Sexually Transmitted Infections: Usefulness of Molecular Methods for Microorganism Detection in Stored Sexual Assault Samples
Abstract
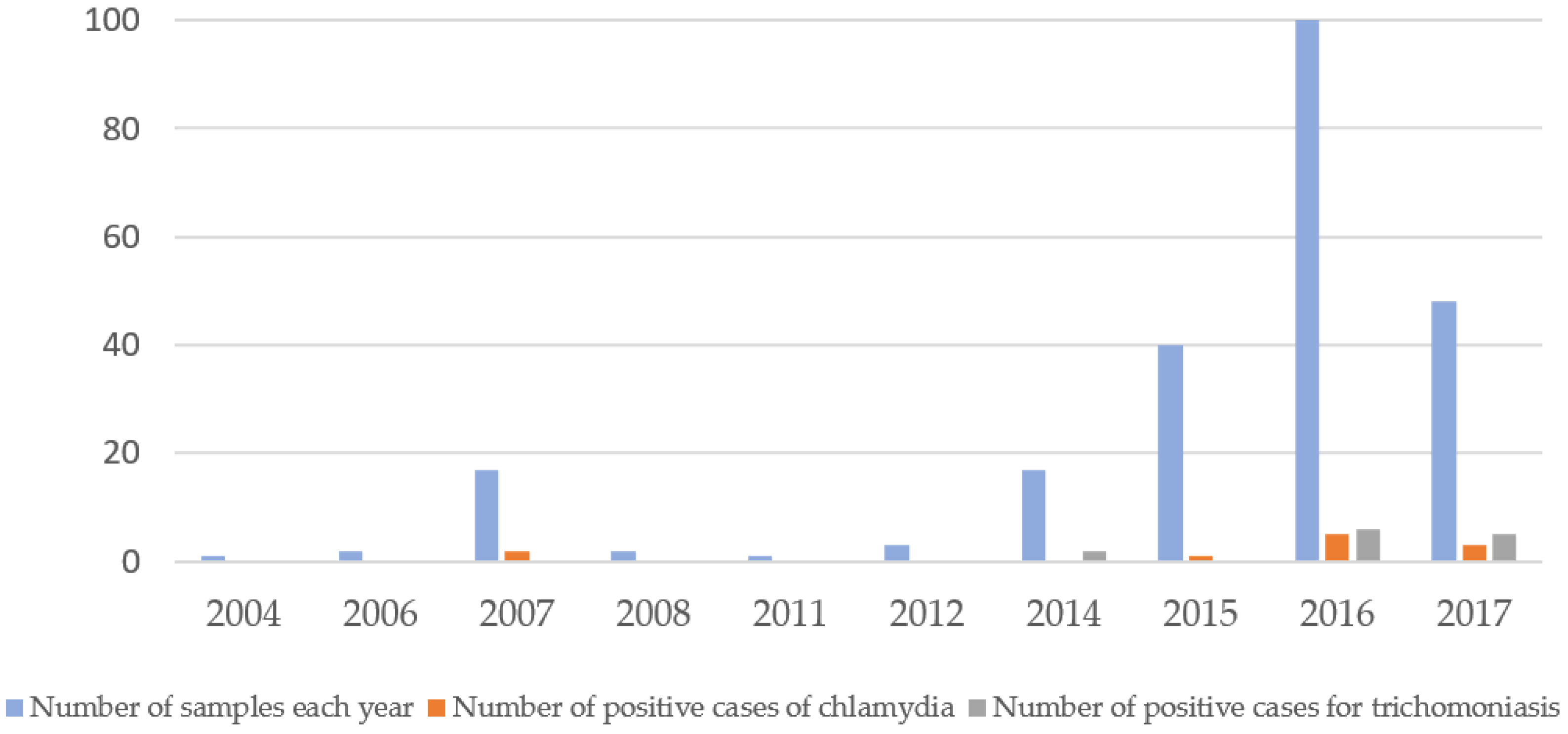
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Extraction and Purification
4.3. Molecular Detection and DNA Amplification
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WHO | World Health Organization |
| STI | Sexually transmitted infections |
| DNA | Deoxyribonucleic acid |
| PCR | Polymerase chain reaction |
| Ct | Cycle threshold |
| RNaseP | Ribonuclease P |
| INMLCF | National Institute of Legal Medicine and Forensic Sciences |
| STR | Short tandem repeat |
| NaCl | Sodium chloride |
| F | Forward primer |
| R | Reverse primer |
| P | Probe |
References
- World Health Organisation. Sexually Transmitted Infections. Available online: https://www.who.int/data/gho/data/themes/topics/global-and-regional-sti-estimates (accessed on 4 August 2025).
