in Cancer. Cancers 2020, 12, 2624. [Google Scholar] [CrossRef] [PubMed]
- Sappington, D.R.; Siegel, E.R.; Hiatt, G.; Desai, A.; Penney, R.B.; Jamshidi-Parsian, A.; Griffin, R.J.; Boysen, G. Glutamine drives glutathione synthesis and contributes to radiation sensitivity of A549 and H460 lung cancer cell lines. Biochim. Biophys. Acta. 2016, 1860, 836–843. [Google Scholar] [CrossRef]
- Shestov, A.A.; Lee, S.-C.; Nath, K.; Guo, L.; Nelson, D.S.; Roman, J.C.; Leeper, D.B.; Wasik, M.A.; Blair, I.A.; Glickson, J.D. 13C MRS and LC–MS Flux Analysis of Tumor Intermediary Metabolism. Front. Oncol. 2016, 6, 135. [Google Scholar] [CrossRef]
- van Geldermalsen, M.; Wang, Q.; Nagarajah, R.; Marshall, A.D.; Thoeng, A.; Gao, D.; Ritchie, W.; Feng, Y.; Bailey, C.G.; Deng, N.; et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 2016, 35, 3201–3208. [Google Scholar] [CrossRef]
- Katt, W.P.; Cerione, R.A. Glutaminase regulation in cancer cells: A druggable chain of events. Drug Discov. Today 2014, 19, 450–457. [Google Scholar] [CrossRef]
- Curthoys, N.P.; Watford, M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995, 15, 133–159. [Google Scholar] [CrossRef]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Yu, W.; Yang, X.; Zhang, Q.; Sun, L.; Yuan, S.; Xin, Y. Targeting GLS1 to cancer therapy through glutamine metabolism. Clin. Transl. Oncol. 2021, 23, 2253–2268. [Google Scholar] [CrossRef]
- Elgadi, K.M.; Meguid, R.A.; Qian, M.; Souba, W.W.; Abcouwer, S.F. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genom. 1999, 1, 51–62. [Google Scholar] [CrossRef]
- Cassago, A.; Ferreira, A.P.S.; Ferreira, I.M.; Fornezari, C.; Gomes, E.R.M.; Greene, K.S.; Pereira, H.M.; Garratt, R.C.; Dias, S.M.G.; Ambrosio, A.L.B. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. USA 2012, 109, 1092–1097. [Google Scholar] [CrossRef]
- Matés, J.M.; Campos-Sandoval, J.A.; de Los Santos-Jiménez, J.; Márquez, J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett. 2019, 467, 29–39. [Google Scholar] [CrossRef]
- Matés, J.M.; Campos-Sandoval, J.A.; Márquez, J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2018, 1870, 158–164. [Google Scholar] [CrossRef]
- Choi, Y.K.; Park, K.G. Targeting Glutamine Metabolism for Cancer Treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Stalnecker, C.; Zhang, C.; McDermott, L.A.; Iyer, P.; O’Neill, J.; Reimer, S.; Cerione, R.A.; Katt, W.P. Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism. J. Biol. Chem. 2018, 293, 3535–3545. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Feng, X.; Chen, Y.; Selfridge, J.E.; Gorityala, S.; Du, Z.; Wang, J.M.; Hao, Y.; Cioffi, G.; Conlon, R.A.; et al. 5-fluorouracil enhances the anti-tumor activity of the glutaminase inhibitor CB-839 against PIK3CA-mutant colorectal cancers. Cancer Res. 2020, 80, 4815–4827. [Google Scholar] [CrossRef]
- Cohen, A.S.; Geng, L.; Zhao, P.; Fu, A.; Schulte, M.L.; Graves-Deal, R.; Washington, M.K.; Berlin, J.; Coffey, R.J.; Manning, H.C. Combined blockade of EGFR and glutamine metabolism in preclinical models of colorectal cancer. Transl. Oncol. 2020, 13, 100828. [Google Scholar] [CrossRef]
- Su, C.; Li, M.; Yang, Y.; Wang, Z.; Wang, Q.; Wang, W.; Ma, X.; Jie, R.; Chen, H.; Li, X.; et al. Targeting Glutamine Metabolism through Glutaminase Inhibition Suppresses Cell Proliferation and Progression in Nasopharyngeal Carcinoma. Anti-Cancer Agents Med. Chem. 2023, 23, 1944–1957. [Google Scholar] [CrossRef] [PubMed]
