Integrative Analysis of Fungal and Bacterial Microbiomes Across Skin, Blood, and Stool in Rosacea Patients
Abstract
1. Introduction
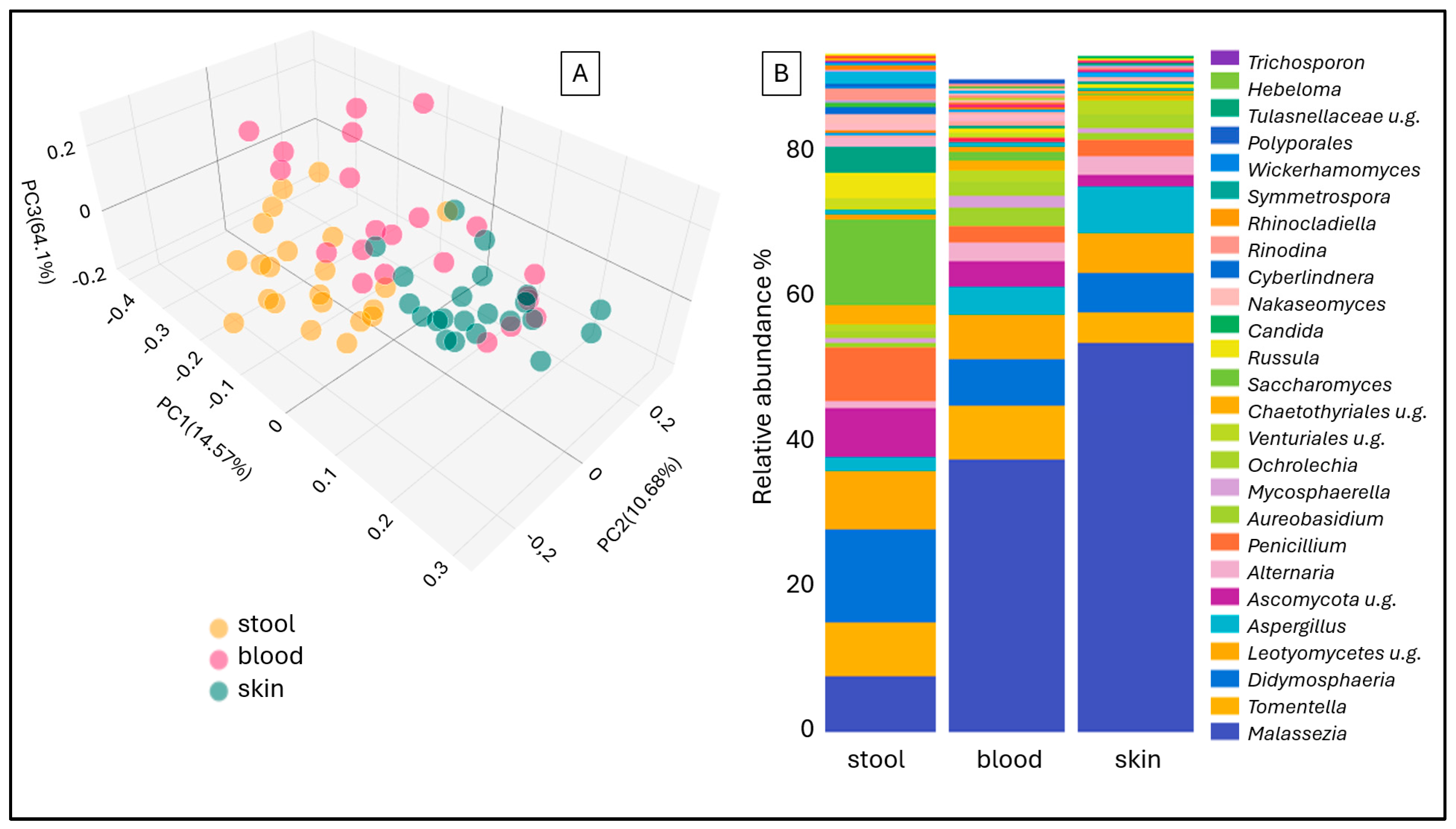
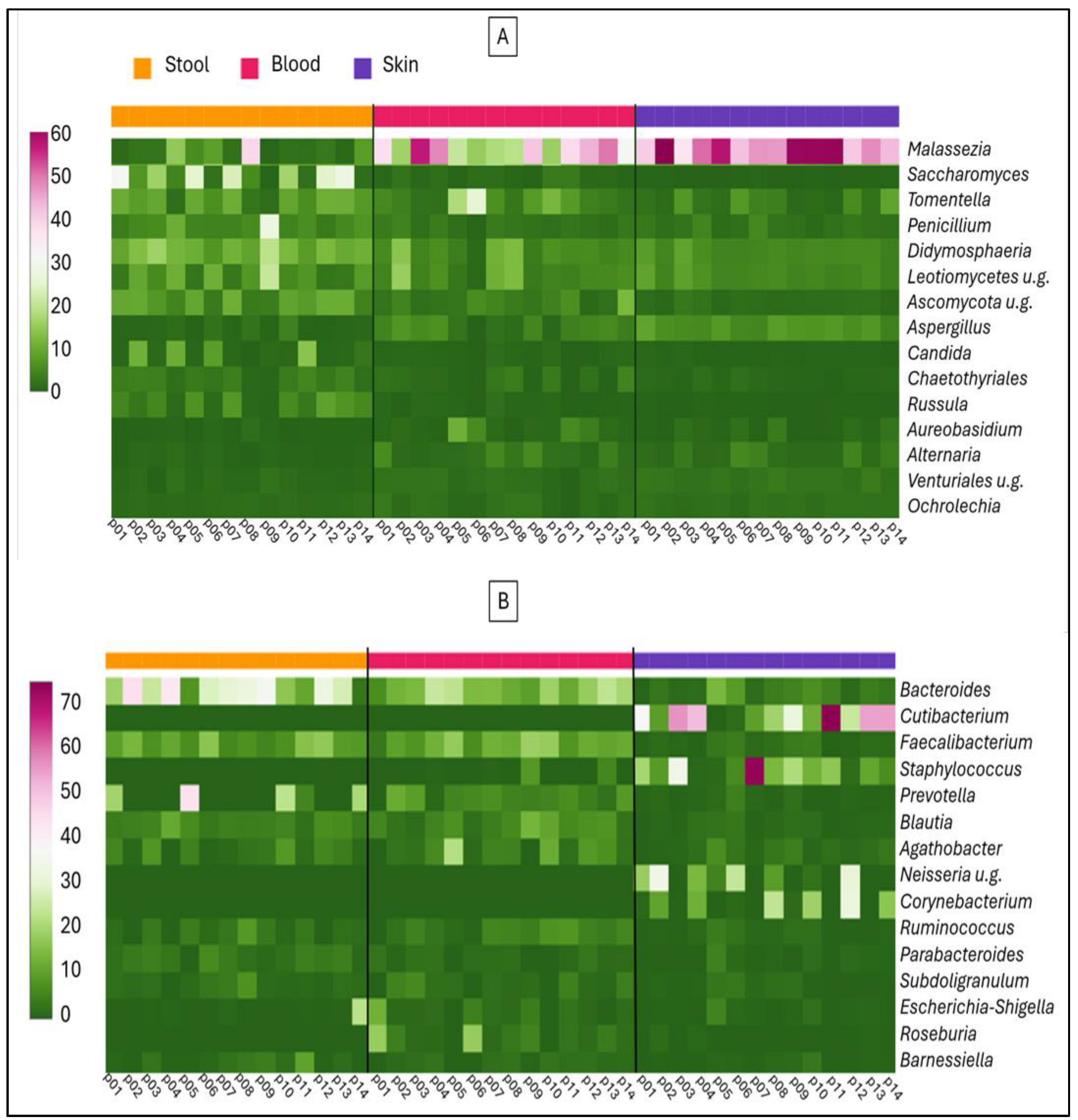
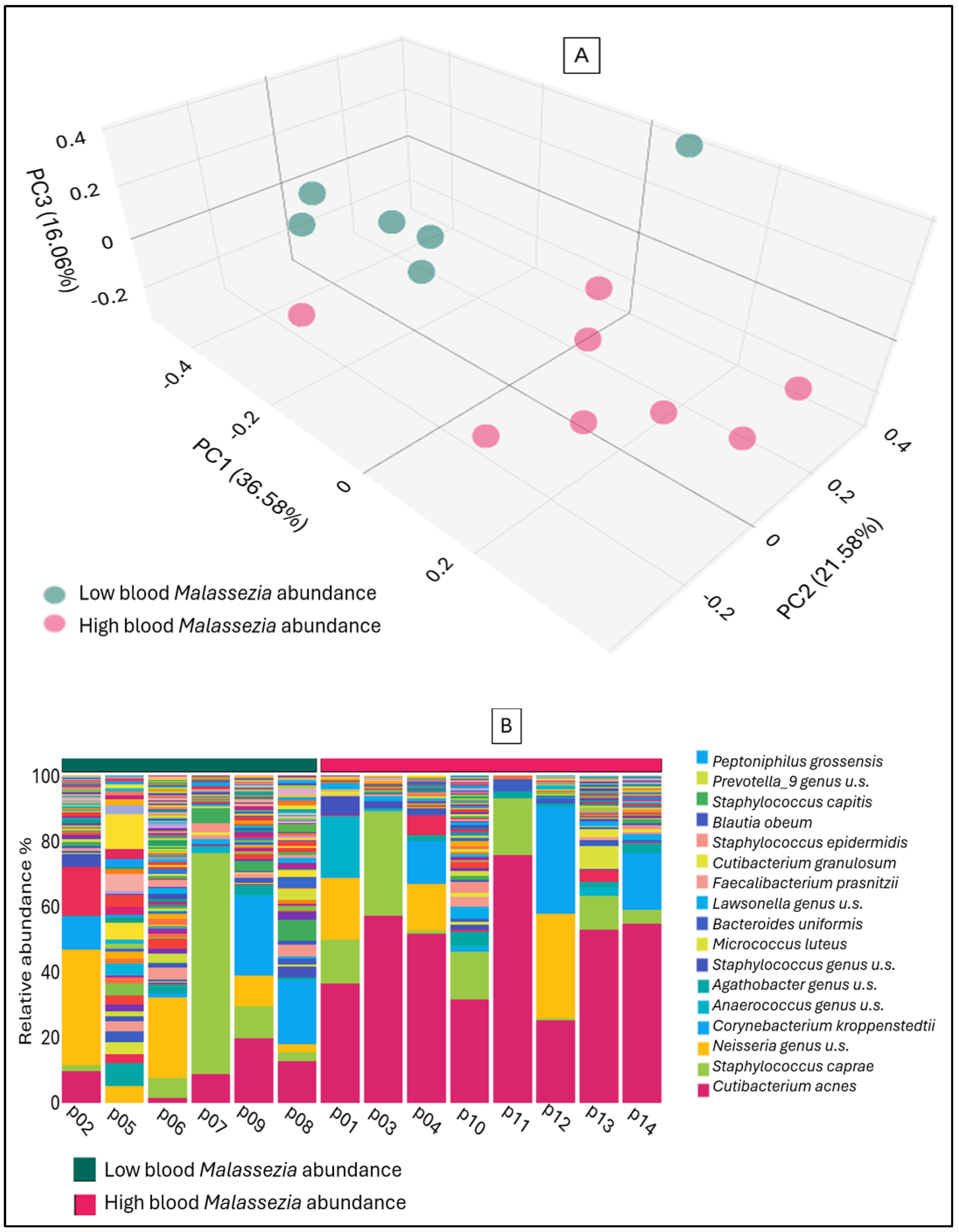
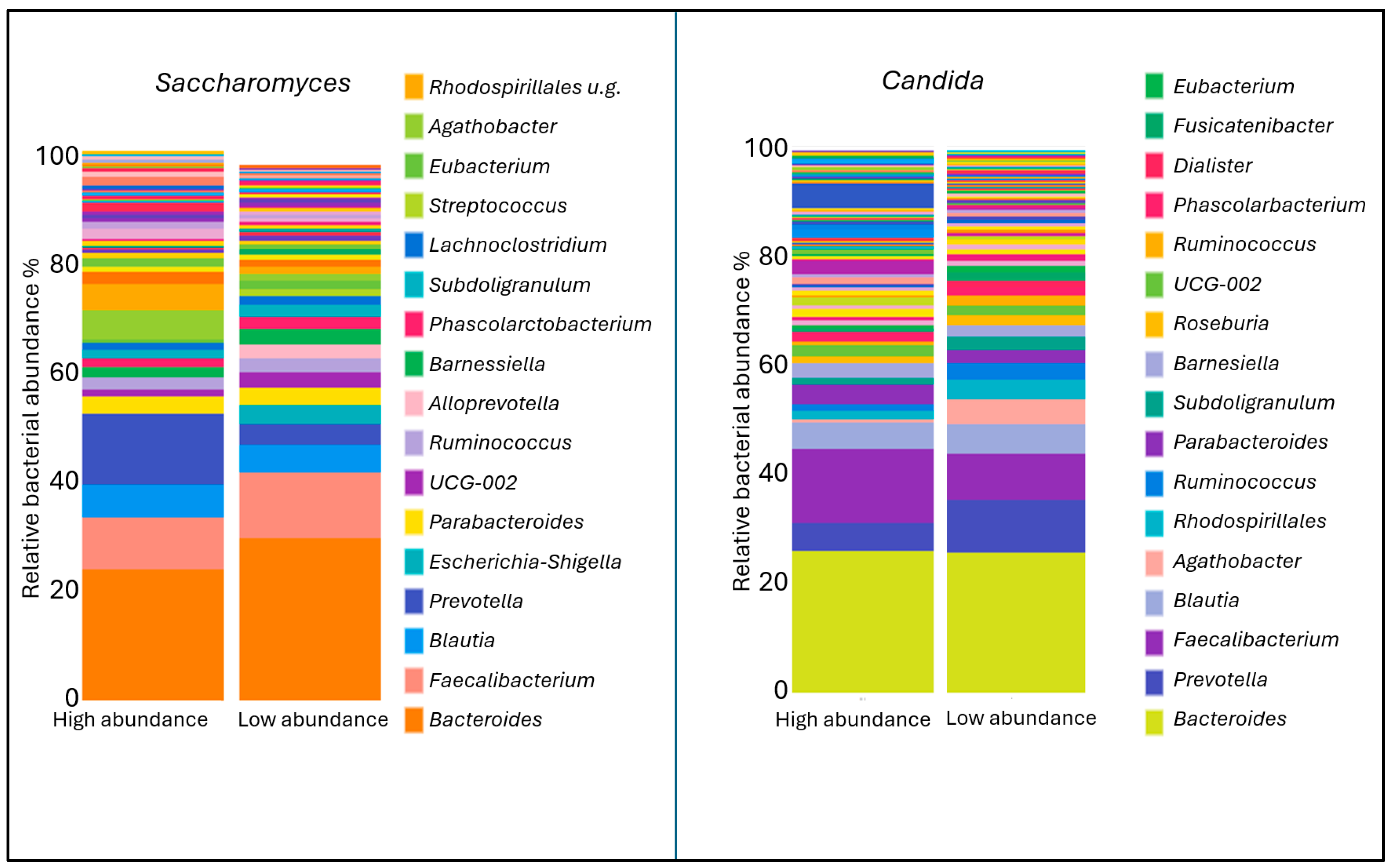
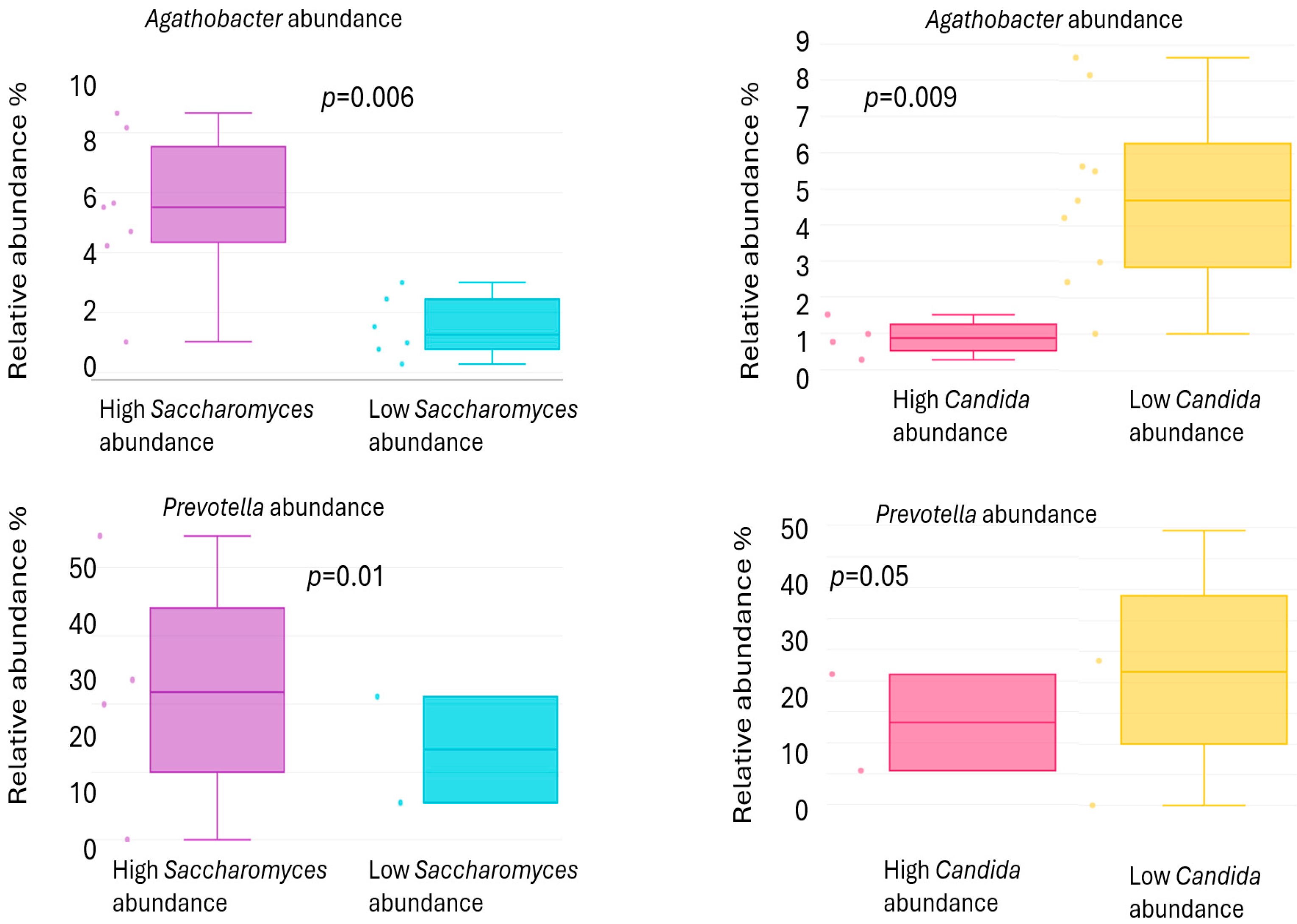
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Isolation
4.3. ITS Mycobiome Analysis
4.4. Statistical Analysis
4.5. Ethical Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ETR | Erythematotelangiectatic rosacea |
| ITS | Internal transcribed spacer |
| IQR | interquartile range |
| PCoA | Principal Coordinate Analysis |
| PCR | Polymerase Chain Reaction |
| PPR | Papulopustular rosacea |
| SIBO | Small Intestine Bacterial Overgrowth |
| TLR | Toll-like Receptor |
References
- Wilkin, J.; Dahl, M.; Detmar, M.; Drake, L.; Feinstein, A.; Odom, R.; Powell, F. Standard classification of rosacea: Report of the national rosacea society expert committee on the classification and staging of rosacea. J Am. Acad. Dermatol. 2002, 46, 584–587. [Google Scholar] [CrossRef]
- Searle, T.; Al-Niaimi, F.; Ali, F.R. Rosacea. Br. J. Hosp. Med. 2021, 82, 1–8. [Google Scholar] [CrossRef]
- Aedo, G.; Chahuan, M.; Gatica, E.; Herrera, I.; Parada, L.F.; Seguel, A.; Murray, N.P.; Aedo, S.; Aragon-Caqueo, D. Managing a Burning Face: Clinical Manifestations and Therapeutic Approaches for Neurogenic Rosacea. Int. J. Mol. Sci. 2025, 26, 2366. [Google Scholar] [CrossRef]
