In Silico Analysis of Possible microRNAs Involved in the Pathogenesis of White-Nose Syndrome in Myotis lucifugus
Abstract
1. Introduction
2. Results
2.1. Putative miRNA Targets Identified by Literature Review
2.2. Pathway Enrichment Analysis of miRNA Targets Using Online Databases
2.3. miRNAs Involved in Fat Metabolism
2.4. miRNAs Involved in Insulin Metabolism
2.5. miRNAs Involved in Skeletal Muscle Maintenance
2.6. miRNAs Involved in Immune System Regulation
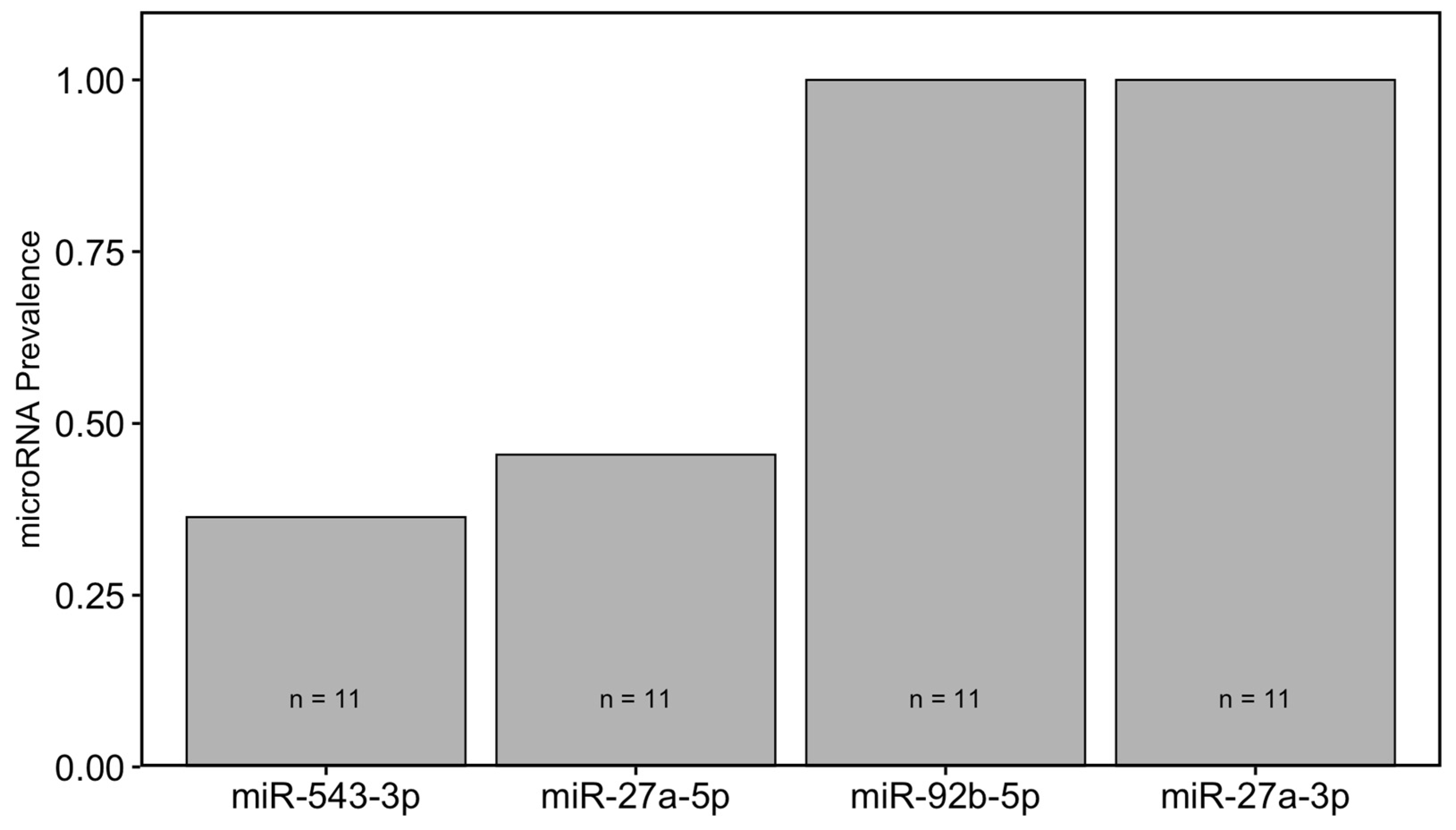
2.7. Prevalence of miRNAs in Blood Plasma of Wild Bats
3. Discussion
4. Materials and Methods
4.1. Literature Search Strategy
4.2. Pathway Enrichment Analysis of miRNA Targets
4.3. Study Site and Animal Collection
4.4. Plasma Sampling
4.5. RNA Extraction and cDNA Synthesis Using Plasma Samples
4.6. Identification of Homologous Sequence for RT-qPCR Primers
4.7. Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging Fungal Threats to Animal, Plant and Ecosystem Health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Blehert, D.S.; Hicks, A.C.; Behr, M.; Meteyer, C.U.; Berlowski-Zier, B.M.; Buckles, E.L.; Coleman, J.T.H.; Darling, S.R.; Gargas, A.; Niver, R.; et al. Bat White-Nose Syndrome: An Emerging Fungal Pathogen? Science 2009, 323, 227. [Google Scholar] [CrossRef]
- Lorch, J.M.; Meteyer, C.U.; Behr, M.J.; Boyles, J.G.; Cryan, P.M.; Hicks, A.C.; Ballmann, A.E.; Coleman, J.T.H.; Redell, D.N.; Reeder, D.M.; et al. Experimental Infection of Bats with Geomyces destructans Causes White-Nose Syndrome. Nature 2011, 480, 376–378. [Google Scholar] [CrossRef]
- Cheng, T.L.; Reichard, J.D.; Coleman, J.T.H.; Weller, T.J.; Thogmartin, W.E.; Reichert, B.E.; Bennett, A.B.; Broders, H.G.; Campbell, J.; Etchison, K.; et al. The Scope and Severity of White-nose Syndrome on Hibernating Bats in North America. Conserv. Biol. 2021, 35, 1586–1597. [Google Scholar] [CrossRef]
- Frick, W.F.; Pollock, J.F.; Hicks, A.C.; Langwig, K.E.; Reynolds, D.S.; Turner, G.G.; Butchkoski, C.M.; Kunz, T.H. An Emerging Disease Causes Regional Population Collapse of a Common North American Bat Species. Science 2010, 329, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Meteyer, C.U.; Buckles, E.L.; Blehert, D.S.; Hicks, A.C.; Green, D.E.; Shearn-Bochsler, V.; Thomas, N.J.; Gargas, A.; Behr, M.J. Histopathologic Criteria to Confirm White-Nose Syndrome in Bats. J. Vet. Diagn. Investig. 2009, 21, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Verant, M.L.; Boyles, J.G.; Waldrep, W.; Wibbelt, G.; Blehert, D.S. Temperature-Dependent Growth of Geomyces destructans, the Fungus That Causes Bat White-Nose Syndrome. PLoS ONE 2012, 7, e46280. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, C.C.; Carey, H.V. Seasonal Changes in the Intestinal Immune System of Hibernating Ground Squirrels. Dev. Comp. Immunol. 2007, 31, 415–428. [Google Scholar] [CrossRef]
- Bouma, H.R.; Carey, H.V.; Kroese, F.G.M. Hibernation: The Immune System at Rest? J. Leukoc. Biol. 2010, 88, 619–624. [Google Scholar] [CrossRef]
- Reeder, S.M.; Palmer, J.M.; Prokkola, J.M.; Lilley, T.M.; Reeder, D.A.M.; Field, K.A. Pseudogymnoascus destructans Transcriptome Changes during White-Nose Syndrome Infections. Virulence 2017, 8, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Field, K.A.; Johnson, J.S.; Lilley, T.M.; Reeder, S.M.; Rogers, E.J.; Behr, M.J.; Reeder, D.M. The White-Nose Syndrome Transcriptome: Activation of Anti-Fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis. PLoS Pathog. 2015, 11, e1005168. [Google Scholar] [CrossRef]
- Ruf, T.; Geiser, F. Daily Torpor and Hibernation in Birds and Mammals. Biol. Rev. 2015, 90, 891–926. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, L.; Turner, J.M.; Bollinger, T.K.; Misra, V.; Cryan, P.M.; Blehert, D.S.; Wibbelt, G.; Willis, C.K.R. Pathophysiology of White-Nose Syndrome in Bats: A Mechanistic Model Linking Wing Damage to Mortality. Biol. Lett. 2013, 9, 20130177. [Google Scholar] [CrossRef] [PubMed]
- Verant, M.L.; Meteyer, C.U.; Speakman, J.R.; Cryan, P.M.; Lorch, J.M.; Blehert, D.S. White-Nose Syndrome Initiates a Cascade of Physiologic Disturbances in the Hibernating Bat Host. BMC Physiol. 2014, 14, 10. [Google Scholar] [CrossRef]
- McGuire, L.P.; Mayberry, H.W.; Willis, C.K.R. White-Nose Syndrome Increases Torpid Metabolic Rate and Evaporative Water Loss in Hibernating Bats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R680–R686. [Google Scholar] [CrossRef]
- Cryan, P.M.; Meteyer, C.U.; Boyles, J.G.; Blehert, D.S. Wing Pathology of White-Nose Syndrome in Bats Suggests Life-Threatening Disruption of Physiology. BMC Biol. 2010, 8, 135. [Google Scholar] [CrossRef]