- Gouda, M.A.; Voss, M.H.; Tawbi, H.; Gordon, M.; Tykodi, S.S.; Lam, E.T.; Vaishampayan, U.; Tannir, N.M.; Chaves, J.; Nikolinakos, P.; et al. A phase I/II study of the safety and efficacy of telaglenastat (CB-839) in combination with nivolumab in patients with metastatic melanoma, renal cell carcinoma, and non-small-cell lung cancer. Clinical trial. 2025, 10, 104536. [Google Scholar] [CrossRef] [PubMed]
- Corazao-Rozas, P.; Guerreschi, P.; André, F.; Gabert, P.-E.; Lancel, S.; Dekiouk, S.; Fontaine, D.; Tardivel, M.; Savina, A.; Quesnel, B.; et al. Mitochondrial oxidative phosphorylation controls cancer cell’s life and death decisions upon exposure to MAPK inhibitors. Oncotarget 2016, 7, 39473–39485. [Google Scholar] [CrossRef]
- Carpenter, E.L.; Becker, A.L.; Indra, A.K. NRF2 and Key Transcriptional Targets in Melanoma Redox Manipulation. Cancers 2022, 14, 1531. [Google Scholar] [CrossRef]
- Khamari, R.; Trinh, A.; Gabert, P.E.; Corazao-Rozas, P.; Riveros-Cruz, S.; Balayssac, S.; Malet-Martino, M.; Dekiouk, S.; Joncquel Chevalier Curt, M.; Maboudou, P.; et al. Glucose metabolism and NRF2 coordinate the antioxidant response in melanoma resistant to MAPK inhibitors. Cell Death Dis. 2018, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Araujo, L.F.; Siena, A.D.D.; Plaça, J.R.; Brotto, D.B.; Barros, I.I.; Muys, B.R.; Biagi, C.a.O.; Peronni, K.C.; Sousa, J.F.; Molfetta, G.A.; et al. Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci. Rep. 2018, 8, 14190. [Google Scholar] [CrossRef]
- Soumoy, L.; Genbauffe, A.; Mouchart, L.; Sperone, A.; Trelcat, A.; Mukeba-Harchies, L.; Wells, M.; Blankert, B.; Najem, A.; Ghanem, G.; et al. ATP1A1 is a promising new target for melanoma treatment and can be inhibited by its physiological ligand bufalin to restore targeted therapy efficacy. Cancer Cell Int. 2024, 24, 8. [Google Scholar] [CrossRef]
- Najem, A.; Krayem, M.; Sabbah, S.; Pesetti, M.; Journe, F.; Awada, A.; Désaubry, L.; Ghanem, G.E. Targeting Prohibitins to Inhibit Melanoma Growth and Overcome Resistance to Targeted Therapies. Cells 2023, 12, 1855. [Google Scholar] [CrossRef]
- Varghese, S.; Pramanik, S.; Williams, L.J.; Hodges, H.R.; Hudgens, C.W.; Fischer, G.M.; Luo, C.K.; Knighton, B.; Tan, L.; Lorenzi, P.L.; et al. The glutaminase inhibitor CB-839 (Telaglenastat) enhances the anti-melanoma activity of T cell mediated immunotherapies. Mol. Cancer Ther. 2021, 20, 500–511. [Google Scholar] [CrossRef]
- Tiersma, J.F.; Evers, B.; Bakker, B.M.; Jalving, M.; de Jong, S. Pyruvate Dehydrogenase Kinase Inhibition by Dichloroacetate in Melanoma Cells Unveils Metabolic Vulnerabilities. Int. J. Mol.Sci. 2022, 23, 3745. [Google Scholar] [CrossRef]
- Guo, L.; Zhou, B.; Liu, Z.; Xu, Y.; Lu, H.; Xia, M.; Guo, E.; Shan, W.; Chen, G.; Wang, C. Blockage of glutaminolysis enhances the sensitivity of ovarian cancer cells to PI3K/mTOR inhibition involvement of STAT3 signaling. Tumor. Biol. 2016, 37, 11007–11015. [Google Scholar] [CrossRef] [PubMed]
- Lampa, M.; Arlt, H.; He, T.; Ospina, B.; Reeves, J.; Zhang, B.; Murtie, J.; Deng, G.; Barberis, C.; Hoffmann, D.; et al. Glutaminase is essential for the growth of triple-negative breast cancer cells with a deregulated glutamine metabolism pathway and its suppression synergizes with mTOR inhibition. PLoS ONE 2017, 12, e0185092. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Guan, F.; Li, S.; Zhang, Q.; Zhang, X.; Qin, Y.; Sun, Z.; Peng, S.; Cheng, J.; Li, Y.; et al. Glutaminase potentiates the glycolysis in esophageal squamous cell carcinoma by interacting with PDK1. Mol. Carcinog. 2024, 63, 897–911. [Google Scholar] [CrossRef]