- Moran, E.M.; Foley, R.; Powell, F.C. Demodex and rosacea revisited. Clin. Dermatol. 2017, 35, 195–200. [Google Scholar] [CrossRef]
- Yuan, C.; Ma, Y.; Wang, Y.; Wang, X.; Qian, C.; Hocquet, D.; Zheng, S.; Mac-Mary, S.; Humbert, P. Rosacea is associated with conjoined interactions between physical barrier of the skin and microorganisms: A pilot study. J. Clin. Lab. Anal. 2020, 34, e23363. [Google Scholar] [CrossRef]
- Mylonas, A.; Hawerkamp, H.C.; Wang, Y.; Chen, J.; Messina, F.; Demaria, O.; Meller, S.; Homey, B.; Di Domizio, J.; Mazzolai, L.; et al. Type I IFNs link skin-associated dysbiotic commensal bacteria to pathogenic inflammation and angiogenesis in rosacea. JCI. Insight. 2023, 8, e151846. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xiao, Y.; Zhang, X.; Zhu, Z.; Zhang, H.; Wei, J.; Zhao, Z.; Li, J.; Chen, T. Probiotics suppress LL37 generated rosacea-like skin inflammation by modulating the TLR2/MyD88/NF-kappaB signaling pathway. Food. Funct. 2024, 15, 8916–8934. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, F.; Liu, Y.; Jiang, X. Causal relationship between gut microbiota and rosacea: A two-sample Mendelian randomization study. Front. Med. 2024, 11, 1322685. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.K.; Spaunhurst, K.; Sprockett, D.; Thomason, Y.; Mann, M.W.; Fu, P.; Ammons, C.; Gerstenblith, M.; Tuttle, M.S.; Popkin, D.L. Characterization of the facial microbiome in twins discordant for rosacea. Exp. Dermatol. 2018, 27, 295–298. [Google Scholar] [CrossRef]