- Kunz, T.H.; Wrazen, J.A.; Burnett, C.D. Changes in Body Mass and Fat Reserves in Pre-Hibernating Little Brown Bats (Myotis lucifugus). Ecoscience 1998, 5, 8–17. [Google Scholar] [CrossRef]
- McGuire, L.P.; Muise, K.A.; Shrivastav, A.; Willis, C.K.R. No Evidence of Hyperphagia during Prehibernation in a Northern Population of Little Brown Bats (Myotis lucifugus). Can. J. Zool. 2016, 94, 821–827. [Google Scholar] [CrossRef]
- Cheng, T.L.; Gerson, A.; Moore, M.S.; Reichard, J.D.; DeSimone, J.; Willis, C.K.R.; Frick, W.F.; Kilpatrick, A.M. Higher Fat Stores Contribute to Persistence of Little Brown Bat Populations with White-Nose Syndrome. J. Anim. Ecol. 2019, 88, 591–600. [Google Scholar] [CrossRef]
- Langwig, K.E.; Hoyt, J.R.; Parise, K.L.; Frick, W.F.; Foster, J.T.; Kilpatrick, A.M. Resistance in Persisting Bat Populations after White-Nose Syndrome Invasion. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160044. [Google Scholar] [CrossRef]
- Maslo, B.; Valent, M.; Gumbs, J.F.; Frick, W.F. Conservation Implications of Ameliorating Survival of Little Brown Bats with White-nose Syndrome. Ecol. Appl. 2015, 25, 1832–1840. [Google Scholar] [CrossRef]
- Langwig, K.E.; Frick, W.F.; Bried, J.T.; Hicks, A.C.; Kunz, T.H.; Marm Kilpatrick, A. Sociality, Density-Dependence and Microclimates Determine the Persistence of Populations Suffering from a Novel Fungal Disease, White-Nose Syndrome. Ecol. Lett. 2012, 15, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, I.; Bashmakov, Y.; Ikemoto, S.; Horton, J.D.; Brown, M.S.; Goldstein, J.L. Insulin Selectively Increases SREBP-1c MRNA in the Livers of Rats with Streptozotocin-Induced Diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 13656–13661. [Google Scholar] [CrossRef]
- Florant, G.L.; Lawrence, A.K.; Williams, K.; Bauman, W.A. Seasonal Changes in Pancreatic B-Cell Function in Euthermic Yellow-Bellied Marmots. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1985, 249, R159–R165. [Google Scholar] [CrossRef]
- Kronfeld-Schor, N.; Richardson, C.; Silvia, B.A.; Kunz, T.H.; Widmaier, E.P. Dissociation of Leptin Secretion and Adiposity during Prehibernatory Fattening in Little Brown Bats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 279, R1277–R1281. [Google Scholar] [CrossRef] [PubMed]
- Bauman, W.A. Seasonal Changes in Pancreatic Insulin and Glucagon in the Little Brown Bat (Myotis lucifugus). Pancreas 1990, 5, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.F.; Storey, K.B. Differential Expression of Akt, PPARγ, and PGC-1 during Hibernation in Bats. Biochem. Cell Biol. 2003, 81, 269–274. [Google Scholar] [CrossRef]
- Yecies, J.L.; Zhang, H.H.; Menon, S.; Liu, S.; Yecies, D.; Lipovsky, A.I.; Gorgun, C.; Kwiatkowski, D.J.; Hotamisligil, G.S.; Lee, C.-H.; et al. Akt Stimulates Hepatic SREBP1c and Lipogenesis through Parallel MTORC1-Dependent and Independent Pathways. Cell Metab. 2011, 14, 21–32. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and Regulation of Akt/PKB by the Rictor-MTOR Complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Guertin, D.A.; Stevens, D.M.; Thoreen, C.C.; Burds, A.A.; Kalaany, N.Y.; Moffat, J.; Brown, M.; Fitzgerald, K.J.; Sabatini, D.M. Ablation in Mice of the MTORC Components Raptor, Rictor, or MLST8 Reveals That MTORC2 Is Required for Signaling to Akt-FOXO and PKCα, but Not S6K1. Dev. Cell 2006, 11, 859–871. [Google Scholar] [CrossRef]
- Andres-Mateos, E.; Brinkmeier, H.; Burks, T.N.; Mejias, R.; Files, D.C.; Steinberger, M.; Soleimani, A.; Marx, R.; Simmers, J.L.; Lin, B.; et al. Activation of Serum/Glucocorticoid-Induced Kinase 1 (SGK1) Is Important to Maintain Skeletal Muscle Homeostasis and Prevent Atrophy. EMBO Mol. Med. 2013, 5, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, S.F.; Biggar, K.K.; Storey, K.B. Differential Expression of Mature MicroRNAs Involved in Muscle Maintenance of Hibernating Little Brown Bats, Myotis lucifugus: A Model of Muscle Atrophy Resistance. Genom. Proteom. Bioinform. 2012, 10, 295–301. [Google Scholar] [CrossRef]
- Kollias, H.D.; Perry, R.L.S.; Miyake, T.; Aziz, A.; McDermott, J.C. Smad7 Promotes and Enhances Skeletal Muscle Differentiation. Mol. Cell. Biol. 2006, 26, 6248–6260. [Google Scholar] [CrossRef]
- Choi, M.-C.; Cohen, T.J.; Barrientos, T.; Wang, B.; Li, M.; Simmons, B.J.; Yang, J.S.; Cox, G.A.; Zhao, Y.; Yao, T.-P. A Direct HDAC4-MAP Kinase Crosstalk Activates Muscle Atrophy Program. Mol. Cell 2012, 47, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Rapin, N.; Johns, K.; Martin, L.; Warnecke, L.; Turner, J.M.; Bollinger, T.K.; Willis, C.K.R.; Voyles, J.; Misra, V. Activation of Innate Immune-Response Genes in Little Brown Bats (Myotis lucifugus) Infected with the Fungus Pseudogymnoascus destructans. PLoS ONE 2014, 9, e112285. [Google Scholar] [CrossRef]
