Molecular Basis of Intron Retention in PI-PLC γ1 mRNA from Osteoarthritis Synoviocytes
Abstract
1. Introduction
2. Results
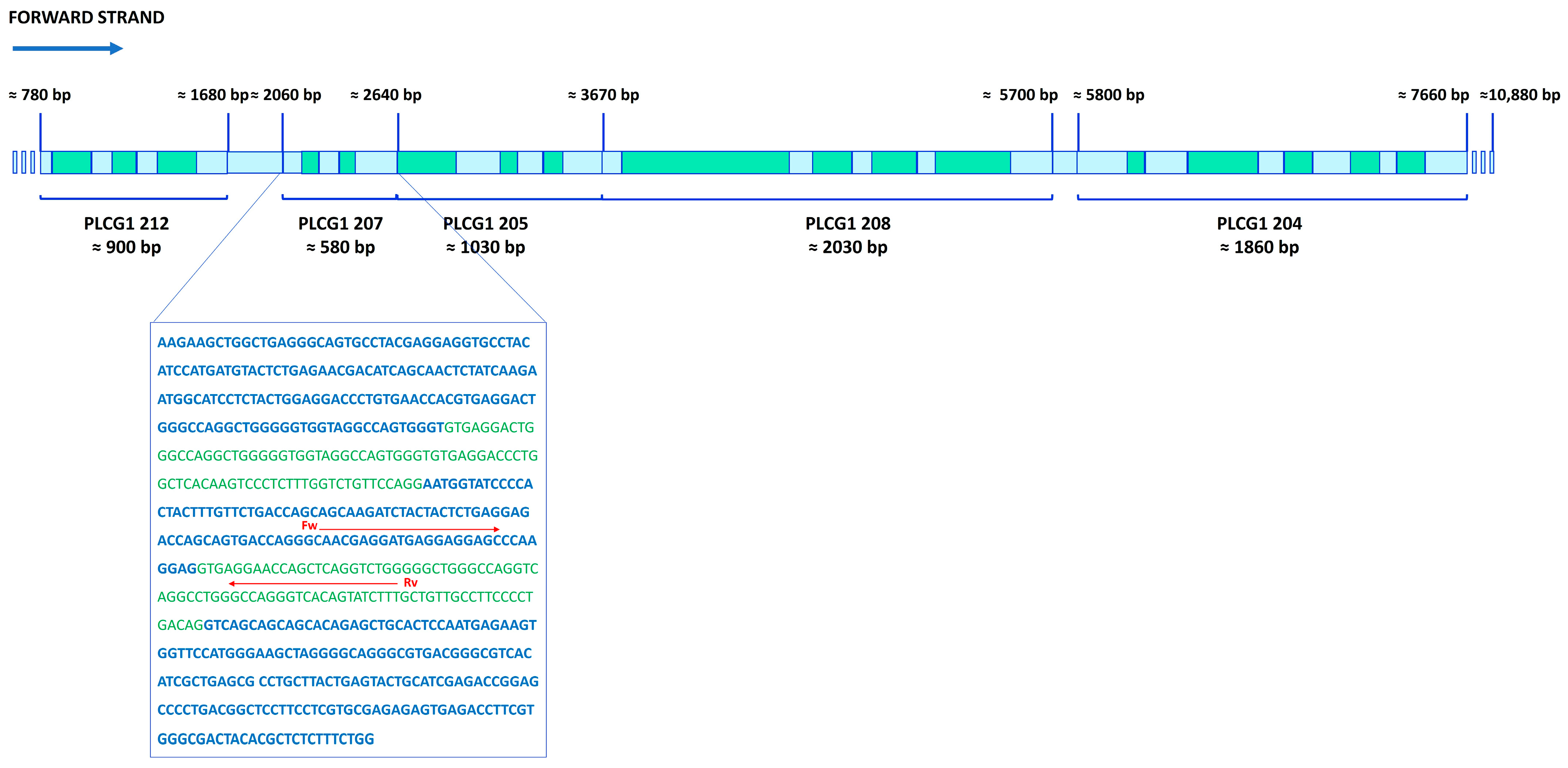
2.1. PI-PLC γ1 Intron Retention: Background
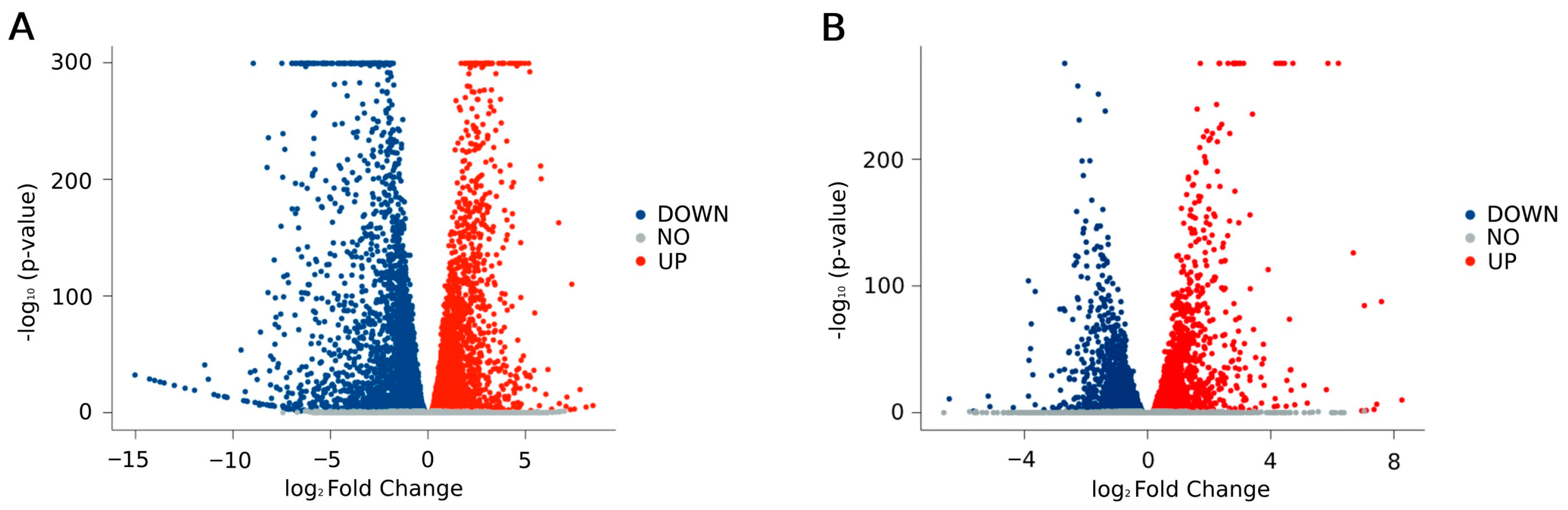
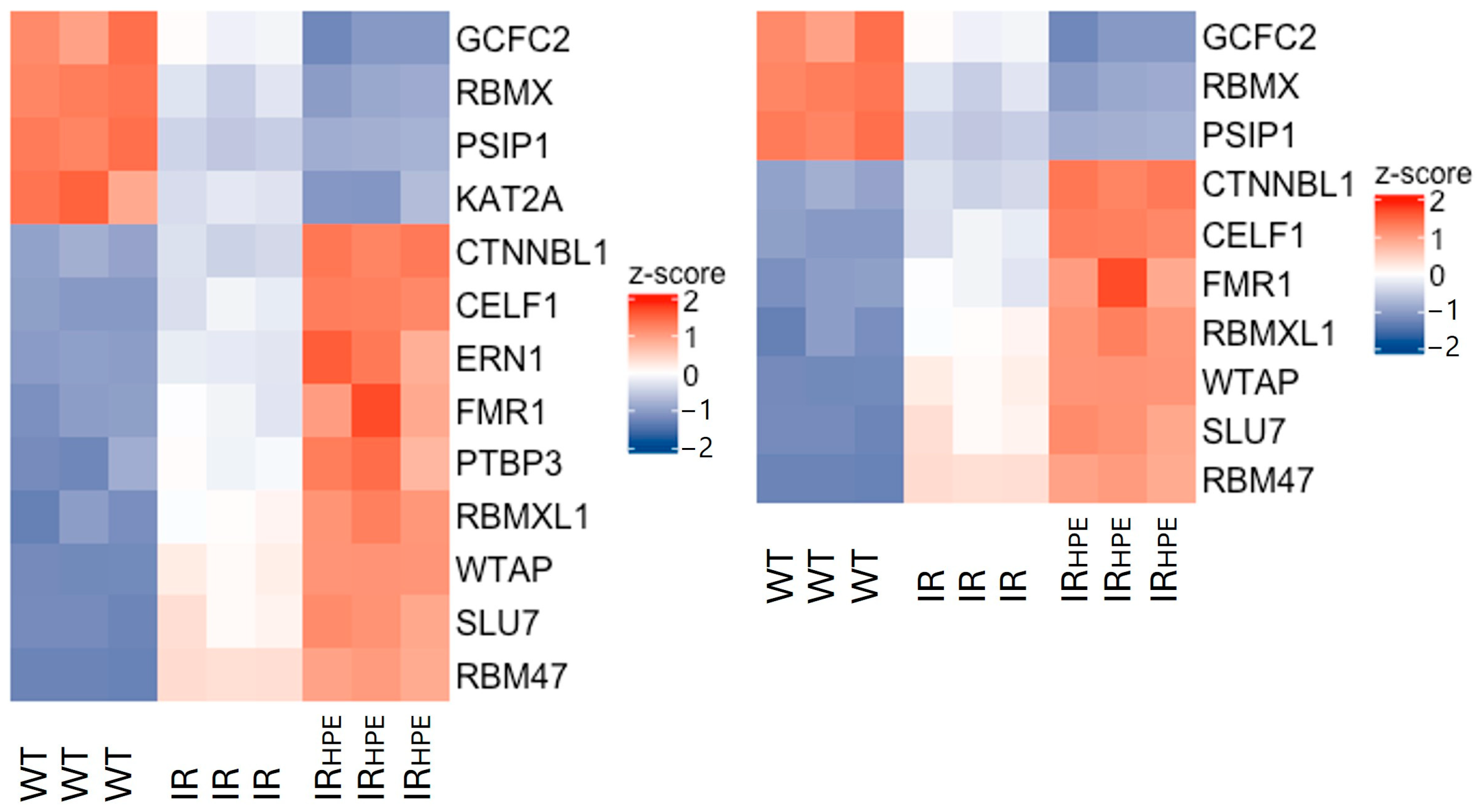
2.2. RNA-Seq Analysis
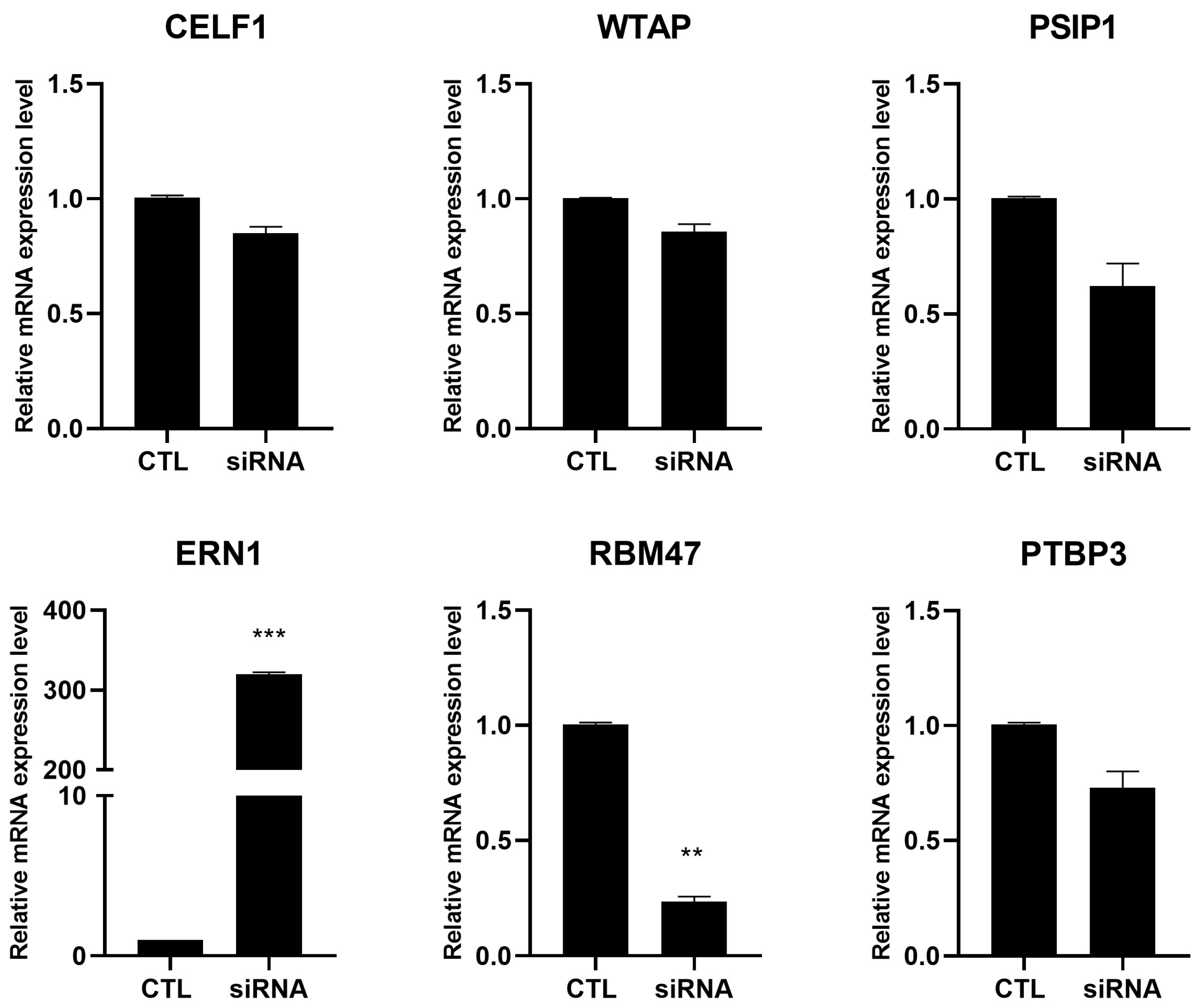
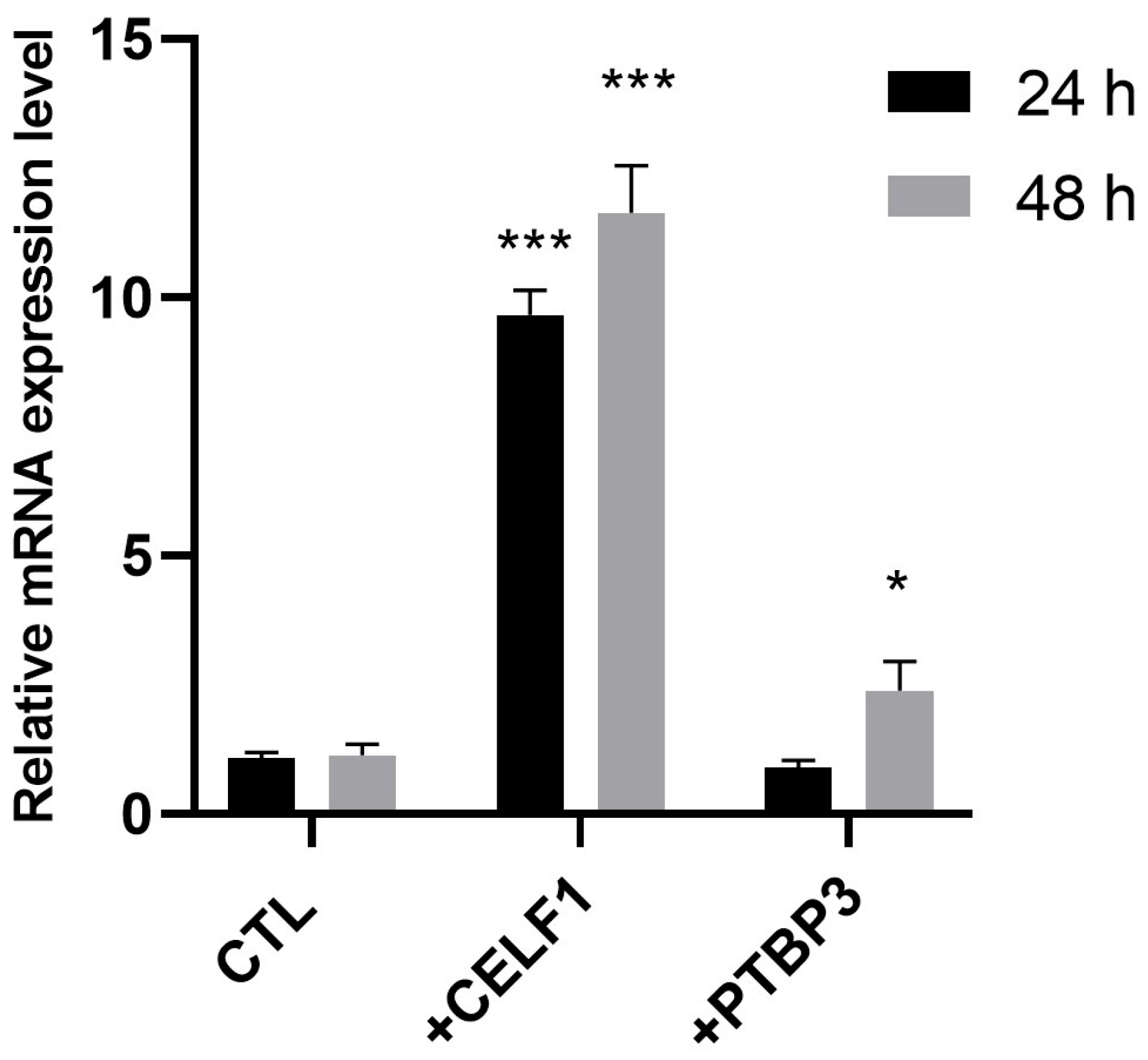
2.3. RNA-Seq Validation Through RT-PCR
3. Discussion
4. Materials and Methods
4.1. Synoviocyte Isolation and Cell Culture
4.2. Characterization for PI-PLC γ1 Intron Retention
4.3. Cell Treatment
4.4. RNA Extraction
4.5. RNA-Seq Library Preparation and Sequencing
4.6. Bioinformatic Analysis Process
4.7. Validation of Gene Expression Data by Quantitative-Real Time-PCR
4.8. siRNA Transfections
4.9. CELF1 and PTBP3 Transfections
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, J.J.L.; Schmitz, U. Intron retention: Importance, challenges, and opportunities. Trends Genet. 2022, 38, 789–792. [Google Scholar] [CrossRef]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef]
- Naro, C.; Jolly, A.; Di Persio, S.; Bielli, P.; Setterblad, N.; Alberdi, A.J.; Vicini, E.; Geremia, R.; De la Grange, P.; Sette, C. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev. Cell 2017, 41, 82–93.E4. [Google Scholar] [CrossRef]