- Thompson, K.G.; Rainer, B.M.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Comparison of the skin microbiota in acne and rosacea. Exp. Dermatol. 2020, 30, 1375–1380. [Google Scholar] [CrossRef]
- Rainer, B.M.; Thompson, K.G.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Bui, J.; Fischer, A.H.; Pasieka, H.B.; Garza, L.A.; Kang, S.; et al. Characterization and analysis of the skin microbiota in rosacea: A case-control study. Am. J. Clin. Dermatol. 2020, 21, 139–147. [Google Scholar] [CrossRef]
- Nam, J.H.; Yun, Y.; Kim, H.S.; Kim, H.N.; Jung, H.J.; Chang, Y.; Ryu, S.; Shin, H.; Kim, H.L.; Kim, W.S. Rosacea and its association with enteral microbiota in Korean females. Exp. Dermatol. 2018, 27, 37–42. [Google Scholar] [CrossRef]
- Chen, Y.J.; Lee, W.H.; Ho, H.J.; Tseng, C.H.; Wu, C.Y. An altered fecal microbial profiling in rosacea patients compared to matched controls. J. Formos. Med. Assoc. 2021, 120, 256–264. [Google Scholar] [CrossRef]
- Yun, Y.; Kim, H.N.; Chang, Y.; Lee, Y.; Ryu, S.; Shin, H.; Kim, W.S.; Kim, H.L.; Nam, J.H. Characterization of the Blood Microbiota in Korean Females with Rosacea. Dermatology 2019, 235, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Joura, M.I.; Jobbagy, A.; Dunai, Z.A.; Makra, N.; Banvolgyi, A.; Kiss, N.; Sardy, M.; Sandor, S.E.; Hollo, P.; Ostorhazi, E. Characteristics of the Stool, Blood and Skin Microbiome in Rosacea Patients. Microorganisms 2024, 12, 2667. [Google Scholar] [CrossRef]
- Jung, W.H. Alteration in skin mycobiome due to atopic dermatitis and seborrheic dermatitis. Biophys. Rev. 2023, 4, 011309. [Google Scholar] [CrossRef] [PubMed]