- Filler, S.G.; Yeaman, M.R.; Sheppard, D.C. Tumor Necrosis Factor Inhibition and Invasive Fungal Infections. Clin. Infect. Dis. 2005, 41, S208–S212. [Google Scholar] [CrossRef]
- Lilley, T.M.; Prokkola, J.M.; Johnson, J.S.; Rogers, E.J.; Gronsky, S.; Kurta, A.; Reeder, D.M.; Field, K.A. Immune Responses in Hibernating Little Brown Myotis (Myotis lucifugus) with White-Nose Syndrome. Proc. R. Soc. B Biol. Sci. 2017, 284, 20162232. [Google Scholar] [CrossRef]
- Wei, W.-C.; Liu, C.-P.; Yang, W.-C.; Shyur, L.-F.; Sheu, J.-H.; Chen, S.-S.; Yang, N.-S. Mammalian Target of Rapamycin Complex 2 (MTORC2) Regulates LPS-Induced Expression of IL-12 and IL-23 in Human Dendritic Cells. J. Leukoc. Biol. 2015, 97, 1071–1080. [Google Scholar] [CrossRef]
- Ohtani, M.; Nagai, S.; Kondo, S.; Mizuno, S.; Nakamura, K.; Tanabe, M.; Takeuchi, T.; Matsuda, S.; Koyasu, S. Mammalian Target of Rapamycin and Glycogen Synthase Kinase 3 Differentially Regulate Lipopolysaccharide-Induced Interleukin-12 Production in Dendritic Cells. Blood 2008, 112, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.E.; Weldon, C.B.; Melnik, L.I.; Duong, B.N.; Collins-Burow, B.M.; Beckman, B.S.; McLachlan, J.A. PI3-K/AKT Regulation of NF-κB Signaling Events in Suppression of TNF-Induced Apoptosis. Biochem. Biophys. Res. Commun. 2000, 271, 342–345. [Google Scholar] [CrossRef]
- Juntilla, M.M.; Wofford, J.A.; Birnbaum, M.J.; Rathmell, J.C.; Koretzky, G.A. Akt1 and Akt2 Are Required for αβ Thymocyte Survival and Differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 12105–12110. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Tili, E.G.; Dose, M.; Haks, M.C.; Bear, S.E.; Maroulakou, I.; Horie, K.; Gaitanaris, G.A.; Fidanza, V.; Ludwig, T.; et al. Unequal Contribution of Akt Isoforms in the Double-Negative to Double-Positive Thymocyte Transition. J. Immunol. 2007, 178, 5443–5453. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-Dependent Human RISC Assembly Pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef]
- Meijer, H.A.; Smith, E.M.; Bushell, M. Regulation of MiRNA Strand Selection: Follow the Leader? Biochem. Soc. Trans. 2014, 42, 1135–1140. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent Deletions and Down-Regulation of Micro-RNA Genes MiR15 and MiR16 at 13q14 in Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and Validation of Plasma MiRNA Biomarker Signature Panel for the Detection of Early HIV-1 Infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Hu, H.; Guo, H.; Xu, P.; Shi, Z.; Huan, X.; Zhu, Z.; Zhou, M.; Cui, L. MicroRNA Profiling in Plasma of HIV-1 Infected Patients: Potential Markers of Infection and Immune Status. J. Public Health Emerg. 2017, 1, 65. [Google Scholar] [CrossRef]
- Gupta, S.K.; Maclean, P.H.; Ganesh, S.; Shu, D.; Buddle, B.M.; Wedlock, D.N.; Heiser, A. Detection of MicroRNA in Cattle Serum and Their Potential Use to Diagnose Severity of Johne’s Disease. J. Dairy Sci. 2018, 101, 10259–10270. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, R.; Zhang, G.; Zhai, Z.; Deng, Y.; Li, J.; Cui, S. Analysis of the MicroRNA Expression Profiles in Feline Kidney Cell Line Infected with Feline Panleukopenia Virus. Infect. Genet. Evol. 2019, 75, 103945. [Google Scholar] [CrossRef]
- Luu, B.E.; Lefai, E.; Giroud, S.; Swenson, J.E.; Chazarin, B.; Gauquelin-Koch, G.; Arnemo, J.M.; Evans, A.L.; Bertile, F.; Storey, K.B. MicroRNAs Facilitate Skeletal Muscle Maintenance and Metabolic Suppression in Hibernating Brown Bears. J. Cell. Physiol. 2020, 235, 3984–3993. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Storey, K.B. Identification and Expression of MicroRNA in the Brain of Hibernating Bats, Myotis lucifugus. Gene 2014, 544, 67–74. [Google Scholar] [CrossRef]
- Iwanowicz, D.D.; Iwanowicz, L.R.; Hitt, N.P.; King, T.L. Differential Expression Profiles of MicroRNA in the Little Brown Bat (Myotis lucifugus) Associated with White Nose Syndrome Affected and Unaffected Individuals; Open-File Report; US Geological Survey: Reston, VA, USA, 2013. [Google Scholar]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A Brief Review on the Mechanisms of MiRNA Regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of MicroRNA-Mediated Gene Regulation in Animal Cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef]
- Hwang, H.; Chang, H.R.; Baek, D. Determinants of Functional MicroRNA Targeting. Mol. Cells 2023, 46, 21–32. [Google Scholar] [CrossRef]