- Edwards, C.R.; Ritchie, W.; Wong, J.J.-L.; Schmitz, U.; Middleton, R.; An, X.; Mohandas, N.; Rasko, J.E.J.; Blobel, G.A. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood 2016, 127, e24–e34. [Google Scholar] [CrossRef] [PubMed]
- Mauger, O.; Lemoine, F.; Scheiffele, P. Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 2016, 92, 1266–1278. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef]
- Gill, J.; Park, Y.; McGinnis, J.P.; Perez-Sanchez, C.; Blanchette, M.; Si, K. Regulated Intron Removal Integrates Motivational State and Experience. Cell 2017, 169, 836–848.E15. [Google Scholar] [CrossRef]
- Mariano, A.; Ammendola, S.; Migliorini, A.; Leopizzi, M.; Raimondo, D.; Scotto d’Abusco, A. Intron retention in PI-PLC γ1 mRNA as a key mechanism affecting MMP expression in human primary fibroblast-like synovial cells. Cell Biochem. Funct. 2024, 42, e4091. [Google Scholar] [CrossRef] [PubMed]
- Pulik, Ł.; Łęgosz, P.; Motyl, G. Matrix metalloproteinases in rheumatoid arthritis and osteoarthritis: A state of the art review. Reumatologia 2023, 61, 191–201. [Google Scholar] [CrossRef]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef]
- Scanzello, C.R. Role of low-grade inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2017, 29, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.J.; Driban, J.B.; McAlindon, T.E. Pharmaceutical treatment of osteoarthritis. Osteoarthr. Cartil. 2023, 31, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, D.; Wu, Q.; Wu, G.; Zhou, Y.; Zhang, Y.; Zhang, L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim. Biophys. Acta-Gene Regul. Mech. 2017, 1860, 911–921. [Google Scholar] [CrossRef]