- Farahi, N.; Mceachern, M. Definitions and Epidemiology Sexual Assault of Women. Am. Fam. Physician 2021, 103, 168–176. Available online: https://www.aafp.org/afp/2021/0201/p168-s1.html (accessed on 1 August 2025).
- Sigurdardottir, S.; Halldorsdottir, S. Persistent suffering: The serious consequences of sexual violence against women and girls, their search for inner healing and the significance of the #metoo movement. Int. J. Environ. Res. Public Health 2021, 18, 1849. [Google Scholar] [CrossRef]
- Unemo, M.; Bradshaw, C.S.; Hocking, J.S.; de Vries, H.J.C.; Francis, S.C.; Mabey, D.; Marrazzo, J.M.; Sonder, G.J.B.; Schwebke, J.R.; Hoornenborg, E.; et al. Sexually transmitted infections: Challenges ahead. Lancet Infect. Dis. 2017, 17, e235–e279. [Google Scholar] [CrossRef] [PubMed]
- Fasciana, T.; Capra, G.; Lipari, D.; Firenze, A.; Giammanco, A. Sexually Transmitted Diseases: Diagnosis and Control. Int. J. Environ. Res. Public Health 2022, 19, 5293. [Google Scholar] [CrossRef] [PubMed]
- Pair, L.S.; Somerall, W.E. Sexually Transmitted Infections Update. Adv. Fam. Pract. Nurs. 2024, 6, 117–135. [Google Scholar] [CrossRef]
- Aslan, K. Rapid Whole Blood Bioassays using Microwave-Accelerated Metal-Enhanced Fluorescence. Nano Biomed. Eng. 2010, 2, 1–7. [Google Scholar] [CrossRef][Green Version]
- Rodrigues, R.; Sousa, C.; Vale, N. Chlamydia trachomatis as a Current Health Problem: Challenges and Opportunities. Diagnostics 2022, 12, 1795. [Google Scholar] [CrossRef]
- Petrin, D.; Delgaty, K.; Bhatt, R.; Garber, G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 1998, 11, 300–317. [Google Scholar] [CrossRef]
- Mielczarek, E.; Blaszkowska, J. Trichomonas vaginalis: Pathogenicity and potential role in human reproductive failure. Infection 2016, 44, 447–458. [Google Scholar] [CrossRef]
- Rogstad, K.E. Sexually Transmitted Infections Following Sexual Assault. In Forensic and Legal Medicine: Clinical and Pathological Aspects; CRC Press: Boca Raton, FL, USA, 2024; pp. 787–793. [Google Scholar] [CrossRef]
- Magalhães, T.; Dinis-Oliveira, R.J.; Silva, B.; Corte-Real, F.; Nuno Vieira, D. Biological Evidence Management for DNA Analysis in Cases of Sexual Assault. Sci. World J. 2015, 2015, 365674. [Google Scholar] [CrossRef]
- Murphy, C.; Alexander, K.; Stark, M.M.; Davidson, G. Novel recovery methods for biological materials in cases of alleged sexual assault: A word of caution. Sci. Justice 2022, 62, 621–623. [Google Scholar] [CrossRef]
- Newton, M. The forensic aspects of sexual violence. Best Pract. Res. Clin. Obstet. Gynaecol. 2013, 27, 77–90. [Google Scholar] [CrossRef]
- Cătălin, M.; Andrei, A.; Mitraşca, O. Modern Methods of Collection and Preservation of Biological Evidence for Human Identification by DNA Analysis. 2011. Available online: https://www.abacusdiagnostics.com/Modern_Methods_of_Collection.pdf (accessed on 12 April 2025).
- Artika, I.M.; Dewi, Y.P.; Nainggolan, I.M.; Siregar, J.E.; Antonjaya, U. Real-Time Polymerase Chain Reaction: Current Techniques, Applications, and Role in COVID-19 Diagnosis. Genes 2022, 13, 2387. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Sachs, C.J.; Ladd, M.; Thomas, B. Sexual Assault Infectious Disease Prophylaxis. [Updated 2023 September 9]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025; Available online: https://www.ncbi.nlm.nih.gov/books/NBK482239/ (accessed on 4 May 2025).