- Szczepanska, M.; Blicharz, L.; Nowaczyk, J.; Makowska, K.; Goldust, M.; Waskiel-Burnat, A.; Czuwara, J.; Samochocki, Z.; Rudnicka, L. The role of the cutaneous mycobiome in atopic dermatitis. J. Fungi. 2022, 8, 1153. [Google Scholar] [CrossRef]
- Wang, R.; Farhat, M.; Na, J.; Li, R.; Wu, Y. Bacterial and fungal microbiome characterization in patients with rosacea and healthy controls. Br. J. Dermatol. 2020, 183, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Koike, Y.; Kuwatsuka, S.; Motooka, D.; Murota, H. Dysbiosis of the human skin mycobiome in patients receiving systemic IL-23 inhibitors. Allergol. Int. 2025, 74, 72–77. [Google Scholar] [CrossRef]
- Koike, Y.; Kuwatsuka, S.; Nishimoto, K.; Motooka, D.; Murota, H. Skin mycobiome of psoriasis patients is retained during treatment with TNF and IL-17 inhibitors. Int. J. Mol. Sci. 2020, 21, 3892. [Google Scholar] [CrossRef]
- Tao, R.; Li, R.; Wang, R. Dysbiosis of skin mycobiome in atopic dermatitis. Mycoses 2022, 65, 285–293. [Google Scholar] [CrossRef]
- Tao, R.; Wang, R.; Wan, Z.; Song, Y.; Wu, Y.; Li, R. Ketoconazole 2% cream alters the skin fungal microbiome in seborrhoeic dermatitis: A cohort study. Clin. Exp. Dermatol. 2022, 47, 1088–1096. [Google Scholar] [CrossRef]
- Wang, X.; Sun, J.; Zhang, X.; Chen, W.; Cao, J.; Hu, H. Metagenomics reveals unique gut mycobiome biomarkers in psoriasis. Skin. Res. Technol. 2024, 30, e13822. [Google Scholar] [CrossRef]