- Huang, Q.; Mo, J.; Liao, Z.; Chen, X.; Zhang, B. The RNA m6A writer WTAP in diseases: Structure, roles, and mechanisms. Cell Death Dis. 2022, 13, 852. [Google Scholar] [CrossRef]
- Jayakumar, S.; Patel, M.; Boulet, F.; Aziz, H.; Brooke, G.N.; Tummala, H.; Pradeepa, M.M. PSIP1/LEDGF reduces R-loops at transcription sites to maintain genome integrity. Nat. Commun. 2024, 15, 361. [Google Scholar] [CrossRef]
- Radine, C.; Peters, D.; Reese, A.; Neuwahl, J.; Budach, W.; Jänicke, R.U.; Sohn, D. The RNA-binding protein RBM47 is a novel regulator of cell fate decisions by transcriptionally controlling the p53-p21-axis. Cell Death Differ. 2020, 27, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Le Goupil, S.; Laprade, H.; Aubry, M.; Chevet, E. Exploring the IRE1 interactome: From canonical signaling functions to unexpected roles. J. Biol. Chem. 2024, 300, 107169. [Google Scholar] [CrossRef]
- Yi, Q.; Deng, Z.; Yue, J.; He, J.; Xiong, J.; Sun, W.; Sun, W. RNA binding proteins in osteoarthritis. Front. Cell Dev. Biol. 2022, 10, 954376. [Google Scholar] [CrossRef]
- Kajdasz, A.; Niewiadomska, D.; Sekrecki, M.; Sobczak, K. Distribution of alternative untranslated regions within the mRNA of the CELF1 splicing factor affects its expression. Sci. Rep. 2022, 12, 190. [Google Scholar] [CrossRef]