- Kebbi-Beghdadi, C.; Aeby, S.; Baud, D.; Greub, G. Evaluation of a Multiplex Real-Time PCR Assay for Detecting Chlamydia trachomatis in Vaginal Samples. Diagnostics 2022, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Schirm, J.; Bos, P.A.J.; Roozeboom-Roelfsema, I.K.; Luijt, D.S.; Möller, L.V. Trichomonas vaginalis detection using real-time TaqMan PCR. J. Microbiol. Methods 2007, 68, 243–247. [Google Scholar] [CrossRef]
- Caliendo, A.M.; Jordan, J.A.; Green, A.M.; Ingersoll, J.; Diclemente, R.J.; Wingood, G.M. Real-time PCR improves detection of Trichomonas vaginalis infection compared with culture using self-collected vaginal swabs. Infect. Dis. Obstet. Gynecol. 2005, 13, 145–150. [Google Scholar] [CrossRef]
- Šoba, B.; Skvarč, M.; Matičič, M. Trichomoniasis: A brief review of diagnostic methods and our experience with real-time PCR for detecting infection. Acta Dermatovenerol. Alp. Pannonica et Adriat. 2015, 24, 7–10. [Google Scholar] [CrossRef]
- Hamed Elsherif, R.; Fatah Youssef, M.A. Real-time PCR improve detection of Trichomonas vaginalis compared to conventional techniques. Comp. Clin. Pathol. 2013, 22, 295–300. [Google Scholar] [CrossRef]
- Dhawan, B.; Rawre, J.; Ghosh, A.; Malhotra, N.; Ahmed, M.M.; Sreenivas, V.; Chaudhry, R. Diagnostic efficacy of a real time-PCR assay for Chlamydia trachomatis infection in infertile women in north India. Indian J. Med. Res. 2014, 140, 252–261. Available online: https://pubmed.ncbi.nlm.nih.gov/25297359/ (accessed on 4 May 2025). [PubMed] [PubMed Central]
- Silva, J.; Cerqueira, F.; Teixeira, A.L.; Campainha, R.; Amorim, J.; Medeiros, R. Prevalence of Neisseria gonorrhoeae and Trichomonas vaginalis in Portuguese women of childbearing age. J. Obstet. Gynaecol. 2020, 41, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Jervoe-Storm, P.-M.; Koltzscher, M.; Falk, W.; Dorfler, A.; Jepsen, S. Comparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque samples. J. Clin. Periodontol. 2005, 32, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Omar, B.A.; Atif, H.A.; Mogahid, M.E. Comparison of three DNA extraction methods for polymerase chain reaction (PCR) analysis of bacterial genomic DNA. Afr. J. Microbiol. Res. 2014, 8, 598–602. [Google Scholar] [CrossRef]
- Uwiringiyeyezu, T.; El Khalfi, B.; Saile, R.; Belhachmi, J.; Soukri, A. Comparability of CMV DNA Extraction Methods and Validation of Viral Load. Methods Protoc. 2022, 5, 6. [Google Scholar] [CrossRef]
- Mackay, I.M. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef]
- Sellami, H.; Znazen, A.; Sellami, A.; Mnif, H.; Louati, N.; Ben Zarrouk, S.; Keskes, L.; Rebai, T.; Gdoura, R.; Hammami, A. Molecular detection of Chlamydia trachomatis and other sexually transmitted bacteria in semen of male partners of infertile couples in Tunisia: The effect on semen parameters and spermatozoa apoptosis markers. PLoS ONE 2014, 9, e98903. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heymans, R.; van der Helm, J.J.; de Vries, H.J.; Fennema, H.S.; Coutinho, R.A.; Bruisten, S.M. Clinical value of Treponema pallidum real-time PCR for diagnosis of syphilis. J. Clin. Microbiol. 2010, 48, 497–502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koek, A.G.; Bruisten, S.M.; Dierdorp, M.; van Dam, A.P.; Templeton, K. Specific and sensitive diagnosis of syphilis using a real-time PCR for Treponema pallidum. Clin. Microbiol. Infect. 2006, 12, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Alleged Victim | Year of Collection | Victim’s Age | Sample ID | Swab Type | STI Detected | Genetic Profiles Obtained |
|---|---|---|---|---|---|---|
| Individual 1 | 2007 | 22 | IST015 | Vulvar | Chlamydia trachomatis | No information |