- Tsukerman, P.; Enk, J.; Mandelboim, O. Metastamir-Mediated Immune Evasion. Oncoimmunology 2013, 2, e22245. [Google Scholar] [CrossRef] [PubMed]
- Tsukerman, P.; Stern-Ginossar, N.; Gur, C.; Glasner, A.; Nachmani, D.; Bauman, Y.; Yamin, R.; Vitenshtein, A.; Stanietsky, N.; Bar-Mag, T.; et al. MiR-10b Downregulates the Stress-Induced Cell Surface Molecule MICB, a Critical Ligand for Cancer Cell Recognition by Natural Killer Cells. Cancer Res. 2012, 72, 5463–5472. [Google Scholar] [CrossRef]
- Singh, R.; Ha, S.E.; Wei, L.; Jin, B.; Zogg, H.; Poudrier, S.M.; Jorgensen, B.G.; Park, C.; Ronkon, C.F.; Bartlett, A.; et al. MiR-10b-5p Rescues Diabetes and Gastrointestinal Dysmotility. Gastroenterology 2021, 160, 1662–1678.e18. [Google Scholar] [CrossRef]
- Hou, R.; Wang, D.; Lu, J. MicroRNA-10b Inhibits Proliferation, Migration and Invasion in Cervical Cancer Cells via Direct Targeting of Insulin-like Growth Factor-1 Receptor. Oncol. Lett. 2017, 13, 5009–5015. [Google Scholar] [CrossRef]
- Aryal, B.; Singh, A.K.; Rotllan, N.; Price, N.; Fernández-Hernando, C. MicroRNAs and Lipid Metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef]
- Wang, L.; Sinnott-Armstrong, N.; Wagschal, A.; Wark, A.R.; Camporez, J.-P.; Perry, R.J.; Ji, F.; Sohn, Y.; Oh, J.; Wu, S.; et al. A MicroRNA Linking Human Positive Selection and Metabolic Disorders. Cell 2020, 183, 684–701.e14. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Lane, T.; Venugopal, R.; Parthasarathy, P.T.; Cho, Y.; Galam, L.; Lockey, R.; Kolliputi, N. MicroRNA-133a-1 Regulates Inflammasome Activation through Uncoupling Protein-2. Biochem. Biophys. Res. Commun. 2013, 439, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Hu, M.; Zhang, R.; Shen, Z.; Flatow, L.; You, M. MicroRNA-217 Promotes Ethanol-Induced Fat Accumulation in Hepatocytes by Down-Regulating SIRT1. J. Biol. Chem. 2012, 287, 9817–9826. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, J.; He, Q.; Chao, Z.; Li, X.; Chen, M. MicroRNA 217 Is Involved in the Progression of Atherosclerosis through Regulating Inflammatory Responses by Targeting Sirtuin 1. Mol. Med. Rep. 2019, 20, 3182–3190. [Google Scholar] [CrossRef]
- Tay, H.L.; Kaiko, G.E.; Plank, M.; Li, J.; Maltby, S.; Essilfie, A.-T.; Jarnicki, A.; Yang, M.; Mattes, J.; Hansbro, P.M.; et al. Antagonism of MiR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-Typeable Haemophilus influenzae (NTHi) from Infected Lung. PLoS Pathog. 2015, 11, e1004549. [Google Scholar] [CrossRef]
- Guo, L.; Qiu, Z.; Wei, L.; Yu, X.; Gao, X.; Jiang, S.; Tian, H.; Jiang, C.; Zhu, D. The MicroRNA-328 Regulates Hypoxic Pulmonary Hypertension by Targeting at Insulin Growth Factor 1 Receptor and L-Type Calcium Channel-α1C. Hypertension 2012, 59, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kang, L.; Han, J.; Wang, Y.; Shen, C.; Yan, Z.; Tai, Y.; Zhao, C. MiR-342-3p Suppresses Hepatocellular Carcinoma Proliferation through Inhibition of IGF-1R-Mediated Warburg Effect. Onco Targets Ther. 2018, 11, 1643–1653. [Google Scholar] [CrossRef]
- Galleggiante, V.; De Santis, S.; Liso, M.; Verna, G.; Sommella, E.; Mastronardi, M.; Campiglia, P.; Chieppa, M.; Serino, G. Quercetin-Induced MiR-369-3p Suppresses Chronic Inflammatory Response Targeting C/EBP-β. Mol. Nutr. Food Res. 2019, 63, 1801390. [Google Scholar] [CrossRef]
- Peng, J.; Wu, Y.; Deng, Z.; Zhou, Y.; Song, T.; Yang, Y.; Zhang, X.; Xu, T.; Xia, M.; Cai, A.; et al. MiR-377 Promotes White Adipose Tissue Inflammation and Decreases Insulin Sensitivity in Obesity via Suppression of Sirtuin-1 (SIRT1). Oncotarget 2017, 8, 70550–70563. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, C.; Zhao, H.; Liu, G.; Mao, H.; Liu, Y. Elevated Linc00936 or Silenced MicroRNA-425-3p Inhibits Immune Escape of Gastric Cancer Cells via Elevation of ZC3H12A. Int. Immunopharmacol. 2021, 95, 107559. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, Y.; Li, Y.; Zhao, D.; Ma, L.; Tan, L. MiR-483 Is Down-Regulated in Polycystic Ovarian Syndrome and Inhibits KGN Cell Proliferation via Targeting Insulin-Like Growth Factor 1 (IGF1). Med. Sci. Monit. 2016, 22, 3383–3393. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; Visone, R.; Veronese, A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers 2018, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Kuang, W.; Hao, Y.; Zhang, D.; Lei, M.; Du, L.; Jiao, H.; Zhang, X.; Wang, F. Downregulation of MiR-27a* and MiR-532-5p and Upregulation of MiR-146a and MiR-155 in LPS-Induced RAW264.7 Macrophage Cells. Inflammation 2012, 35, 1308–1313. [Google Scholar] [CrossRef]