- Tan, L.Y.; Whitfield, P.; Llorian, M.; Monzon-Casanova, E.; Diaz-Munoz, M.D.; Turner, M.; Smith, C.W.J. Generation of functionally distinct isoforms of PTBP3 by alternative splicing and translation initiation. Nucleic Acids Res. 2015, 43, 5586–5600. [Google Scholar] [CrossRef]
- Xiao, Y.; Xie, S.; Li, H.-D.; Liu, Y.; Zhang, H.; Zuo, X.; Zhu, H.; Li, Y.; Luo, H. Characterised intron retention profiles in muscle tissue of idiopathic inflammatory myopathy subtypes. Ann. Rheum. Dis. 2024, 83, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Z.; Li, Z.; Zeng, Z.; Peng, W.; Zhu, J.; Zhao, J.; Zhu, C.; Zeng, C.; Stearrett, N.; et al. Abnormalities in intron retention characterize patients with systemic lupus erythematosus. Sci. Rep. 2023, 13, 5141. [Google Scholar] [CrossRef]
- El-Seedy, A.; Ladevèze, V. Intron Retention and Alzheimer’s Disease (AD): A Review of Regulation Genes Implicated in AD. Genes 2025, 16, 782. [Google Scholar] [CrossRef]
- Hu, Q.; Ecker, M. Overview of MMP-13 as a Promising Target for the Treatment of Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 1742. [Google Scholar] [CrossRef]
- D’Adamo, S.; Cetrullo, S.; Panichi, V.; Mariani, E.; Flamigni, F.; Borzì, R.M. Nutraceutical Activity in Osteoarthritis Biology: A Focus on the Nutrigenomic Role. Cells 2020, 9, 1232. [Google Scholar] [CrossRef]