- Chantanaskul, T.; Patumcharoenpol, P.; Roytrakul, S.; Kingkaw, A.; Vongsangnak, W. Exploring protein functions of gut bacteriome and mycobiome in Thai infants associated with atopic dermatitis through metaproteomic and host interaction analysis. Int. J. Mol. Sci. 2024, 25, 13533. [Google Scholar] [CrossRef] [PubMed]
- Vanni, P.; Turunen, J.; Aijala, V.K.; Tapiainen, V.V.; Paalanne, M.; Pokka, T.; Paalanne, N.; Tejesvi, M.V.; Ruuska, T.S. Gut mycobiome in atopic dermatitis and in overweight young children: A prospective cohort study in Finland. J. Fungi 2024, 10, 333. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Vongsangnak, W.; Nakphaichit, M. ITS2 sequencing and targeted meta-proteomics of infant gut mycobiome reveal the functional role of Rhodotorula sp. during atopic dermatitis manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef]
- Belvoncikova, P.; Splichalova, P.; Videnska, P.; Gardlik, R. The human mycobiome: Colonization, composition and the role in health and disease. J. Fungi 2022, 8, 1046. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing kingdoms: How the mycobiota and fungal-bacterial interactions impact host health and disease. Infect. Immun. 2021, 89. [Google Scholar] [CrossRef]
- Xiong, J.; Chen, S.; Wang, P.; Chen, A.; Zheng, Q.; Cai, T. Characterisation of the bacterial microbiome in patients with rosacea and healthy controls. Eur. J. Dermatol. 2023, 33, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Hamblin, M.R.; Wen, X. Role of the skin microbiota and intestinal microbiome in rosacea. Front. Microbiol. 2023, 14, 1108661. [Google Scholar] [CrossRef]