- Ji, J.; Zhang, J.; Huang, G.; Qian, J.; Wang, X.; Mei, S. Over-Expressed MicroRNA-27a and 27b Influence Fat Accumulation and Cell Proliferation during Rat Hepatic Stellate Cell Activation. FEBS Lett. 2009, 583, 759–766. [Google Scholar] [CrossRef]
- Kim, T.-D.; Lee, S.U.; Yun, S.; Sun, H.-N.; Lee, S.H.; Kim, J.W.; Kim, H.M.; Park, S.-K.; Lee, C.W.; Yoon, S.R.; et al. Human MicroRNA-27a* Targets Prf1 and GzmB Expression to Regulate NK-Cell Cytotoxicity. Blood 2011, 118, 5476–5486. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, J.; Xu, B.; Chrusciel, M.; Gao, J.; Bazert, M.; Stelmaszewska, J.; Xu, Y.; Zhang, H.; Pawelczyk, L.; et al. Functional Characterization of MicroRNA-27a-3p Expression in Human Polycystic Ovary Syndrome. Endocrinology 2018, 159, 297–309. [Google Scholar] [CrossRef]
- Cao, Y.; Li, R.; Du, Y.; Jin, N.; Fang, T.; Ma, F.; Jin, P. MiR-92b-5p Negatively Regulates IKK through Targeting Its ORF Region in the Innate Immune Responses of Amphioxus (Branchiostoma belcheri). Dev. Comp. Immunol. 2023, 138, 104556. [Google Scholar] [CrossRef]
- Shen, Z.; Yu, Y.; Yang, Y.; Xiao, X.; Sun, T.; Chang, X.; Tang, W.; Zhu, Y.; Han, X. MiR-25 and MiR-92b Regulate Insulin Biosynthesis and Pancreatic β-Cell Apoptosis. Endocrine 2022, 76, 526–535. [Google Scholar] [CrossRef]
- Li, H.-S.; Ning, Y.; Li, S.-B.; Shao, P.-Y.; Chen, S.-J.; Ye, Q.; Heng, X. Expression and Clinical Significance of MiR-181a and MiR-203 in Systemic Lupus Erythematosus Patients. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4790–4796. [Google Scholar]
- Chang, H.; Wang, X.; Yang, S. MiR-350-3p Contributes to Age-Associated Impairment of IL-6 Production by Macrophages. Immunol. Investig. 2018, 47, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Li, X. MiR-375, a MicroRNA Related to Diabetes. Gene 2014, 533, 1–4. [Google Scholar] [CrossRef]
- Hu, F.; Tong, J.; Deng, B.; Zheng, J.; Lu, C. MiR-495 Regulates Macrophage M1/M2 Polarization and Insulin Resistance in High-Fat Diet-Fed Mice via Targeting FTO. Pflug. Arch. 2019, 471, 1529–1537. [Google Scholar] [CrossRef]
- Pan, C.; Xiang, L.; Pan, Z.; Wang, X.; Li, J.; Zhuge, L.; Fang, P.; Xie, Q.; Hu, X. MiR-544 Promotes Immune Escape through Downregulation of NCR1/NKp46 via Targeting RUNX3 in Liver Cancer. Cancer Cell Int. 2018, 18, 52. [Google Scholar] [CrossRef]
- Lee, J.; Hong, B.S.; Ryu, H.S.; Lee, H.-B.; Lee, M.; Park, I.A.; Kim, J.; Han, W.; Noh, D.-Y.; Moon, H.-G. Transition into Inflammatory Cancer-Associated Adipocytes in Breast Cancer Microenvironment Requires MicroRNA Regulatory Mechanism. PLoS ONE 2017, 12, e0174126. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Liu, J.; Wu, P.; Gong, Y.; Xu, X.; Li, W. The Key MicroRNA on Lipid Droplet Formation during Adipogenesis from Human Mesenchymal Stem Cells. J. Cell. Physiol. 2020, 235, 328–338. [Google Scholar] [CrossRef]
- Hu, X.; Chi, L.; Zhang, W.; Bai, T.; Zhao, W.; Feng, Z.; Tian, H. Down-Regulation of the MiR-543 Alleviates Insulin Resistance through Targeting the SIRT1. Biochem. Biophys. Res. Commun. 2015, 468, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Liu, J.; Zhou, J.; Zhang, X.; Li, Z. MiR-27a-5p Inhibits Malignant Progression of Differentiated Thyroid Cancer by Directly Affecting the MiR-27a-5p/SREBP1 Axis. J. Endocrinol. Investig. 2025, 48, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, B.; Yang, Y.; Niu, F.; Lin, C.; Yuan, H.; Wang, J.; Wu, T.; Shao, Y.; Shao, S.; et al. Visceral Adipocyte–Derived Extracellular Vesicle MiR-27a-5p Elicits Glucose Intolerance by Inhibiting Pancreatic β-Cell Insulin Secretion. Diabetes 2024, 73, 1832–1847. [Google Scholar] [CrossRef]
- Choo, K.B.; Soon, Y.L.; Nguyen, P.N.N.; Hiew, M.S.Y.; Huang, C.-J. MicroRNA-5p and -3p Co-Expression and Cross-Targeting in Colon Cancer Cells. J. Biomed. Sci. 2014, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.-L.; Yan, T.-T.; Shen, C.-Q.; Tang, J.-Y.; Kong, X.; Wang, Y.-C.; Chen, J.; Liu, Q.; He, J.; Zhong, M.; et al. The Distinct Role of Strand-Specific MiR-514b-3p and MiR-514b-5p in Colorectal Cancer Metastasis. Cell Death Dis. 2018, 9, 687. [Google Scholar] [CrossRef]
- Guo, Z.-T.; Yu, Q.; Miao, C.; Ge, W.; Li, P. MicroRNA-27a-5p Inhibits Proliferation, Migration, and Invasion and Promotes Apoptosis of Wilms’ Tumor Cell by Targeting PBOV1. Mol. Cell. Biol. 2022, 42, e0039721. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, Z.; Yang, G.; You, L.; Zhang, T.; Zhao, Y. MicroRNA-27a (MiR-27a) in Solid Tumors: A Review Based on Mechanisms and Clinical Observations. Front. Oncol. 2019, 9, 893. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a Promotes Insulin Resistance and Mediates Glucose Metabolism by Targeting PPAR-γ-Mediated PI3K/AKT Signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef]