- Qie, S.; Yoshida, A.; Parnham, S.; Oleinik, N.; Beeson, G.C.; Beeson, C.C.; Ogretmen, B.; Bass, A.J.; Wong, K.-K.; Rustgi, A.K.; et al. Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma. Nat. Commun. 2019, 10, 1296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Li, Y.Q.; Wang, D.Y.; Shen, Y.Q. Natural product piperlongumine inhibits proliferation of oral squamous carcinoma cells by inducing ferroptosis and inhibiting intracellular antioxidant capacity. Transl. Cancer Res. 2023, 12, 2911–2922. [Google Scholar] [CrossRef]
- Rodrigues, M.F.; Obre, E.; de Melo, F.H.M.; Santos, G.C.; Galina, A.; Jasiulionis, M.G.; Rossignol, R.; Rumjanek, F.D.; Amoêdo, N.D. Enhanced OXPHOS, glutaminolysis and β-oxidation constitute the metastatic phenotype of melanoma cells. Biochem. J. 2016, 473, 703–715. [Google Scholar] [CrossRef]
- Shah, R.; Singh, S.J.; Eddy, K.; Filipp, F.V.; Chen, S. Concurrent targeting of glutaminolysis and metabotropic glutamate receptor 1 (GRM1) reduces glutamate bioavailability in GRM1+ melanoma. Cancer Res. 2019, 79, 1799–1809. [Google Scholar] [CrossRef]
- Lyons, S.A.; Chung, W.J.; Weaver, A.K.; Ogunrinu, T.; Sontheimer, H. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res. 2007, 67, 9463–9471. [Google Scholar] [CrossRef]
- Namkoong, J.; Shin, S.-S.; Lee, H.J.; Marín, Y.E.; Wall, B.A.; Goydos, J.S.; Chen, S. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007, 67, 2298–2305. [Google Scholar] [CrossRef]
- Wang, E.S.; Frankfurt, O.; Orford, K.W.; Bennett, M.; Flinn, I.W.; Maris, M.; Konopleva, M. Phase 1 Study of CB-839, a First-in-Class, Orally Administered Small Molecule Inhibitor of Glutaminase in Patients with Relapsed/Refractory Leukemia. Blood 2015, 126, 2566. [Google Scholar] [CrossRef]
- Harding, J.J.; Telli, M.L.; Munster, P.N.; Le, M.H.; Molineaux, C.; Bennett, M.K.; Mittra, E.; Burris, H.A.; Clark, A.S.; Dunphy, M.; et al. Safety and tolerability of increasing doses of CB-839, a first-in-class, orally administered small molecule inhibitor of glutaminase, in solid tumors. J. Clin. Oncol. 2015, 33 (Suppl. 15), 2512. [Google Scholar] [CrossRef]
- Kalinsky, K.; Harding, J.; DeMichele, A.; Infante, J.; Gogineni, K.; Owonikoko, T.; Isakoff, S.; Iliopoulos, O.; Patel, M.; Munster, P.; et al. Abstract PD3–13: Phase 1 study of CB-839, a first-in-class oral inhibitor of glutaminase, in combination with paclitaxel in patients with advanced triple negative breast cancer. Cancer Res. 2018, 78 (Suppl. 4), PD3–PD13. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.; Zhao, Y.; Dinh, T.; Jiang, D.; Selfridge, J.E.; Myers, G.; Wang, Y.; Zhao, X.; Tomchuck, S.; et al. Neutrophil extracellular traps induced by chemotherapy inhibit tumor growth in murine models of colorectal cancer. J. Clin. Invest. 2024, 134, e175031. [Google Scholar] [CrossRef] [PubMed]
- Ciombor, K.K.; Bae, S.-W.; Whisenant, J.G.; Ayers, G.D.; Sheng, Q.; Peterson, T.E.; Smith, G.T.; Lin, K.; Chowdhury, S.; Kanikarla Marie, P.; et al. Results of the Phase I/II Study and Preliminary B-cell Gene Signature of Combined Inhibition of Glutamine Metabolism and EGFR in Colorectal Cancer. Clin. Cancer Res. 2025, 31, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Zhang, X.; Ruan, H.; Wang, J.; Bu, X. Long noncoding RNA OIP5-AS1 acts as a competing endogenous RNA to promote glutamine catabolism and malignant melanoma growth by sponging miR-217. J. Cell. Physiol. 2019, 234, 16609–16618. [Google Scholar] [CrossRef]
- Luan, W.; Zhou, Z.; Zhu, Y.; Xia, Y.; Wang, J.; Xu, B. miR-137 inhibits glutamine catabolism and growth of malignant melanoma by targeting glutaminase. Biochem. Biophys. Res. Commun. 2018, 495, 46–52. [Google Scholar] [CrossRef]
- Saha, S.K.; Islam, S.M.R.; Abdullah-Al-Wadud, M.; Islam, S.; Ali, F.; Park, K.S. Multiomics Analysis Reveals that GLS and GLS2 Differentially Modulate the Clinical Outcomes of Cancer. J. Clin. Med. 2019, 8, 355. [Google Scholar] [CrossRef]