- Hu, Q.; Yu, L.; Zhai, Q.; Zhao, J.; Tian, F. Anti-Inflammatory, barrier maintenance, and gut microbiome modulation effects of saccharomyces cerevisiae QHNLD8L1 on DSS-induced ulcerative colitis in mice. Int. J. Mol. Sci. 2023, 24, 6721. [Google Scholar] [CrossRef]
- Chiaro, T.R.; Soto, R.; Stephens, W.Z.; Kubinak, J.L.; Petersen, C.; Gogokhia, L.; Bell, R.; Delgado, J.C.; Cox, J.; Voth, W.; et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 2017, 9, 380, Erratum in Sci. Transl. Med. 2017, 9, 388. [Google Scholar] [CrossRef]
- Sun, S.; Xu, X.; Liang, L.; Wang, X.; Bai, X.; Zhu, L.; He, Q.; Liang, H.; Xin, X.; Wang, L.; et al. Lactic acid-producing probiotic saccharomyces cerevisiae attenuates ulcerative colitis via suppressing macrophage pyroptosis and modulating gut microbiota. Front. Immunol. 2021, 12, 777665. [Google Scholar] [CrossRef]
- Ramazzotti, M.; Stefanini, I.; Di Paola, M.; De Filippo, C.; Rizzetto, L.; Berna, L.; Dapporto, L.; Rivero, D.; Tocci, N.; Weil, T.; et al. Population genomics reveals evolution and variation of Saccharomyces cerevisiae in the human and insects gut. Environ. Microbiol. 2019, 21, 50–71. [Google Scholar] [CrossRef]
- Di Paola, M.; Rizzetto, L.; Stefanini, I.; Vitali, F.; Massi-Benedetti, C.; Tocci, N.; Romani, L.; Ramazzotti, M.; Lionetti, P.; De Filippo, C.; et al. Comparative immunophenotyping of Saccharomyces cerevisiae and Candida spp. strains from Crohn’s disease patients and their interactions with the gut microbiome. J. Transl. Autoimmun. 2020, 3, 100036. [Google Scholar] [CrossRef]