- Zheng, L.; Lv, G.; Sheng, J.; Yang, Y. Effect of MiRNA-10b in Regulating Cellular Steatosis Level by Targeting PPAR-α Expression, a Novel Mechanism for the Pathogenesis of NAFLD. J. Gastroenterol. Hepatol. 2010, 25, 156–163. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. MiR-27a Is a Negative Regulator of Adipocyte Differentiation via Suppressing PPARγ Expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Urstad, A.P.L.; Semenkovich, C.F. Fatty Acid Synthase and Liver Triglyceride Metabolism: Housekeeper or Messenger? Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Hudgins, L.C.; Hellerstein, M.; Seidman, C.; Neese, R.; Diakun, J.; Hirsch, J. Human Fatty Acid Synthesis Is Stimulated by a Eucaloric Low Fat, High Carbohydrate Diet. J. Clin. Investig. 1996, 97, 2081–2091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, W.; Zhou, M.; Tang, Y. MicroRNA-27a Regulates Hepatic Lipid Metabolism and Alleviates NAFLD via Repressing FAS and SCD1. Sci. Rep. 2017, 7, 14493. [Google Scholar] [CrossRef]
- Lin, X.; Luo, J.; Zhang, L.; Wang, W.; Shi, H.; Zhu, J. MiR-27a Suppresses Triglyceride Accumulation and Affects Gene mRNA Expression Associated with Fat Metabolism in Dairy Goat Mammary Gland Epithelial Cells. Gene 2013, 521, 15–23. [Google Scholar] [CrossRef]
- Wu, H.; Pula, T.; Tews, D.; Amri, E.-Z.; Debatin, K.-M.; Wabitsch, M.; Fischer-Posovszky, P.; Roos, J. MicroRNA-27a-3p but Not -5p Is a Crucial Mediator of Human Adipogenesis. Cells 2021, 10, 3205. [Google Scholar] [CrossRef]
- Wakil, S.J.; Abu-Elheiga, L.A. Fatty Acid Metabolism: Target for Metabolic Syndrome. J. Lipid. Res. 2009, 50, S138–S143. [Google Scholar] [CrossRef]
- Li, B.; Huang, X.; Yang, C.; Ge, T.; Zhao, L.; Zhang, X.; Tian, L.; Zhang, E. MiR-27a Regulates Sheep Adipocyte Differentiation by Targeting CPT1B Gene. Animals 2021, 12, 28. [Google Scholar] [CrossRef]
- Tang, H.; Xu, X.; Xiao, W.; Liao, Y.; Xiao, X.; Li, L.; Li, K.; Jia, X.; Feng, H. Silencing of MicroRNA-27a Facilitates Autophagy and Apoptosis of Melanoma Cells through the Activation of the SYK-dependent MTOR Signaling Pathway. J. Cell. Biochem. 2019, 120, 13262–13274. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Ruan, J.; Zheng, W.; Yang, Z.; Pan, W. Long Non-Coding RNA OIP5-AS1 Suppresses Multiple Myeloma Progression by Sponging MiR-27a-3p to Activate TSC1 Expression. Cancer Cell Int. 2020, 20, 155. [Google Scholar] [CrossRef]
- Barisciano, G.; Colangelo, T.; Rosato, V.; Muccillo, L.; Taddei, M.L.; Ippolito, L.; Chiarugi, P.; Galgani, M.; Bruzzaniti, S.; Matarese, G.; et al. MiR-27a Is a Master Regulator of Metabolic Reprogramming and Chemoresistance in Colorectal Cancer. Br. J. Cancer 2020, 122, 1354–1366. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, M.; Wang, Y.; Liu, A.; Lv, M.; Li, Y.; Yang, X.; Wu, Z. Neuroprotective Effects of MiR-27a against Traumatic Brain Injury via Suppressing FoxO3a-Mediated Neuronal Autophagy. Biochem. Biophys. Res. Commun. 2017, 482, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Ren, F.; Li, L.; Liu, D.; Li, Y.; Wang, A.; Li, W.; Dong, Y.; Guo, W. MiR-328-3p Inhibits Cell Proliferation and Metastasis in Colorectal Cancer by Targeting Girdin and Inhibiting the PI3K/Akt Signaling Pathway. Exp. Cell Res. 2020, 390, 111939. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Yao, M.; Wang, Y.; Zheng, W.; Liu, S.; Hou, Z.; Cheng, X.; Sun, S.; Li, T.; Zhao, H.; et al. Fatty Acid β-Oxidation Promotes Breast Cancer Stemness and Metastasis via the MiRNA-328-3p-CPT1A Pathway. Cancer Gene Ther. 2022, 29, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wu, X.-L. MiR-328-3p Promotes TGF-β1-Induced Proliferation, Migration, and Inflammation of Airway Smooth Muscle Cells by Regulating the PTEN/Akt Pathway. Allergol. Immunopathol. 2023, 51, 151–159. [Google Scholar] [CrossRef]
- Yan, T.; Ye, X.-X. MicroRNA-328-3p Inhibits the Tumorigenesis of Bladder Cancer through Targeting ITGA5 and Inactivating PI3K/AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5139–5148. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Wu, P.; Zhou, Y.; Long, K.; Wang, T. MiR-328-5p Inhibits the Adipogenic Differentiation of HMSCs by Targeting Fatty Acid Synthase. Folia Histochem. Cytobiol. 2022, 60, 292–300. [Google Scholar] [CrossRef]