| IST016 | Vaginal | Chlamydia trachomatis | ||||
| Individual 2 | 2014 | 36 | IST032 | Vulvar | Trichomonas vaginalis | No male profile identified |
| IST033 | Vaginal | Trichomonas vaginalis | ||||
| Individual 3 | 2015 | 37 | IST073 | Vaginal | Chlamydia trachomatis | Female and male admixture |
| Individual 4 | 2016 | 14 | IST092 | Vulvar | Trichomonas vaginalis | No male profile identified |
| IST104 | Vaginal | Trichomonas vaginalis | ||||
| Individual 5 | 2016 | Unknown | IST095 | Vaginal | Chlamydia trachomatis | Female and male admixture |
| Individual 6 | 2017 | 23 | IST142 | Vaginal | Chlamydia trachomatis | Male profile (STR) + 2 different Y haplotype profiles |
| IST147 | Vulvar | Chlamydia trachomatis | ||||
| IST148 | Vaginal | Chlamydia trachomatis | ||||
| Individual 7 | 2017 | 26 | IST154 | Perianal | Trichomonas vaginalis | Female and male admixture profile + Y haplotype |
| IST158 | Vaginal | Trichomonas vaginalis | ||||
| IST197 | Vaginal | Trichomonas vaginalis | ||||
| Individual 8 | 2016 | 15 | IST169 | Vulvar | Chlamydia trachomatis | Female and male admixture profile + Y haplotype |
| IST171 | Vaginal | Chlamydia trachomatis | ||||
| IST196 | Vaginal | Chlamydia trachomatis | ||||
| IST200 | Vulvar | Trichomonas vaginalis and Chlamydia trachomatis | ||||
| Individual 9 | 2017 | 17 | IST190 | Vaginal | Trichomonas vaginalis | No male profile identified |
| Individual 10 | 2016 | 25 | IST198 | Oral | Trichomonas vaginalis | Admixture profile of two Y haplotypes in a sample |
| Individual 11 | 2017 | 31 | IST199 | Vaginal | Trichomonas vaginalis | Male profile (STR) + Y haplotype |
| Individual 12 | 2016 | 37 | IST204 | Vaginal | Trichomonas vaginalis | Male profile identified |
| IST232 | Trichomonas vaginalis |
| Target Gene | Primer Sequences (5′3′) |
|---|---|
| Cryptic plasmid from C. trachomatis | F: AACCAAGGTCGATGTGATAG R: TCAGATAATTGGCGATTCTT P: ROX-CGAACTCATCGGCGATAAGG |
| Por A pseudogene from N. gonorrhoeae | F: CCGGAACTGGTTTCATCTGATT R: GTTTCAGCGGCAGCATTCA P: CGTGAAAGTAGCAGGCGTATAGGCGGACTT |
| polA gene from T. pallidum | F: GGTAGAAGGGAGGGCTAGTA R: CTAAGATCTCTATTTTCTATAGGTATGG P: ACACAGCACTCGTCTTCAACTCC |
| 2 kb repeat sequence from Trichomonas vaginalis | F: AAG ATG GGT GTT TTA AGC TAG ATA AGG T R: CGT CTT CAA GTA TGC CCC AGT AC P: CCG AAG TTC ATG TCC TCT CCA AGC GT |
| Human RNase P gene | F: AGATTTGGACCTGCGAGCG R: GAGCGGCTGTCTCCACAAGT P: TTCTGACCTGAAGGCTCTGCGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cainé, L.; Eira, A.; Fadoni, J.; Franco, M.; Correia Dias, H.; Amorim, A. Sexually Transmitted Infections: Usefulness of Molecular Methods for Microorganism Detection in Stored Sexual Assault Samples. Int. J. Mol. Sci. 2025, 26, 8124. https://doi.org/10.3390/ijms26178124
Cainé L, Eira A, Fadoni J, Franco M, Correia Dias H, Amorim A. Sexually Transmitted Infections: Usefulness of Molecular Methods for Microorganism Detection in Stored Sexual Assault Samples. International Journal of Molecular Sciences. 2025; 26(17):8124. https://doi.org/10.3390/ijms26178124
Chicago/Turabian StyleCainé, Laura, Ana Eira, Jennifer Fadoni, Magda Franco, Helena Correia Dias, and António Amorim. 2025. "Sexually Transmitted Infections: Usefulness of Molecular Methods for Microorganism Detection in Stored Sexual Assault Samples" International Journal of Molecular Sciences 26, no. 17: 8124. https://doi.org/10.3390/ijms26178124
APA StyleCainé, L., Eira, A., Fadoni, J., Franco, M., Correia Dias, H., & Amorim, A. (2025). Sexually Transmitted Infections: Usefulness of Molecular Methods for Microorganism Detection in Stored Sexual Assault Samples. International Journal of Molecular Sciences, 26(17), 8124. https://doi.org/10.3390/ijms26178124