- Mariano, A.; Bigioni, I.; Misiti, F.; Fattorini, L.; Scotto d’Abusco, A.; Rodio, A. The Nutraceuticals as Modern Key to Achieve Erythrocyte Oxidative Stress Fighting in Osteoarthritis. Curr. Issues Mol. Biol. 2022, 44, 3481–3495. [Google Scholar] [CrossRef]
- Mariano, A.; Bigioni, I.; Mattioli, R.; Di Sotto, A.; Leopizzi, M.; Garzoli, S.; Mariani, P.F.; Dalla Vedova, P.; Ammendola, S.; Scotto d’Abusco, A. Harpagophytum procumbens Root Extract Mediates Anti-Inflammatory Effects in Osteoarthritis Synoviocytes through CB2 Activation. Pharmaceuticals 2022, 15, 457. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Lu, Y.; Li, F.; Qiao, L.; Wang, Q.; Li, N.; Borgia, J.A.; Deng, Y.; Lei, G.; Zheng, Q. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 2014, 5, e1469. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yang, Y.; Shen, P.; Ma, J.; Fang, B.; Wang, Q.; Wang, K.; Shi, P.; Fan, S.; Fang, X. CircPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. 2021, 80, 1209–1219. [Google Scholar] [CrossRef]
- Xiao, L.; Hu, B.; Ding, B.; Zhao, Q.; Liu, C.; Öner, F.C.; Xu, H. N(6)-methyladenosine RNA methyltransferase like 3 inhibits extracellular matrix synthesis of endplate chondrocytes by downregulating sex-determining region Y-Box transcription factor 9 expression under tension. Osteoarthr. Cartil. 2022, 30, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, T.; Ladd, A.N. The importance of CELF control: Molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2012, 3, 104–121. [Google Scholar] [CrossRef]