- Chakrabarti, P.; English, T.; Karki, S.; Qiang, L.; Tao, R.; Kim, J.; Luo, Z.; Farmer, S.R.; Kandror, K.V. SIRT1 Controls Lipolysis in Adipocytes via FOXO1-Mediated Expression of ATGL. J. Lipid. Res. 2011, 52, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, G.; Wang, X.; Li, L.; Li, Y.; Xiang, J.; Kang, L.; Liang, Z. MicroRNA-92b in the Skeletal Muscle Regulates Exercise Capacity via Modulation of Glucose Metabolism. J. Cachexia Sarcopenia Muscle 2023, 14, 2925–2938. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Heo, J.; Kang, H. MiR-92b-3p-TSC1 Axis Is Critical for MTOR Signaling-Mediated Vascular Smooth Muscle Cell Proliferation Induced by Hypoxia. Cell Death Differ. 2019, 26, 1782–1795. [Google Scholar] [CrossRef]
- Li, M.; Shan, W.; Hua, Y.; Chao, F.; Cui, Y.; Lv, L.; Dou, X.; Bian, X.; Zou, J.; Li, H.; et al. Exosomal MiR-92b-3p Promotes Chemoresistance of Small Cell Lung Cancer Through the PTEN/AKT Pathway. Front. Cell Dev. Biol. 2021, 9, 661602. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, G.; Zhu, Y.; Xu, Q.; Zhou, H.; Xu, K.; Huang, K.; Zhan, R.; Pan, J. Foxo1-Induced MiR-92b Down-regulation Promotes Blood-Brain Barrier Damage after Ischaemic Stroke by Targeting NOX4. J. Cell. Mol. Med. 2021, 25, 5269–5282. [Google Scholar] [CrossRef]
- Lin, X.L.; Zhu, J.; Wang, L.M.; Yan, F.; Sha, W.P.; Yang, H.L. MiR-92b-5p Inhibitor Suppresses IL-18 Mediated Inflammatory Amplification after Spinal Cord Injury via IL-18BP up-Regulation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1891–1898. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MiRPath v3.0: Deciphering MicroRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Kostoulas, N.; Vergoulis, T.; Georgakilas, G.; Reczko, M.; Maragkakis, M.; Paraskevopoulou, M.D.; Prionidis, K.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA MiRPath v. 2.0: Investigating the Combinatorial Effect of MicroRNAs in Pathways. Nucleic Acids Res. 2012, 40, W498–W504. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MicroT Web Server v5.0: Service Integration into MiRNA Functional Analysis Workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.M.; Baek, D.; Shin, C.; Bell, G.W.; Grimson, A.; Bartel, D.P. Weak Seed-Pairing Stability and High Target-Site Abundance Decrease the Proficiency of Lsy-6 and Other MicroRNAs. Nat. Struct. Mol. Biol. 2011, 18, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.-L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing More than Half a Million Experimentally Supported MiRNA:MRNA Interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Canadian Wildlife Health Cooperative. Canadian National White-Nose Syndrome Decontamination Protocol. 2024. Available online: https://www.cwhc-rcsf.ca/docs/bat_health/WNS%20Decontamination%20protocol%20(English)%2020240306.pdf (accessed on 17 August 2025).
- U.S. Fish and Wildlife Service. National White-Nose Syndrome Decontamination Protocol. 2020. Available online: https://www.whitenosesyndrome.org/mmedia-education/united-states-national-white-nose-syndrome-decontamination-protocol-april-2016-2 (accessed on 17 August 2025).
- Canadian Council on Animal Care. Bats. 2009. Available online: https://ccac.ca/Documents/Standards/Guidelines/Vol2/bats.pdf (accessed on 17 August 2025).
- Davis, W.H.; Hitchcock, H.B. Biology and Migration of the Bat, Myotis lucifugus, in New England. J. Mammal. 1965, 46, 296. [Google Scholar] [CrossRef]
- Kunz, T.H.; Anthony, E.L.P. Age Estimation and Post-Natal Growth in the Bat Myotis lucifugus. J. Mammal. 1982, 63, 23–32. [Google Scholar] [CrossRef]
- Baloun, D.E.; Webber, Q.M.R.; McGuire, L.P.; Boyles, J.G.; Shrivastav, A.; Willis, C.K.R. Testing the “Fasting While Foraging” Hypothesis: Effects of Recent Feeding on Plasma Metabolite Concentrations in Little Brown Bats (Myotis lucifugus). Physiol. Biochem. Zool. 2019, 92, 373–380. [Google Scholar] [CrossRef]
- Sweeney, B.A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, S.E.; Cannone, J.J.; et al. RNAcentral: A Hub of Information for Non-Coding RNA Sequences. Nucleic Acids Res. 2019, 47, D221–D229. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating High Confidence MicroRNAs Using Deep Sequencing Data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]

| miRNAs | Immune System Regulation | Lipid Metabolism | Insulin Signaling Pathway |
|---|---|---|---|
| miR-101c | No | No | No |
| miR-10b | Yes [58,59] | Yes [60] | Yes [61] |
| miR-28b | No | No | No |
| miR-128-1 | No | Yes [62,63] | Yes [63] |
| miR-133a-1//miR-133a-2 | Yes [64] | No | No |
| miR-135b | No | No | No |
| miR-181d | No | No | No |
| miR-202 | No | No | No |
| miR-217 | No | Yes [65,66] | No |
| miR-302a | No | No | No |
| miR-328 | Yes [67] | No | Yes [68] |