- Itano, A.; Maslin, D.; Ramani, K.; Mehraei, G.; Carpenter, N.; Cormack, T.; Saghari, M.; Moerland, M.; Troy, E.; Caffry, W.; et al. Clinical translation of anti-inflammatory effects of Prevotella histicola in Th1, Th2, and Th17 inflammation. Front. Med. 2023, 10, 1070433. [Google Scholar] [CrossRef]
- Laigaard, A.; Krych, L.; Zachariassen, L.F.; Ellegaard-Jensen, L.; Nielsen, D.S.; Hansen, A.K.; Hansen, C.H.F. Dietary prebiotics promote intestinal Prevotella in association with a low-responding phenotype in a murine oxazolone-induced model of atopic dermatitis. Sci. Rep. 2020, 10, 21204. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, F.; Meng, X.; Zhou, L. The associations between gut microbiota and inflammatory skin diseases: A bi-directional two-sample Mendelian randomization study. Front. Immunol. 2024, 15, 1297240. [Google Scholar] [CrossRef] [PubMed]
- Hertz, S.; Anderson, J.M.; Nielsen, H.L.; Schachtschneider, C.; McCauley, K.E.; Ozcam, M.; Larsen, L.; Lynch, S.V.; Nielsen, H. Fecal microbiota is associated with extraintestinal manifestations in inflammatory bowel disease. Ann. Med. 2024, 56, 2338244. [Google Scholar] [CrossRef] [PubMed]

| Rosacea Patients (14) | Healthy Controls (8) | |
|---|---|---|
| Gender: male/female | 4/10 | 2/6 |
| Age, Year (median + IQR) | 48.5, IQR:25 | 41, IQR: 27 |
| Diagnosis | 13 PPR, 1 PPR + ETR | - |
| BMI | 27, IQR: 7 | 26, IQR: 6.5 |
| Consumption of alcohol: yes/no | 7/7 | 4/4 |
| Smoking: yes/no | 6/8 | 2/6 |