- Marquis, J.; Paillard, L.; Audic, Y.; Cosson, B.; Danos, O.; Le Bec, C.; Osborne, H.B. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006, 400, 291–301. [Google Scholar] [CrossRef]
- Mariano, A.; Di Sotto, A.; Leopizzi, M.; Garzoli, S.; Di Maio, V.; Gullì, M.; Vedova, P.D.; Ammendola, S.; Scotto d’Abusco, A. Antiarthritic effects of a root extract from harpagophytum procumbens DC: Novel insights into the molecular mechanisms and possible bioactive phytochemicals. Nutrients 2020, 12, 2545. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC. A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 April 2023).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; Blankenberg, D.; et al. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.L.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.J.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef]

| Gene | Primer Sequences (Fw-Rv) |
|---|---|
| CELF1 NM_001376376 | 5′-ATCCCTGTCTCTTCAGCTTC-3′ 5′-GCATCAAAGGTCAACACAAGG-3′ |
| ERN1 NM_001433 | 5′-AGGTGGTTCTGTCAGAGATC-3′ 5′-AAGTAGCACACGAAGTCGTC-3′ |
| RBM47 NM_001098634 | 5′-TGGAAATCCCTACCGTCAAC-3′ 5′-GGAAACATGGAATACTGAGC-3′ |
| WTAP NM_001270531 | 5′-AGTGCCTGGAAGTTTACGCC-3′ 5′-TAAGCATTCGACACTTCGCC-3′ |
| PTBP3 NM_001163788 | 5′-ATTTATGCCTGCCTGTTCCC-3′ 5′-TGAGCAGAGGCATTCATAGG-3′ |