| miR-342 | No | No | Yes [69] |
| miR-369 | Yes [70] | No | No |
| miR-377 | No | No | Yes [71] |
| miR-425 | No | Yes [72] | No |
| miR-483 | No | No | Yes [73,74] |
| miR-501 | No | No | No |
| miR-675 | No | No | No |
| miR-1306 | No | No | No |
| miR-1957 | No | No | No |
| miR-3967 | No | No | No |
| miR-27a | Yes [75] | Yes [76,77] | Yes [78] |
| miR-92b | Yes [79] | No | Yes [80] |
| miR-132 | No | No | No |
| miR-135a-1//miR-135a-2 | No | No | No |
| miR-154 | No | No | No |
| miR-1843b | No | No | No |
| miR-203 | Yes [81] | No | No |
| miR-219-1 | No | No | No |
| miR-3096 | No | No | No |
| miR-340 | No | No | No |
| miR-350 | Yes [82] | No | No |
| miR-375 | No | No | Yes [83] |
| miR-412 | No | No | No |
| miR-450a-1//miR-450a-2 | No | No | No |
| miR-495 | No | No | Yes [84] |
| miR-539 | No | No | No |
| miR-544 | Yes [85] | No | No |
| miR-758 | No | No | No |
| miR-1895 | No | No | No |
| miR-3966 | No | No | No |
| miR-5112 | Yes [86] | No | No |
| miR-543 | No | Yes [87] | Yes [88] |
| Pathway | p-Value | Number of Genes Regulated by | |||||||
|---|---|---|---|---|---|---|---|---|---|
| mmu-miR-27a-3p | mmu-miR-27a-5p | mmu-miR-328-3p | mmu-miR-328-5p | mmu-miR-92b-3p | mmu-miR-92b-5p | mmu-miR-543-3p | mmu-miR-543-5p | ||
| Fatty acid biosynthesis | <0.001 | 2 | 1 | ||||||
| Fatty acid metabolism | <0.001 | 10 | 2 | 1 | 1 | ||||
| Fatty acid elongation | <0.001 | 1 | |||||||
| FoxO signaling pathway | <0.001 | 31 | 21 | ||||||
| Lysine degradation | <0.001 | 13 | |||||||
| Biosynthesis of unsaturated fatty acids | <0.01 | 2 | |||||||
| PI3K-Akt signaling pathway | 0.02 | 19 | |||||||
| T-cell receptor signaling pathway | 0.31 | 30 | |||||||
| Fc gamma R-mediated phagocytosis | 0.31 | 24 | 9 | ||||||
| Bacterial invasion of epithelial cells | 0.35 | 22 | 8 | ||||||
| Fatty acid degradation | 0.45 | 1 | |||||||
| mTOR signaling pathway | 0.62 | 18 | |||||||
| B-cell receptor signaling pathway | 0.76 | 18 | |||||||
| Pathway | p-Value | Number of Genes Regulated by | ||||||
|---|---|---|---|---|---|---|---|---|
| hsa-miR-27a-3p | hsa-miR-27a-5p | hsa-miR-328-3p | hsa-miR-328-5p | hsa-miR-92b-3p | hsa-miR-92b-5p | hsa-miR-10b-5p | ||
| Fatty acid metabolism | <0.001 | 8 | 3 | 3 | 1 | 1 | ||
| Lysine degradation | <0.001 | 14 | 8 | |||||
| Fatty acid degradation | <0.001 | 2 | 1 | |||||
| Bacterial invasion of epithelial cells | <0.001 | 31 | 10 | |||||
| FoxO signaling pathway | <0.01 | 38 | 19 | |||||
| Insulin signaling pathway | 0.006 | 41 | 9 | 11 | ||||
| Fatty acid biosynthesis | 0.00 | 3 | 2 | 1 | ||||
| mTOR signaling | 0.39 | 22 | ||||||
| Fatty acid elongation | 0.09 | 1 | ||||||
| Insulin secretion | 0.92 | 14 | ||||||
| PI3K-Akt signaling pathway | 0.93 | 48 | ||||||
| Sequence Reported in Myotis lucifugus | Sequence by miRBase | miRNA |
|---|---|---|
| TACCCTGTAGAACCGAATTTGTG | UACCCUGUAGAACCGAAUUUGUG | hsa-miR-10b-5p |
| AGGGCTTAGCTGCTTGTGAGCA | AGGGCUUAGCUGCUUGUGAGCA | hsa-miR-27a-5p |
| AGGGACGGGACGTGGTGCAGTGTT | AGGGACGGGACGUGGUGCAGUGUU | mmu-miR-92b-5p |
| GGGGGGCAGGAGGGGCTCAGGG | GGGGGGCAGGAGGGGCUCAGGG | mmu-miR-328-5p |
| AAGTTGCCCGCGTGTTTTTCG | AAGUUGCCCGCGUGUUUUUCG | mmu-miR-543-5p |
| CGGGGCCGTAGCACTGTCTGA | CGGGGCCGUAGCACUGUCUGA | mmu-miR-128-1-5p |
| Sequence Used for Primer Design | Target miRNA |
|---|---|
| AAACAUUCGCGGUGCACUUCUU | mmu-miR-543-3p |
| AGGGCUUAGCUGCUUGUGAGCA | hsa-miR-27a-5p |
| AGGGACGGGACGCGGUGCAGUG | hsa-miR-92b-5p |
| UUCACAGUGGCUAAGUUCCGC | hsa-miR-27a-3p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, A.; Willis, C.K.R.; Shrivastav, A. In Silico Analysis of Possible microRNAs Involved in the Pathogenesis of White-Nose Syndrome in Myotis lucifugus. Int. J. Mol. Sci. 2025, 26, 8200. https://doi.org/10.3390/ijms26178200
Agarwal A, Willis CKR, Shrivastav A. In Silico Analysis of Possible microRNAs Involved in the Pathogenesis of White-Nose Syndrome in Myotis lucifugus. International Journal of Molecular Sciences. 2025; 26(17):8200. https://doi.org/10.3390/ijms26178200
Chicago/Turabian StyleAgarwal, Anouska, Craig K. R. Willis, and Anuraag Shrivastav. 2025. "In Silico Analysis of Possible microRNAs Involved in the Pathogenesis of White-Nose Syndrome in Myotis lucifugus" International Journal of Molecular Sciences 26, no. 17: 8200. https://doi.org/10.3390/ijms26178200
APA StyleAgarwal, A., Willis, C. K. R., & Shrivastav, A. (2025). In Silico Analysis of Possible microRNAs Involved in the Pathogenesis of White-Nose Syndrome in Myotis lucifugus. International Journal of Molecular Sciences, 26(17), 8200. https://doi.org/10.3390/ijms26178200