| Diet related symptom exacerbation: yes/no | 8/6 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joura, M.I.; Nemes-Nikodém, É.; Jobbágy, A.; Dunai, Z.A.; Makra, N.; Bánvölgyi, A.; Kiss, N.; Sárdy, M.; Sándor, S.E.; Holló, P.; et al. Integrative Analysis of Fungal and Bacterial Microbiomes Across Skin, Blood, and Stool in Rosacea Patients. Int. J. Mol. Sci. 2025, 26, 8127. https://doi.org/10.3390/ijms26178127
Joura MI, Nemes-Nikodém É, Jobbágy A, Dunai ZA, Makra N, Bánvölgyi A, Kiss N, Sárdy M, Sándor SE, Holló P, et al. Integrative Analysis of Fungal and Bacterial Microbiomes Across Skin, Blood, and Stool in Rosacea Patients. International Journal of Molecular Sciences. 2025; 26(17):8127. https://doi.org/10.3390/ijms26178127
Chicago/Turabian StyleJoura, Marie Isolde, Éva Nemes-Nikodém, Antal Jobbágy, Zsuzsanna A Dunai, Nóra Makra, András Bánvölgyi, Norbert Kiss, Miklós Sárdy, Sarolta Eszter Sándor, Péter Holló, and et al. 2025. "Integrative Analysis of Fungal and Bacterial Microbiomes Across Skin, Blood, and Stool in Rosacea Patients" International Journal of Molecular Sciences 26, no. 17: 8127. https://doi.org/10.3390/ijms26178127
APA StyleJoura, M. I., Nemes-Nikodém, É., Jobbágy, A., Dunai, Z. A., Makra, N., Bánvölgyi, A., Kiss, N., Sárdy, M., Sándor, S. E., Holló, P., & Ostorházi, E. (2025). Integrative Analysis of Fungal and Bacterial Microbiomes Across Skin, Blood, and Stool in Rosacea Patients. International Journal of Molecular Sciences, 26(17), 8127. https://doi.org/10.3390/ijms26178127