| PSIP1 NM_033222 | 5′-AGATCAAGGGAAGAAAGGGC-3′ 5′-TCTTGAGCATCAGATCCTCC-3′ |
| PI-PLCG1 NM_182811.2 | 5′-CTACCTGGAGGACCCTGTGAAC-3′ 5′-ATCCTCGTTGCCCTGGTCACTG-3′ |
| 18S NM_003286 | 5′-CGCCGCTAGAGGTGAAATTC-3′ 5′-CATTCTTGGCAAATGCTTTCG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mariano, A.; D’Andrea, D.; Mattioli, R.; Ammendola, S.; Scotto d’Abusco, A. Molecular Basis of Intron Retention in PI-PLC γ1 mRNA from Osteoarthritis Synoviocytes. Int. J. Mol. Sci. 2025, 26, 8123. https://doi.org/10.3390/ijms26178123
Mariano A, D’Andrea D, Mattioli R, Ammendola S, Scotto d’Abusco A. Molecular Basis of Intron Retention in PI-PLC γ1 mRNA from Osteoarthritis Synoviocytes. International Journal of Molecular Sciences. 2025; 26(17):8123. https://doi.org/10.3390/ijms26178123
Chicago/Turabian StyleMariano, Alessia, Daniel D’Andrea, Roberto Mattioli, Sergio Ammendola, and Anna Scotto d’Abusco. 2025. "Molecular Basis of Intron Retention in PI-PLC γ1 mRNA from Osteoarthritis Synoviocytes" International Journal of Molecular Sciences 26, no. 17: 8123. https://doi.org/10.3390/ijms26178123
APA StyleMariano, A., D’Andrea, D., Mattioli, R., Ammendola, S., & Scotto d’Abusco, A. (2025). Molecular Basis of Intron Retention in PI-PLC γ1 mRNA from Osteoarthritis Synoviocytes. International Journal of Molecular Sciences, 26(17), 8123. https://doi.org/10.3390/ijms26178123