- Ying, L.; Cheng, M.; Lu, Y.; Tao, Q.; Chen, X.; Shen, B.; Xiong, F.; Hu, Z.; Wang, D.; Li, X. Glutamine Metabolism Scoring Predicts Prognosis and Therapeutic Resistance in Hepatocellular Carcinoma. Pathol. Oncol. Res. 2021, 27, 1610075. [Google Scholar] [CrossRef] [PubMed]
- Sabbah, M.; Krayem, M.; Najem, A.; Sales, F.; Miller, W.; Del Rincon, S.; Awada, A.; Ghanem, G.E.; Journe, F. Dasatinib Stimulates Its Own Mechanism of Resistance by Activating a CRTC3/MITF/Bcl-2 Pathway in Melanoma with Mutant or Amplified c-Kit. Mol. Cancer Res. 2021, 19, 1221–1233. [Google Scholar] [CrossRef]
- Alonzo, T.A. Standards for reporting prognostic tumor marker studies. J. Clin. Oncol. 2005, 23, 9053–9054. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst. 2005, 97, 1180–1184. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Wahl, R.L.; Jacene, H.; Kasamon, Y.; Lodge, M.A. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. J. Nucl. Med. 2009, 50 (Suppl. 1), 122S–150S. [Google Scholar] [CrossRef] [PubMed]
| A. | ||||||
|---|---|---|---|---|---|---|
| Parameters | N | Median | Range | |||
| Metastatic site (lymph nodes/skin) | 48/36 | |||||
| Gender (female/male) | 50/34 | |||||
| Age at diagnosis of primary melanoma (years) | 84 | 55 | 20–77 | |||
| Stage at sample collection (III/IV) | 54/30 | |||||
| Breslow thickness (<1/≥1) (mm) | 19/65 | 2.6 | 0.3–28.0 | |||
| Ulceration of primary lesion (no/yes) | 29/55 | |||||
| Invaded lymph nodes at primary (0/≥1) | 40/23 | |||||
| Overall survival (years) | 80 | 4.9 | 0.8–45.9 | |||
| Status (alive/dead) | 26/58 | |||||
| B. | ||||||
| Sample Number | Melanoma Type | Metastatic Site | Response to Vemurafenib | Duration of Vemurafenib Treatment (Months) | OS (Months) | OS Status |
| #1 | NM | LN | Responder | 4 | 65 | Dead |
| #2 | SSM | LN | Responder | 9 | 70 | Dead |
| #3 | SSM | LN | Responder | 39 | 101 | Dead |
| #4 | SSM | LN | Responder | 89 | 92 | Alive |
| #5 | SSM | LN | Non-responder | 4 | 23 | Dead |
| #6 | NM | SK | Non-responder | 3 | 79 | Dead |
| #7 | SSM | SK | Non-responder | 4 | 28 | Dead |
| #8 | NM | LN | Non-responder | 3 | 30 | Dead |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soumoy, L.; Genbauffe, A.; Sant’Angelo, D.; Everaert, M.; Mukeba-Harchies, L.; Sarry, J.-E.; Declèves, A.-E.; Journe, F. Therapeutic Potential of Glutaminase Inhibition Targeting Metabolic Adaptations in Resistant Melanomas to Targeted Therapy. Int. J. Mol. Sci. 2025, 26, 8241. https://doi.org/10.3390/ijms26178241
Soumoy L, Genbauffe A, Sant’Angelo D, Everaert M, Mukeba-Harchies L, Sarry J-E, Declèves A-E, Journe F. Therapeutic Potential of Glutaminase Inhibition Targeting Metabolic Adaptations in Resistant Melanomas to Targeted Therapy. International Journal of Molecular Sciences. 2025; 26(17):8241. https://doi.org/10.3390/ijms26178241
Chicago/Turabian StyleSoumoy, Laura, Aline Genbauffe, Dorianne Sant’Angelo, Maude Everaert, Léa Mukeba-Harchies, Jean-Emmanuel Sarry, Anne-Emilie Declèves, and Fabrice Journe. 2025. "Therapeutic Potential of Glutaminase Inhibition Targeting Metabolic Adaptations in Resistant Melanomas to Targeted Therapy" International Journal of Molecular Sciences 26, no. 17: 8241. https://doi.org/10.3390/ijms26178241
APA StyleSoumoy, L., Genbauffe, A., Sant’Angelo, D., Everaert, M., Mukeba-Harchies, L., Sarry, J.-E., Declèves, A.-E., & Journe, F. (2025). Therapeutic Potential of Glutaminase Inhibition Targeting Metabolic Adaptations in Resistant Melanomas to Targeted Therapy. International Journal of Molecular Sciences, 26(17), 8241. https://doi.org/10.3390/ijms26178241






